Introduction
Ovarian cancer is a common malignant tumor of the
female reproductive organs, with an incidence ranked third of all
malignant tumors, and the majority of these tumors are epitheliomas
(1). With not only non-typical early
symptoms but also complex ovarian embryonic development, tissue
dissection and endocrine functions, only 19% of all ovarian cancers
can be diagnosed at an early stage, and the mortality rate for this
disease is highest in the field of gynecological oncology (2).
In recent years, a number of studies have been
performed to investigate the molecular mechanisms of ovarian
cancer. For example, as a member of the protein kinase B family,
v-akt murine thymoma viral oncogene homolog 2 (AKT2) can be
frequently activated and induce the apoptosis of human primary
ovarian cancer by inhibiting the phosphoinositide 3-kinase
(PI3K)/Akt pathway (3). With elevated
expression levels in patients with epithelial ovarian cancer,
ovarian cancer carbohydrate antigen 125 can be used for monitoring
patients with ovarian cancer (4). As
a chemokine receptor, chemokine (C-X-C motif) receptor 4 can be
expressed on a subset of ovarian cancer cells and may have
important roles in the development of site-specific metastasis
(5,6).
Chemokine (C-X-C motif) ligand 12 may affect ovarian cancer through
several methods, such as stimulating cell migration and invasion,
inducing establishment of a cytokine network and DNA synthesis in
conditions that are suboptimal for tumor cell growth (7). PI3Ks are lipid kinases associated with
neoplasia (8), and
phosphoinositide-3-kinase, catalytic subunit α (which encodes the
p110alpha catalytic subunit of PI3-kinase) is an oncogene that
plays a critical role in ovarian cancer (9).
In 2012, Zhao et al (10) used a whole-genome microarray approach
to analyze genes that differed between ovarian cancer cases and
healthy controls, and obtained a total of 10,435 mRNA genes that
could be used for downstream analysis. Using the data obtained by
Zhao et al (10), the present
study aimed to obtain the differentially-expressed genes (DEGs) and
investigate their possible functions by Gene Ontology (GO) and
pathway enrichment analyses. In addition, the interaction
associations between these DEGs were searched using the
protein-protein interaction (PPI) network and modules of the PPI
network.
Materials and methods
Microarray data
The expression profile of GSE37582, deposited by
Zhao et al (10), was
downloaded from Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and was based on the
platform of the GPL6947 Illumina HumanHT-12 V3.0 expression
beadchip. GSE37582 included data from 74 ovarian cancer cases and
47 healthy controls.
DEG screening and functional
annotation
Once GSE37582 had been downloaded, microarray data
was preprocessed by robust multi-array average (11) background correction. Next, quantile
normalization was conducted. The average value of multiple probes
mapped with one gene was obtained as the ultimate gene expression
value. The linear models for microarray data package in R (12) was used to analyze the DEGs between
ovarian cancer cases and healthy controls. A false discovery rate
(FDR) of <0.05 and |log2fold-change|>1 were used
as the cut-off criteria.
To predict genes with functions of transcription
factors, the DEGs were screened in combination with the
transcription factors database. In combination with the tumor
suppressor gene (TSG) (13) and
tumor-associated gene (14)
databases, the TSGs and oncogenes were then further screened from
the DEGs.
Functional and pathway enrichment
analysis
Through use of controlled and structured
vocabularies, GO (www.geneontology.org/) can act as a community-based
bioinformatics resource and classify gene product functions
(15). As an integrated database
resource, the Kyoto Encyclopedia of Genes and Genomes (KEGG;
www.genome.jp/kegg/) consists of systems
information, genomic information and chemical information (16). GO and KEGG pathway enrichment analyses
were performed for the DEGs. P<0.05 was used as the cut-off
criterion.
PPI network and module
construction
The Search Tool for the Retrieval of Interacting
Genes online software (17) was used
to search interaction associations of the proteins encoded by the
DEGs, and the required confidence (combined score) of >0.4 was
used as the cut-off criterion. Cytoscape (18) was used to visualize the PPI network.
Next, connectivity degree analysis was conducted and key nodes
(i.e., hub proteins) (19) were
searched. The proteins in the network were defined as the nodes.
Next, the BioNet (20) analysis tool
in R was used to screen modules of the PPI network. The FDR was set
to 3.
Results
DEG analysis and functional
annotation
A total of 284 DEGs were screened, consisting of 145
upregulated genes and 139 downregulated genes. Only 1 transcription
factor was significantly upregulated, while 8 transcription factors
were significantly downregulated. For the upregulated genes, there
was 1 oncogene, 4 TSGs and 3 ‘other’ genes [the influence of
‘other’ genes on tumors was unclear, and could therefore not be
distinguished as oncogenes or tumor suppressor genes, including
cluster of differentiation 44 (CD44)]. For the downregulated genes,
there were 4 oncogenes [such as FBJ murine osteosarcoma viral
oncogene homolog (FOS)], 11 TSGs [such as cyclin-dependent kinase
inhibitor 1A (CDKN1A)] and 5 ‘other’ genes (Table I).
 | Table I.Functional annotations of the
differentially-expressed genes. |
Table I.
