Introduction
The insulin-like growth factor (IGF) axis has been
reported by several studies to be among the most deregulated
signaling pathways in several types of neoplasm, including
hepatocellular carcinoma (HCC) (1,2). The IGF
axis and principally the gate-keeper of this pathway, IGF-1
receptor (IGF-1R), has recently been identified as a potential
target for the treatment of HCC. To achieve this, several IGF-1R
inhibitors were developed, including small molecules, monoclonal
antibodies, antisense oligonucleotides and small interfering
(si)RNAs (3–7). However, an unpredictable mechanism of
resistance to IGF-1R inhibitors was observed (8). Compensatory activation of associated
downstream signaling pathways, including the phosphoinositide
3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR), Janus
kinase (JAK)/signal transducer and activator of transcription
(STAT) pathway and RAS/RAF/mitogen-activated protein kinase (MAPK)
signaling cascades (Fig. 1), emerged
as a critical mechanism of drug resistance in HCC cells (9–11). Hence,
targeting a single point in this pivotal signaling pathway was
found to induce compensatory up- and/or downstream activation,
resulting in drug resistance (12,13).
However, a vertical and/or horizontal blockage of this axis may
solve this problem.
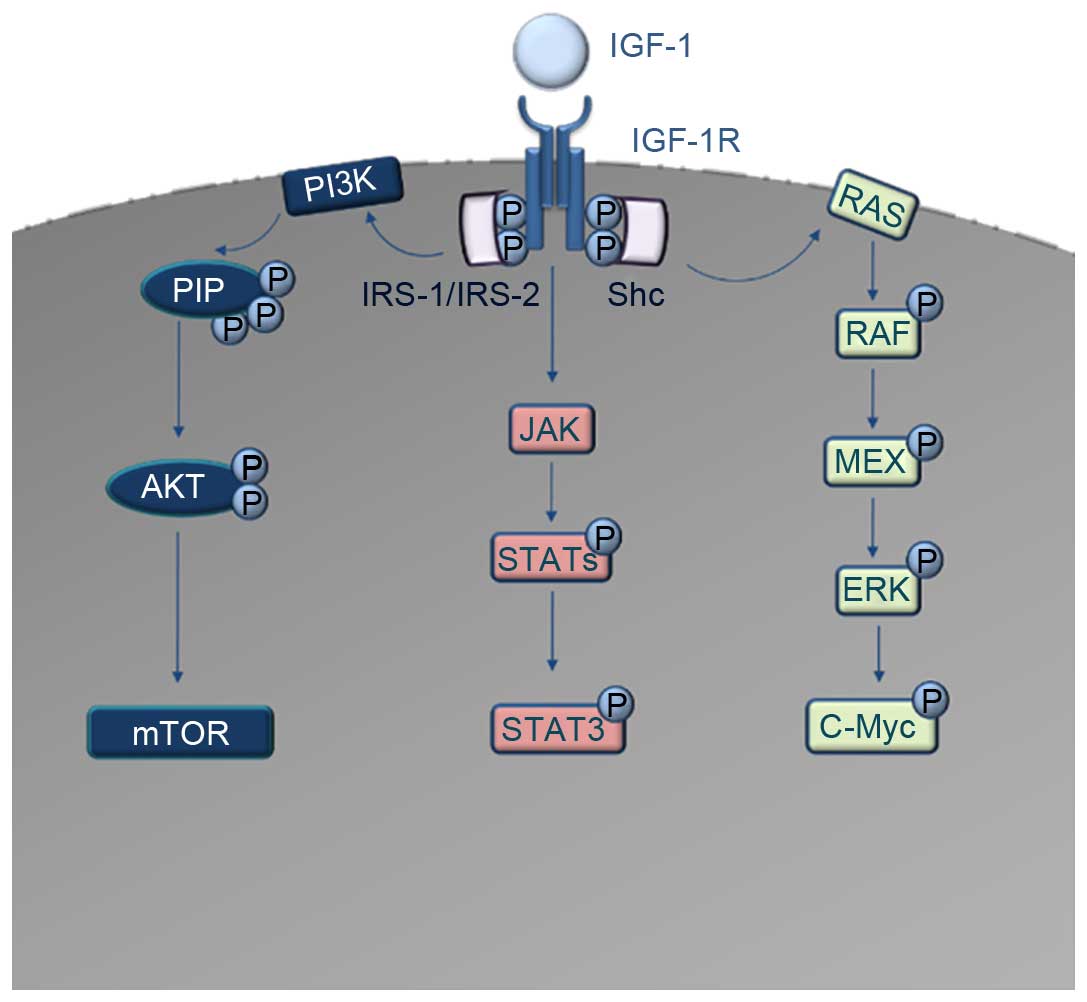 | Figure 1.Schematic diagram of the IGF-1R
downstream signaling cascades. IGF-1 acts as a ligand to IGF-1R.