Functional annotations of the
differentially-expressed genes.
| Regulation | TF numbers | TF symbols | TAG numbers | TAG symbols |
|---|
| Up | 1 | KLF12 | 8 | TCL1B, CTNND1,
MUC1, PTPRF, STARD13, CD44, FES, MIAT |
| Down | 8 | ASCL1, ETV4, HSF1,
LMO3, PML, RUNX3, TCF7, USF2 | 20 | AKT2, FOS, GNA12,
PVT1, CD82, CDKN1A, FBXO32, GADD45G, MAL, PML, PPP1R3C, RUNX3,
SCGB3A1, SOCS1, VHL, ASCL1, BCAS1, OGG1, PMS2, RHOB |
Functional and pathway enrichment
analysis
The enriched GO functions for the DEGs are listed in
Table II. For the upregulated genes,
the enriched GO functions included the collagen catabolic process
(P=1.86×10−2), extracellular matrix disassembly
(P=4.50×10−3) and negative regulation of intrinsic
apoptotic signaling pathway in response to DNA damage by p53 class
mediator (P=3.07×10−3). For the downregulated genes, the
enriched GO functions included the negative regulation of cell
growth (P=1.39×10−4), stress-activated mitogen-activated
protein kinases (MAPK) cascade (P=1.73×10−3) and insulin
receptor signaling pathway (P=1.19×10−2).
 | Table II.Enriched GO functions for the
differentially-expressed genes. |
Table II.
Enriched GO functions for the
differentially-expressed genes.
| Gene
regulation | GO term | Description | Gene number | Gene symbol | P-value |
|---|
| Upregulated
genes | GO:0042158 | Lipoprotein
biosynthetic process | 5 | ABCA1, LCAT, MTTP,
PIGF, PPM1B |
1.30×10−4 |
|
| GO:0007204 | Elevation of
cytosolic calcium ion concentration | 5 | CCR1, CXCR3, GALR2,
LIME1, TPCN2 |
1.34×10−2 |
|
| GO:0022617 | Extracellular
matrix disassembly | 4 | CTSK, KLKB1, MMP3,
MMP7 |
4.50×10−3 |
|
| GO:0051592 | Response to calcium
ion | 4 | ANXA11, KCNMB1,
MTTP, SPARC |
5.28×10−3 |
|
| GO:0030574 | Collagen catabolic
process | 3 | CTSK, MMP3,
MMP7 |
1.86×10−2 |
|
| GO:0010875 | Positive regulation
of cholesterol efflux | 2 | ABCA1, ABCG1 |
3.07×10−3 |
|
| GO:1902166 | Negative regulation
of intrinsic apoptotic signaling pathway in response to DNA damage
by p53 class mediator | 2 | CD44, MUC1 |
3.07×10−3 |
|
| GO:003443 | Cholesterol
esterification | 2 | ABCG1, LCAT |
3.67×10−3 |
|
| GO:0034616 | Response to laminar
fluid shear stress | 2 | ABCA1, TGFB3 |
3.67×10−3 |
|
| GO:0034375 | High-density
lipoprotein particle remodeling | 2 | ABCG1, LCAT |
5.01×10−3 |
| Downregulated
genes | GO:0006469 | Negative regulation
of protein kinase activity | 8 | CDKN1A, DUSP1,
DUSP10, DUSP4, DVL1, GADD45G, PDPK1, SOCS1 |
4.97×10−5 |
|
| GO:0030308 | Negative regulation
of cell growth | 7 | BCL2, CDKN1A,
HSPA1B, PML, PPP1R9B, SCGB3A1, SERTAD3 |
1.39×10−4 |
|
| GO:0051403 | Stress-activated
MAPK cascade | 7 | AKT2, CCM2,
CDC42EP5, DUSP10, DUSP4, FOS, MAP2K3 |
1.73×10−3 |
|
| GO:0046777 | Protein
autophosphorylation | 6 | CAMKK2, ERN1,
PDGFA, PDPK1, PIM3, STK24 |
3.65×10−3 |
|
| GO:0006986 | Response to
unfolded protein | 5 | DNAJB1, ERN1,
HSPA1B, HSPA2, HSPA6 |
4.61×10−3 |
|
| GO:0008286 | Insulin receptor
signaling pathway | 5 | AKT2, EIF4EBP1,
PDPK1, SOCS1, TCIRG1 |
1.19×10−2 |
|
| GO:0048146 | Positive regulation
of fibroblast proliferation | 3 | CDKN1A, PDGFA,
S100A6 |
4.29×10−3 |
|
| GO:0045069 | Regulation of viral
genome replication | 3 | BCL2, OASL,
PPID |
9.26×10−3 |
|
| GO:0030833 | Regulation of actin
filament polymerization | 3 | ARPC1B, CDC42EP5,
PFN2 |
3.98×10−2 |
|
| GO:0008295 | Spermidine
biosynthetic process | 2 | AGMAT, AMD1 |
3.78×10−4 |
The enriched KEGG pathways for the DEGs are listed
in Table III. For the upregulated
genes, the enriched KEGG pathways included the pentose phosphate
pathway (P=2.43×10−2), hematopoietic cell lineage
(P=4.46×10−2) and rheumatoid arthritis
(P=4.84×10−2). Meanwhile, for the downregulated genes,
the enriched KEGG pathways included the MAPK signaling pathway
(P=3.77×10−6), pathways in cancer
(P=7.47×10−3) and toxoplasmosis
(P=7.24×10−5).