Autophosphorylation and recruitment of the adaptor proteins IRS-1,
IRS-2 and Shc occurs. The interaction of IRS-1 and IRS-2 with
IGF-1R induces the activation of PI3K. PI3K converts PIP2 to PIP3,
which codes for the activation of AKT and consequently the
dis-inhibition of mTOR oncoprotein. In parallel, Shc activation
induces the activation of the RAS/RAF/MAPK pathway, which results
in activation and phosphorylation of c-Myc transcription factor.
Another signaling cascade downstream of IGF-1R is the JAK/STAT
pathway, where direct activation of Jak kinases occurs, leading to
the activation and phosphorylation of several STAT molecules. The
most important of these, STAT3, acts as a transcription factor for
numerous oncogenes. IGF-1, insulin-like growth factor 1; IGF-1R,
IGF-1 receptor; IRS, insulin receptor substrate; PI3K,
phosphatidylinositol 3-kinase; PIP2, phosphatidylinositol
4,5-bisphosphate; PIP3, phosphatidylinositol 3,4,5-trisphosphate;
mTOR, mammalian target of rapamycin; MAPK, mitogen-activate protein
kinase; JAK, Janus kinase; STAT, signal transducer and activator of
transcription. |
MicroRNAs (miRNAs/miRs) are small non-coding RNAs
(~22 nucleotides long) that act post-transcriptionally to regulate
gene expression (14). The idea of
using such epigenetic modulators as therapeutic tools is highly
compelling due to the fact that they can affect hundreds of targets
rather than a single one, a trait that gives them the power to shut
down an entire deregulated pathway (15). In addition, they have the ability to
tune the expression level of their targets instead of blunting it,
which is basically less damaging to healthy cells (16). In HCC, the deregulated miRNAs and
their role in HCC development have become a focus of interest, and
a set of miRNAs have been implicated in HCC carcinogenesis
(17,18). Currently, an miRNA drug, MRX34, is
undergoing a phase 1 clinical trial for patients with advanced HCC.
This drug is based around miRNA-34a, which is one of the most
prominent tumor suppressor miRNAs in HCC (19). The miRNA of other potential tumor
suppressors has been found to target several components of the
IGF-signaling pathway; for example, miR-615-5p, which was
previously reported by our group to directly target IGF-2 (20). Focusing on the gatekeeper of the
IGF-axis, IGF-1R, several miRNAs, including miR-145 and miR-99a,
have been found to act as tumor suppressors in HCC through direct
targeting of IGF-1R (21,22). Notably, it was recently reported that
miR-486-5p directly targets IGF-1R in lung cancer (23). However, this has not previously been
investigated in HCC. Therefore, the main aim of the present study
was to investigate the impact of miR-486-5p on IGF-1R and its
downstream signaling mediators, mTOR, STAT3 and c-Myc, in an
approach that aids in our understanding of the molecular mechanism
of miR-486-5p in HCC.
Materials and methods
Patients and tissue samples
The present study included 20 hepatitis C virus
(HCV)-induced HCC patients who underwent liver transplant surgery
in the Kasr El Einy Hospital, Cairo University (Cairo, Egypt). Due
to the scarcity of healthy liver donors, only 10 liver biopsies
were obtained from 10 healthy donors during liver transplantation
operation. All HCC patients were HCV-positive, with no metastases
or extrahepatic manifestations, and no invasion of the vascular
system. The healthy donors did not suffer from diabetes or
hypertension, and were negative for the hepatitis B virus (HBV) and
HCV, as shown in Table I. In total,
70% of the patients presented with >1 focal lesion, as
determined by the pathology report, and were subjected to clinical
assessment according to the Milan criteria (24), as shown in Table II. All patients provided written
informed consent, and the study was approved by the Ethical Review
Committee of Cairo University. The study complies with the
principles set forth in the Declaration of Helsinki and the
International Ethical Guidelines For Biomedical Research Involving
Human Subjects issued by the Council For International
Organizations Of Medical Sciences, and as previously described
(20,25).
 | Table I.Characteristic features of
HCV-induced HCC patients (n=20) and healthy controls (n=10). |
Table I.