 | Table III.Enriched KEGG pathways for the
differentially-expressed genes. |
Table III.
Enriched KEGG pathways for the
differentially-expressed genes.
| Gene
regulation | Term | Description | Gene number | Gene symbol | P-value |
|---|
| Upregulated
genes | 30 | Pentose phosphate
pathway | 2 | H6PD, RBKS |
2.43×10−2 |
|
| 4640 | Hematopoietic cell
lineage | 3 | CD1C, CD44,
CR1 |
4.46×10−2 |
|
| 5323 | Rheumatoid
arthritis | 3 | CTSK, MMP3,
TGFB3 |
4.84×10−2 |
| Downregulated
genes | 4010 | MAPK signaling
pathway | 13 | AKT2, DUSP1,
DUSP10, DUSP4, FOS, GADD45G, GNA12, HSPA1A, HSPA1B, HSPA2, HSPA6,
MAP2K3, PDGFA |
3.77×10−6 |
|
| 5200 | Pathways in
cancer | 9 | AKT2, BCL2, CDKN1A,
DVL1, FOS, PDGFA, PML, TCF7, VHL |
7.47×10−3 |
|
| 5145 | Toxoplasmosis | 8 | AKT2, BCL2, HSPA1A,
HSPA1B, HSPA2, HSPA6, MAP2K3, SOCS1 |
7.24×10−5 |
|
| 4141 | Protein processing
in endoplasmic reticulum | 8 | BCL2, DNAJB1, ERN1,
HSPA1A, HSPA1B, HSPA2, HSPA6, HSPBP1 |
3.41×10−4 |
|
| 5215 | Prostate
cancer | 6 | AKT2, BCL2, CDKN1A,
PDGFA, PDPK1, TCF7 |
3.48×10−4 |
|
| 4910 | Insulin signaling
pathway | 6 | AKT2, EIF4EBP1,
HK1, PDPK1, PPP1R3C, SOCS1 |
3.42×10−3 |
|
| 4144 | Endocytosis | 6 | CHMP6, HSPA1A,
HSPA1B, HSPA2, HSPA6, PML |
2.01×10−2 |
|
| 3040 | Spliceosome | 5 | HSPA1A, HSPA1B,
HSPA2, HSPA6, PRPF6 |
1.13×10−2 |
|
| 5221 | Acute myeloid
leukemia | 4 | AKT2, EIF4EBP1,
PML, TCF7 |
3.11×10−3 |
|
| 5210 | Colorectal
cancer | 4 | AKT2, BCL2, FOS,
TCF7 |
4.22×10−3 |
PPI network and module
construction
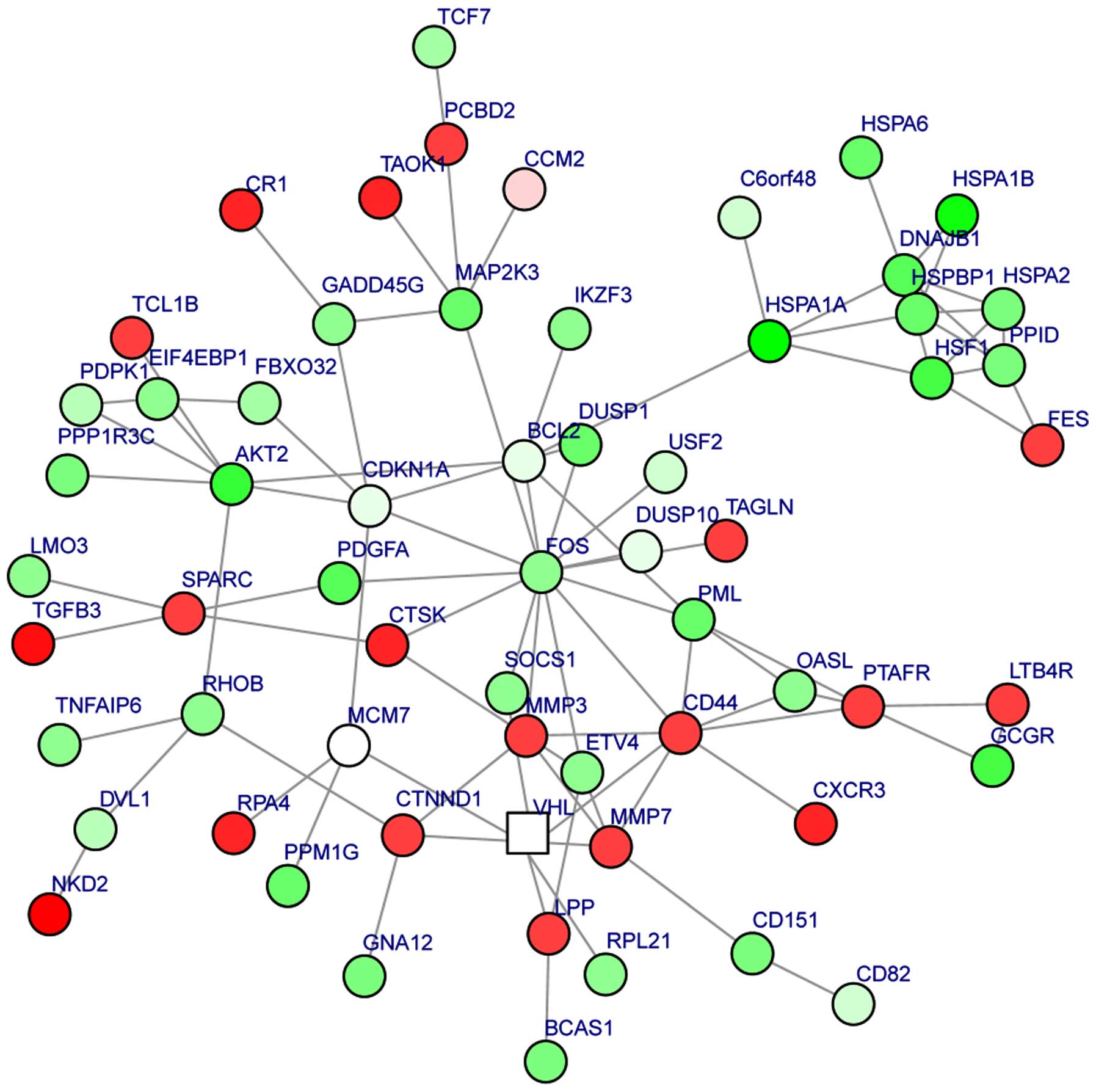
The PPI network consisted of 110 nodes and 136
interactions. There were 45 upregulated genes and 65 downregulated
genes in the PPI network of the DEGs (Fig. 1). The number of upregulated genes was
less than that of the downregulated genes. The DEGs, which encoded
proteins with connectivity degrees of ≥5, included FOS (degree,
15), CD44 (degree, 9), B-cell CLL/lymphoma 2 (BCL-2; degree, 7),
CDKN1A (degree, 7), DnaJ heat shock protein family (Hsp40) member
B1 (degree, 7), AKT2 (degree, 7) and matrix metalloproteinase 3
(MMP3; degree, 6).
A module was obtained from the PPI network of the
DEGs (Fig. 2). The module had 62
nodes and 93 interactions. In this module, the number of
upregulated genes was less than the number of downregulated genes.
Moreover, BCL2 could interact with FOS and CDKN1A, FOS could
interact with CDKN1A and CD44, and MMP7 could interact with MMP3
and CD44. The enriched GO functions for the DEGs in the module
included extracellular matrix organization
(P=2.86×10−6), response to drug (P=3.36×10−4)
and regulation of cell motility (P=1.37×10−3) (Table IV). The enriched KEGG pathways for
the DEGs in the module are listed in Table V, including pathways in cancer
(P=9.01×10−5), prostate cancer (P=3.98×10−5),
colorectal cancer (P=7.89×10−5) and acute myeloid
leukemia (P=7.44×10−4).
 | Table IV.Top 10 enriched GO functions for the
differentially-expressed genes in the module. |
Table IV.