Characteristic features of
HCV-induced HCC patients (n=20) and healthy controls (n=10).
| Characteristic | Value |
|---|
| HCC patients |
|
| Age
(range), yearsa | 48.8±7.8
(40.0–63.0) |
|
Males/females, n | 19/1 |
| Alcohol
abuse | None |
| AST,
U/la | 100.5±65.8 |
| ALT,
U/la |
85.6±95.6 |
| ALP,
U/la | 110.2±60.7 |
| Serum
albumin, g/dla |
4.6±1.5 |
| Serum
AFP, ng/mla | 155.7±22.3 |
| HCV Ab,
% (n) | 100.0 (20) |
| HBV Ab,
% (n) | 15.0 (3) |
| Healthy controls
(liver donors) |
|
| Mean ±
SD age (range), years | 32.1±7.1
(21.0–42.0) |
|
Males/females, n | 7/3 |
| Alcohol
abuse | None |
| HCV Ab,
% (n) | 0.0 (0) |
| HBV Ab,
% (n) | 0.0 (0) |
 | Table II.Number/sizes of lesions according to
the Milan criteria (24). |
Table II.
Number/sizes of lesions according to
the Milan criteria (24).
| Patient no. | Number of
lesions | Size of lesions,
cm |
|---|
| 1 | 3 focal
lesions | 1.5, 1 and 1 |
| 2 | Unifocal | 2.5 |
| 3 | 3 focal
lesions | 2, 2.5, 3 |
| 4 | 3 focal
lesions | 2, 2, 3.5 |
| 5 | Unifocal | 1.5×2 |
| 6 | 3 focal
lesions | 3×4, 1 and 1 |
| 7 | Unifocal | 4 |
| 8 | 3 focal
lesions | 4, 1 and 1 |
| 9 | 3 focal
lesions | 1, 1 and 1.5 |
| 10 | Unifocal | 2.5 |
| 11 | 2 focal
lesions | 1 and 1.7 |
| 12 | 3 focal
lesions | 1, 1 and 1 |
| 13 | Unifocal | 3 |
| 14 | 3 focal
lesions | 3, 1.5 and 2 |
| 15 | 3 focal
lesions | 1, 1 and 4 |
| 16 | 2 focal
lesions | 3 and 1.5 |
| 17 | 2 focal
lesions | 1.5 and 3 |
| 18 | 3 focal
lesions | 2.5, 2.5 and
1.5 |
| 19 | 3 focal
lesions | 1.5, 1 and 1 |
| 20 | Unifocal | 2 |
Cell culture and transfection of
oligonucleotides
Huh-7 cells (kindly provided by the Institute of
Pathology, Heidelberg University, Heidelberg, Germany) were
maintained in Dulbecco's modified Eagle's medium (DMEM; Lonza,
Basel, Switzerland), supplemented with 4.5 g/l glucose, 4 mmol/l
L-glutamine, 10% fetal bovine serum (Lonza) and Mycozap (1:500;
Lonza) (full DMEM) at 37°C, in a 5% CO2 atmosphere. The
Huh-7 cells were transfected with miR-486-5p mimics and IGF-1R
siRNAs as positive controls in this study (Qiagen GmbH, Hilden,
Germany). All transfection experiments were performed in
triplicates using HiPerfect Transfection Reagent (Qiagen GmbH)
according to the manufacturer's instructions, and experiments were
repeated three times. Cells that were only exposed to transfection
reagent were designated as mock cells, cells transfected with
miR-486-5p mimics were designated as miR-486-5p cells and cells
transfected with IGF-1R siRNAs were designated as IGF-1R siRNA
cells.