Top 10 enriched GO functions for the
differentially-expressed genes in the module.
| GO term | Description | Gene number | Gene symbol | P-value |
|---|
| GO:0030198 | Extracellular
matrix organization | 9 | CTSK, MMP3, CD44,
CD151, SPARC, PDGFA, MMP7, VHL, TGFB3 |
2.86×10−6 |
| GO:0006954 | Inflammatory
response | 9 | LTB4R, CD44,
MAP2K3, FOS, CR1, DUSP10, PTAFR, CXCR3, TNFAIP6 |
1.41×10−4 |
| GO:0022612 | Gland
morphogenesis | 7 | BCL2, CD44, CTNND1,
ETV4, PML, PDGFA, TGFB3 |
2.37×10−7 |
| GO:0006469 | Negative regulation
of protein kinase activity | 7 | DVL1, CDKN1A,
GADD45G, SOCS1, DUSP10, DUSP1, PDPK1 |
2.82×10−6 |
| GO:0042493 | Response to
drug | 7 | BCL2, CDKN1A, MCM7,
FOS, PDGFA, MMP7, GNA12 |
3.36×10−4 |
| GO:2000145 | Regulation of cell
motility | 7 | BCL2, MMP3, FES,
AKT2, PDGFA, CXCR3, PDPK1 |
1.37×10−3 |
| GO:0043583 | Ear
development | 6 | BCL2, DVL1, CCM2,
SPARC, PDGFA, TGFB3 |
9.35×10−5 |
| GO:0051403 | Stress-activated
MAPK cascade | 6 | MAP2K3, AKT2, FOS,
CCM2, DUSP10, TAOK1 |
1.69×10−4 |
| GO:0043068 | Positive regulation
of programmed cell death | 6 | CDKN1A, PML, RHOB,
DUSP1, PPID, TGFB3 |
1.71×10−3 |
| GO:0060333 |
Interferon-γ-mediated signaling
pathway | 5 | CD44, OASL, SOCS1,
PML, PTAFR |
9.81×10−6 |
 | Table V.Enriched KEGG pathways for the
differentially-expressed genes in the module. |
Table V.
Enriched KEGG pathways for the
differentially-expressed genes in the module.
| Term | Description | Gene number | Gene symbol | P-value |
|---|
| 4010 | MAPK signaling
pathway | 14 | GADD45G, MAP2K3,
HSPA2, AKT2, HSPA1A, FOS, HSPA1B, PDGFA, DUSP10, DUSP1, GNA12,
TAOK1, HSPA6, TGFB3 |
2.91×10−9 |
| 5200 | Pathways in
cancer | 10 | BCL2, DVL1, CDKN1A,
AKT2, FOS, PML, PDGFA, VHL, TCF7, TGFB3 |
9.01×10−5 |
| 5145 | Toxoplasmosis | 9 | BCL2, MAP2K3,
HSPA2, AKT2, HSPA1A, SOCS1, HSPA1B, HSPA6, TGFB3 |
3.30×10−7 |
| 4141 | Protein processing
in endoplasmic reticulum | 7 | BCL2, DNAJB1,
HSPA2, HSPA1A, HSPA1B, HSPBP1, HSPA6 |
1.68×10−4 |
| 5215 | Prostate
cancer | 6 | BCL2, CDKN1A, AKT2,
PDGFA, TCF7, PDPK1 |
3.98×10−5 |
| 4144 | Endocytosis | 6 | HSPA2, HSPA1A,
HSPA1B, PML, HSPA6, TGFB3 |
3.17×10−3 |
| 5210 | Colorectal
cancer | 5 | BCL2, AKT2, FOS,
TCF7, TGFB3 |
7.89×10−5 |
| 4910 | Insulin signaling
pathway | 5 | AKT2, SOCS1,
EIF4EBP1, PPP1R3C, PDPK1 |
3.14×10−3 |
| 5221 | Acute myeloid
leukemia | 4 | AKT2, PML,
EIF4EBP1, TCF7 |
7.44×10−4 |
| 4612 | Antigen processing
and presentation | 4 | HSPA2, HSPA1A,
HSPA1B, HSPA6 |
2.18×10−3 |
Discussion
In the present study, a total of 284 DEGs were
screened, consisting of 145 upregulated genes and 139 downregulated
genes. In particular, downregulated FOS was an oncogene, while
downregulated CDKN1A was a tumor suppressor gene and upregulated
CD44 was classed as an ‘other’ gene. The enriched functions
included the collagen catabolic process, stress-activated MAPK
cascade and insulin receptor signaling pathway. Meanwhile, FOS
(degree, 15), CD44 (degree, 9), BCL2 (degree, 7), CDKN1A (degree,
7) and MMP3 (degree, 6) had higher connectivity degrees in the PPI
network for the DEGs. Also, a module was obtained from the PPI
network of the DEGs.
Bcl-2 belongs to an family of pro- and antiapoptotic
proteins (21), able to enhance cell
survival by inhibiting apoptosis (22). BCL2 is frequently expressed in primary
ovarian carcinoma and may be an important determinant of
drug-induced apoptosis in ovarian cancer (23,24). By
deregulating programmed cell death and changing sensitivity to
chemotherapy, alteration of the Bax/Bcl-2 balance may affect the
clinical course of ovarian cancer (25). As a member of the Bcl-2 family,
ABT-737 can significantly improve the activity of carboplatin and
thus may be of great use in the treatment of patients with ovarian
cancer (26). These data may indicate
that the expression level of BCL2 is associated with ovarian
cancer. The c-fos protein may act as a tumor suppressor and may be
involved in apoptosis (27).