Total RNA extraction from liver
biopsies and HCC cell lines
mRNAs and miRNAs were extracted from the liver
biopsies and the HCC cells. Fresh liver samples (HCC and healthy
tissues) were collected during surgery and snap-frozen in liquid
nitrogen. The specimens were manually pulverized in liquid nitrogen
and ~100 mg of tissue powder was used for large and small RNA
extraction using the mirVana miRNA Isolation Kit (Ambion; Thermo
Fisher Scientific Inc., Waltham, MA, USA) according to the
manufacturer's instructions. The HCC cells were harvested at 48 h
post-transfection according to the HiPerfect Transfection Reagent
protocol: 37.5 ng oligonucleotides were used for Huh-7 cell
transfection in a 24-well plate. A spectrophotometer was used to
quantify the yield of RNA, and RNA integrity was tested by 18s rRNA
band detection using 1% agarose gel electrophoresis. The ratio of
absorbance at 260 and 280 nm was used to assess the purity and
quantity of RNA yield. A ratio of ~2.0 is generally accepted as
‘pure’ for RNA. Thus, RNA samples with absorbance values outside
the range of 1.8–2 were excluded from the study, as previously
described (20,25).
miRNA and mRNA quantification
The extracted miRNAs were reverse transcribed into
single-stranded complementary DNA (cDNA) using a TaqMan MicroRNA
Reverse Transcription (RT) kit (Applied Biosystems; Thermo Fisher
Scientific Inc.) and specific primers ((Applied Biosystems; Thermo
Fisher Scientific Inc.) for hsa-miR-486-5p and RNU6B. IGF-1R,
STAT3, mTOR and c-Myc mRNAs were reverse transcribed into cDNA
using the High-capacity cDNA RT kit (Applied Biosystems; Thermo
Fisher Scientific Inc.) according to the manufacturer's
instructions. The relative expression of miR-486-5p and RNU6B (as a
housekeepong gene for normalization), as well as IGF-1R, STAT3,
mTOR, c-Myc and β-2-microglobulin (β2M; as a housekeeping gene for
normalization) was quantified using TaqMan RT-quantitative
polymerase chain reaction (qPCR; Applied Biosystems Assay IDs:
0002470, 001093, Hs00609566_m1, Hs00374280_m1, Hs00234508_m1,
Hs00153408_m1 and Hs00984230_m1, respectively) on a StepOne™ Real
Time PCR instrument (Applied Biosystems; Thermo Fisher Scientific
Inc.). For every sample, a reaction mix was prepared according to
the manufacturer's instructions, and 4 µl of the respective cDNA
was added. The RT-qPCR run was performed in the standard mode,
consisting of two stages: A first 10-min stage at 95°C where the
Taq-polymerase enzyme was activated, followed by a second stage of
40 amplification cycles (15 sec at 95°C and 60 sec at 60°C).
Relative expression was calculated using the 2−ΔΔCq
method, as previously described (20,25).
All PCR reactions including controls were run in
triplicate reactions.
Cellular viability
In order to determine cellular viability, using the
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay, 10,000 Huh-7 cells were seeded in 200 µl of culture media
(full DMEM) per well in a 96-well plate and incubated 24 h prior to
transfection with 12.5 ng miR-486-5p mimics or IGF-1R siRNAs
(according to the HiPerfect protocol; Qiagen GmbH). At 48 h
post-transfection, 20 µl MTT solution (5 mg/ml MTT in
phosphate-buffered saline) was added to each well. Subsequent to
incubation for 5 h, formazan (MTT metabolic product) was
re-suspended in 200 µl dimethyl sulfoxide. Colorimetric
measurements and absorbance were performed using a Wallac 1420
Victor2 Multilabel Counter (Perkin Elmer Inc., Waltham, MA, USA),
as previously described (20,25).
All cell viability experiments were performed in
quadrate and repeated 5 times.
Cellular proliferation
In order to determine cellular proliferation, a BrdU
incorporation assay was used. The Huh-7 cells were seeded 24 h
prior to transfection into black 96-well plates and transfected
with 12.5 ng oligonucleotides (according to the HiPerfect protocol;
Qiagen GmbH) with an initial constant cell count of
5×104 cells/well. At 48 h post-oligonucleotide
transfection, the cells were labeled with BrdU labeling reagent for
4 h (with a final concentration of 100 µM) using the Cell
Proliferation ELISA kit (Roche Applied Science, Penzberg, Germany).
The cells were then fixed using FixDenate for 30 min and incubated
with Anti-BrdU POD (with a final concentration of 10 µM) for 90
min, as previously described (20,25).
All cellular proliferation experiments were
performed in quadrate and repeated 5 times.
Growth assays
Colony-forming assay
Huh-7 cells (1,500 cells/well) were seeded and left
to adhere overnight. At 48 h post-transfection, the cells were
detached and imbedded in soft agarose; the bottom layer was
prepared with 0.76% agarose and the top layer with 0.36% agarose,
in culture media (full DMEM). The cells were incubated at 37°C for
2 weeks to allow for colonization. The colonies were then stained
with crystal violet dye and their numbers were counted per well.