Expression of Fos can affect the progression of ovarian carcinoma
and may function as a prognostic factor (28). Thus, we speculated that FOS may play a
role in ovarian cancer. In the module, the present study found that
BCL2 exhibited interaction associations with FOS, indicating that
BCL2 may also play a role in ovarian cancer by mediating FOS.
As one of the most important CDK inhibitors, p27Kip1
can function in the G1 cell cycle arrest induced by
anti-human epidermal growth factor receptor type 2 antibody and
inhibit tumor growth through post-translational regulation
(29). As the product of the CDKN2A
gene at chromosome 9p2, the CDK inhibitor p16 is associated with
the progression and unfavorable prognosis of ovarian cancer
(30,31). p21, also known as CDKN1A, regulates
several processes, including cell growth (32), differentiation (33) and apoptosis (34). Overexpressed CDK inhibitor p21 can
reduce the phosphorylation of retinoblastoma protein in
trichostatin A-treated ovarian cancer cells, and therefore p21 may
be involved in the cell cycle blockade and the differentiation of
the cells (35). These data may
indicate that the expression level of CDKN1A exhibits an
association with ovarian cancer. In the module in the present
study, it was found that CDKN1A could interact with FOS and BCL2,
indicating that CDKN1A may also affect ovarian cancer by mediating
the two genes.
CD44 is regarded as the main cell surface receptor
for hyaluronan (36), an
extracellular polysaccharide associated with the progression of
ovarian cancer (37). Significant
overexpression of CD44 in ovarian cancer cells can induce strong
adhesion to the peritoneal mesothelium (38,39), and
CD44 has a close association with the implantation of ovarian
cancer metastasis (40). The
CD44+/CD117+ status can be a molecular marker
of ovarian cancer-initiating cells (41). As a stem cell marker, CD44 combined
with higher aldehyde dehydrogenase 1 family member A1 activity may
be used to define ovarian cancer stem cells (42). These data indicate that CD44 may play
a role in ovarian cancer. In the module, the present study found
that CD44 had interaction associations with FOS, indicating that
CD44 may also play a role in ovarian cancer by mediating FOS.
MMPs are a family of zinc-dependent neutral
endopeptidases that are overexpressed in the tumor microenvironment
and are able to degrade essentially all extracellular matrix
components (43,44). Members of the MMP family are involved
in tumor cell metastasis and invasion, including the progression of
ovarian cancer (45). As the smallest
member of the MMP family, MMP7 can be overexpressed in malignant
ovarian epithelium and may assist tumor cell invasion by inducing
progelatinase activation (46).
Meanwhile, MMP7 may act as a marker for detection and as a target
for treatment in ovarian cancer (47). These data suggest that MMP7 may have a
close association with ovarian cancer. In the module, the present
study found there were several interaction associations (e.g.,
MMP3-MMP7 and MMP7-CD44), indicating that MMP7 may also play a role
in ovarian cancer by mediating MMP3 and CD44.
In conclusion, a comprehensive bioinformatics
analysis of DEGs that may affect ovarian cancer was conducted in
the present study. A total of 284 DEGs were screened, and it was
found that BCL2, FOS, CDKN1A, CD44, MMP3 and MMP7 may have a
correlation with ovarian cancer. However, further research is
required to unravel their roles in ovarian cancer.
References
|
1
|
Scully RE: Pathology of ovarian cancer
precursors. J Cell Biochem Suppl. 23:208–218. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Iorio MV and Croce CM: MicroRNA profiling
in ovarian cancer. Methods Mol Biol. 1049:187–197. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yuan ZQ, Sun M, Feldman RI, Wang G, Ma X,
Jiang C, Coppola D, Nicosia SV and Cheng JQ: Frequent activation of
AKT2 and induction of apoptosis by inhibition of
phosphoinositide-3-OH kinase/Akt pathway in human ovarian cancer.
Oncogene. 19:2324–2330. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yin BW and Lloyd KO: Molecular cloning of
the CA125 ovarian cancer antigen: Identification as a new mucin,
Muc16. J Biol Chem. 276:27371–27375. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Scotton CJ, Wilson JL, Milliken D, Stamp G
and Balkwill FR: Epithelial cancer cell migration: A role for
chemokine receptors? Cancer Res. 61:4961–4965. 2001.PubMed/NCBI
|
|
6
|
Strieter RM: Chemokines: Not just
leukocyte chemoattractants in the promotion of cancer. Nat Immunol.
2:285–286. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Scotton CJ, Wilson JL, Scott K, Stamp G,
Wilbanks GD, Fricker S, Bridger G and Balkwill FR: Multiple actions
of the chemokine CXCL12 on epithelial tumor cells in human ovarian
cancer. Cancer Res. 62:5930–5938. 2002.PubMed/NCBI
|
|
8
|
Campbell IG, Russell SE, Choong DY,
Montgomery KG, Ciavarella ML, Hooi CS, Cristiano BE, Pearson RB and
Phillips WA: Mutation of the PIK3CA gene in ovarian and breast
cancer. Cancer Res. 64:7678–7681. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Karakas B, Bachman K and Park B: Mutation
of the PIK3CA oncogene in human cancers. Br J Cancer. 94:455–459.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhao H, Shen J, Wang D, Guo Y, Gregory S,
Medico L, Hu Q, Yan L, Odunsi K, Lele S and Liu S: Associations
between gene expression variations and ovarian cancer risk alleles
identified from genome wide association studies. PloS One.
7:e479622012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Smyth GK: Limma: Linear models for
microarray data. Bioinformatics and Computational Biology Solutions
using R and Bioconductor. Springer. (New York). 397–420. 2005.
View Article : Google Scholar
|
|
13
|
Zhao M, Sun J and Zhao Z: TSGene: A web
resource for tumor suppressor genes. Nucleic Acids Res.
41:D970–D976. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen JS, Hung WS, Chan HH, Tsai SJ and Sun
HS: In silico identification of oncogenic potential of fyn-related
kinase in hepatocellular carcinoma. Bioinformatics. 29:420–427.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Palla G, Derényi I, Farkas I and Vicsek T:
Uncovering the overlapping community structure of complex networks
in nature and society. Nature. 435:814–818. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lu M, Zhang Q, Deng M, Miao J, Guo Y, Gao
W and Cui Q: An analysis of human microRNA and disease
associations. PloS One. 3:e34202008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Von Mering C, Huynen M, Jaeggi D, Schmidt
S, Bork P and Snel B: STRING: A database of predicted functional
associations between proteins. Nucleic Acids Res. 31:258–261. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
He X and Zhang J: Why do hubs tend to be
essential in protein networks? PLoS Genet. 2:e882006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Beisser D, Klau GW, Dandekar T, Müller T
and Dittrich MT: BioNet: An R-Package for the functional analysis
of biological networks. Bioinformatics. 26:1129–1130. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Baekelandt M, Kristensen GB, Nesland JM,
Tropé CG and Holm R: Clinical significance of apoptosis-related
factors p53, Mdm2 and Bcl-2 in advanced ovarian cancer. J Clin
Oncol. 17:20611999.PubMed/NCBI
|
|
22
|
White E: Life, death and the pursuit of
apoptosis. Genes Dev. 10:1–15. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Eliopoulos AG, Kerr DJ, Herod J, Hodgkins
L, Krajewski S, Reed JC and Young LS: The control of apoptosis and
drug resistance in ovarian cancer: Influence of p53 and Bcl-2.
Oncogene. 11:1217–1228. 1995.PubMed/NCBI
|
|
24
|
Miyashita T and Reed JC: Bcl-2 gene
transfer increases relative resistance of S49. 1 and WEHI7. 2
lymphoid cells to cell death and DNA fragmentation induced by
glucocorticoids and multiple chemotherapeutic drugs. Cancer Res.
52:5407–5411. 1992.PubMed/NCBI
|
|
25
|
Marx D, Binder C, Meden H, Lenthe T,
Ziemek T, Hiddemann T, Kuhn W and Schauer A: Differential
expression of apoptosis associated genes bax and bcl-2 in ovarian
cancer. Anti Cancer Res. 17:2233–2240. 1997.
|
|
26
|
Witham J, Valenti MR, De-Haven-Brandon AK,
Vidot S, Eccles SA, Kaye SB and Richardson A: The Bcl-2/Bcl-XL
family inhibitor ABT-737 sensitizes ovarian cancer cells to
carboplatin. Clin Cancer Res. 13:7191–7198. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Teng CS: Protooncogenes as mediators of
apoptosis. Int Rev Cytol. 197:137–202. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mahner S, Baasch C, Schwarz J, Hein S,
Wölber L, Jänicke F and Milde-Langosch K: C-Fos expression is a
molecular predictor of progression and survival in epithelial
ovarian carcinoma. Br J Cancer. 99:1269–1275. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Le XF, Claret FX, Lammayot A, Tian L,
Deshpande D, LaPushin R, Tari AM and Bast RC Jr: The role of
cyclin-dependent kinase inhibitor p27Kip1 in anti-HER2
antibody-induced G1 cell cycle arrest and tumor growth inhibition.