All colony-forming assays were performed in triplicate (3
wells/test) and repeated 5 times, as described previously (20,25).
Cell scratch wound healing assay
Huh-7 cells were left in 24-well plates until they
had reached 85–95% confluence. At 48 h post-transfection, 3
scratches/well were made in each plate with a 20-µl pipette tip.
Serum-free medium was used to wash off detached cells. Culture
medium (full DMEM) was then added, and the culture plates were
incubated at 37°C, in a 5% CO2 atmosphere. At 24 h post-scratching,
migration was documented, and the quantification of wound closure
was performed using Zen2012 software by measuring the surface area
of the scratch. All scratch assays were performed in triplicate (3
wells/test, representing 9 scratches/test) and repeated 3 times, as
described previously (20,25).
Statistical analysis
A Mann-Whitney U test was performed to compare gene
expression between two independent groups. P<0.05 was considered
to indicate a statistically significant difference. All the data
were statistically analyzed using GraphPad Prism 5.0 (Graphpad
Software Inc., La Jolla, CA, USA).
Results
Screening of miR-486-5p in healthy
liver tissues, HCV-induced HCC liver tissues and Huh-7 cell
lines
miR-486-5p exhibited significant downregulation in
the HCV-induced HCC tissues (P=0.0022) and Huh-7 cell lines
(P=0.0121) when compared with the liver tissues obtained from
healthy donors (Fig. 2A).
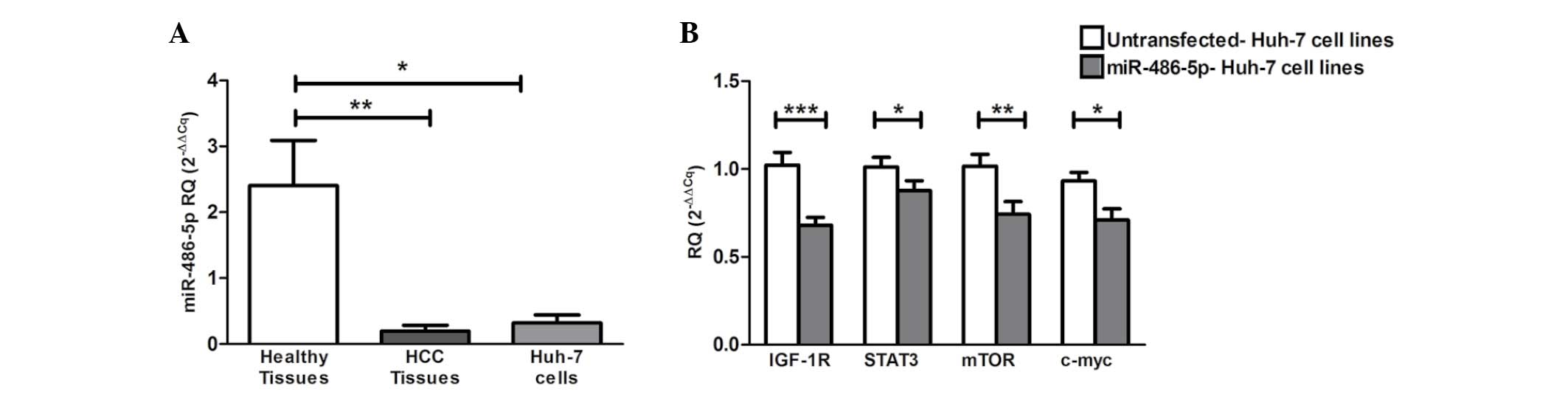 | Figure 2.Screening of miR-486-5p in HCC
tissues and Huh-7 cells, and the impact of ectopic expression of
miR-486-5p on IGF-1R and its downstream mediators in Huh-7 cells.