J Biol Chem. 278:23441–23450. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kamb A, Gruis NA, Weaver-Feldhaus J, Liu
Q, Harshman K, Tavtigian SV, Stockert E, Day RS III, Johnson BE and
Skolnick MH: A cell cycle regulator potentially involved in genesis
of many tumor types. Science. 264:436–440. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dong Y, Walsh MD, McGuckin MA, Gabrielli
BG, Cummings MC, Wright RG, Hurst T, Khoo SK and Parsons PG:
Increased expression of cyclin-dependent kinase inhibitor 2
(CDKN2A) gene product P16INK4A in ovarian cancer is associated with
progression and unfavourable prognosis. Int J Cancer. 74:57–63.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gartel AL, Serfas MS and Tyner AL:
p21–negative regulator of the cell cycle. Proc Soc Exp Biol Med.
213:138–149. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu H, Wade M, Krall L, Grisham J, Xiong Y
and Van Dyke T: Targeted in vivo expression of the cyclin-dependent
kinase inhibitor p21 halts hepatocyte cell-cycle progression,
postnatal liver development and regeneration. Genes Dev.
10:245–260. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ostrovsky O and Bengal E: The
mitogen-activated protein kinase cascade promotes myoblast cell
survival by stabilizing the cyclin-dependent kinase inhibitor,
p21WAF1 protein. J Biol Chem. 278:21221–21231. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Strait KA, Dabbas B, Hammond EH, Warnick
CT, Ilstrup SJ and Ford CD: Cell cycle blockade and differentiation
of ovarian cancer cells by the histone deacetylase inhibitor
trichostatin A are associated with changes in p21, Rb and Id
proteins. Mol Cancer Ther. 1:1181–1190. 2002.PubMed/NCBI
|
|
36
|
Aruffo A, Stamenkovic I, Melnick M,
Underhill CB and Seed B: CD44 is the principal cell surface
receptor for hyaluronate. Cell. 61:1303–1313. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fraser JR, Laurent TC and Laurent UB:
Hyaluronan: Its nature, distribution, functions and turnover. J
Intern Med. 242:27–33. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cannistra SA, Kansas GS, Niloff J,
DeFranzo B, Kim Y and Ottensmeier C: Binding of ovarian cancer
cells to peritoneal mesothelium in vitro is partly mediated by
CD44H. Cancer Res. 53:3830–3838. 1993.PubMed/NCBI
|
|
39
|
Cannistra SA, Abu-Jawdeh G, Niloff J,
Strobel T, Swanson L, Andersen J and Ottensmeier C: CD44 variant
expression is a common feature of epithelial ovarian cancer: Lack
of association with standard prognostic factors. J Clin Oncol.
13:1912–1921. 1995.PubMed/NCBI
|
|
40
|
Bourguignon LY, Zhu H, Chu A, Iida N,
Zhang L and Hung MC: Interaction between the adhesion receptor,
CD44 and the oncogene product, p185 HER2, promotes human ovarian
tumor cell activation. J Biol Chem. 272:27913–27918. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang S, Balch C, Chan MW, Lai HC, Matei
D, Schilder JM, Yan PS, Huang TH and Nephew KP: Identification and
characterization of ovarian cancer-initiating cells from primary
human tumors. Cancer Res. 68:4311–4320. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang YC, Yo YT, Lee HY, Liao YP, Chao TK,
Su PH and Lai HC: ALDH1-bright epithelial ovarian cancer cells are
associated with CD44 expression, drug resistance and poor clinical
outcome. Am J Pathol. 180:1159–1169. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Overall CM and López-Otín C: Strategies
for MMP inhibition in cancer: Innovations for the post-trial era.
Nat Rev Cancer. 2:657–672. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
44
|
Vihinen P and Kähäri VM: Matrix
metalloproteinases in cancer: Prognostic markers and therapeutic
targets. Int J Cancer. 99:157–166. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Schmalfeldt B, Prechtel D, Härting K,
Späthe K, Rutke S, Konik E, Fridman R, Berger U, Schmitt M, Kuhn W
and Lengyel E: Increased expression of matrix metalloproteinases
(MMP)-2, MMP-9 and the urokinase-type plasminogen activator is
associated with progression from benign to advanced ovarian cancer.
Clin Cancer Res. 7:2396–2404. 2001.PubMed/NCBI
|
|
46
|
Wang FQ, So J, Reierstad S and Fishman DA:
Matrilysin (MMP-7) promotes invasion of ovarian cancer cells by
activation of progelatinase. Int J Cancer. 114:19–31. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tanimoto H, Underwood LJ, Shigemasa K,
Parmley TH, Wang Y, Yan Y, Clarke J and O'Brien TJ: The matrix
metalloprotease pump-1 (MMP-7, Matrilysin): A candidate
marker/target for ovarian cancer detection and treatment. Tumor
Biol. 20:88–98. 1999. View Article : Google Scholar
|