(A) Screening of miR-486-5p in liver tissues showed significant
downregulation in the liver tissues obtained from the HCC patients
(P=0.0022) and in the Huh-7 cells (P=0.0121) when compared with
normal liver tissues. (B) Forcing the expression of miR-486-5p in
Huh-7 cells resulted in the repression of IGF-1R mRNA expression
compared with that of the mock cells (P=0.0003). Similarly, STAT3,
mTOR and c-Myc mRNA levels were also found to be significantly
downregulated compared with those of the mock cells (P=0.0472,
P=0.0086 and P=0.0159, respectively). *P<0.05, **P<0.01 and
***P<0.001. HCC, hepatocellular carcinoma; IGF-1, insulin-like
growth factor 1; IGF-1R, IGF-1 receptor; mTOR, mammalian target of
rapamycin; STAT, signal transducer and activator of transcription;
miR, microRNA; RQ, relative quantification. |
Impact of miR-486-5p on its target
IGF-1R and its downstream mediators in Huh-7 cells
The ectopic expression of miR-486-5p in the Huh-7
cells resulted in significant downregulation of the mRNA levels of
IGF-1R and its downstream mediators mTOR, STAT3 and c-Myc compared
with those of the mock cells (P=0.0003, P=0.0472, P=0.0086 and
P=0.0159, respectively) (Fig.
2B).
Functional analysis of miR-486-5p in
Huh-7 cell lines
After proving the impact of miR-486-5p on IGF-1R and
its downstream signaling cascades, given that miR-486-5p has been
rarely investigated in HCC and has never been functionally analyzed
in Huh-7 cell lines, several functional analysis experiments were
performed to evaluate the overall effect of miR-486-5p on various
characteristic properties of tumor cells.
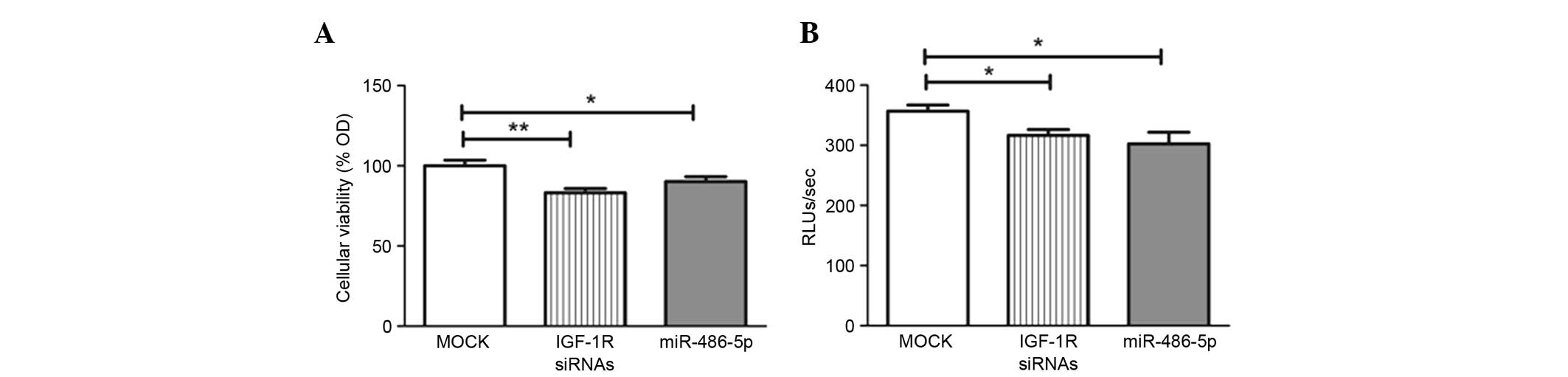
Impact of miR-486-5p on cellular
viability and proliferation
An MTT assay was used for investigating cell
viability and growth. Transfection with miR-486-5p mimics led to a
significant decline in cellular viability compared with that in the
mock cells (P=0.0301), in the same manner as the positive control
IGF-1R siRNAs used in the study (Fig.
3A).
A BrdU incorporation assay was used for
investigating cell proliferation and growth. Forcing the expression
of miR-486-5p significantly reduced cellular proliferation compared
with the mock Huh-7 cells (P=0.0325), in the same manner as the
IGF-1R siRNAs (Fig. 3B).
Impact of miR-486-5p on cellular
migration in Huh-7 cells
Images for the two-dimensional scratch-migration
assay were documented at 10-fold magnification. Cellular migration
was calculated as the percentage of wound closure. Representative
pictures of the scratches are shown in Fig. 4A. The transfection of the Huh-7 cells
with miR-486-5p mimics led to a marked reduction in tumor cell
migration compared with that in the mock cells (P=0.0286), which
was comparable to the effect of IGF-1R siRNAs (Fig. 4B).
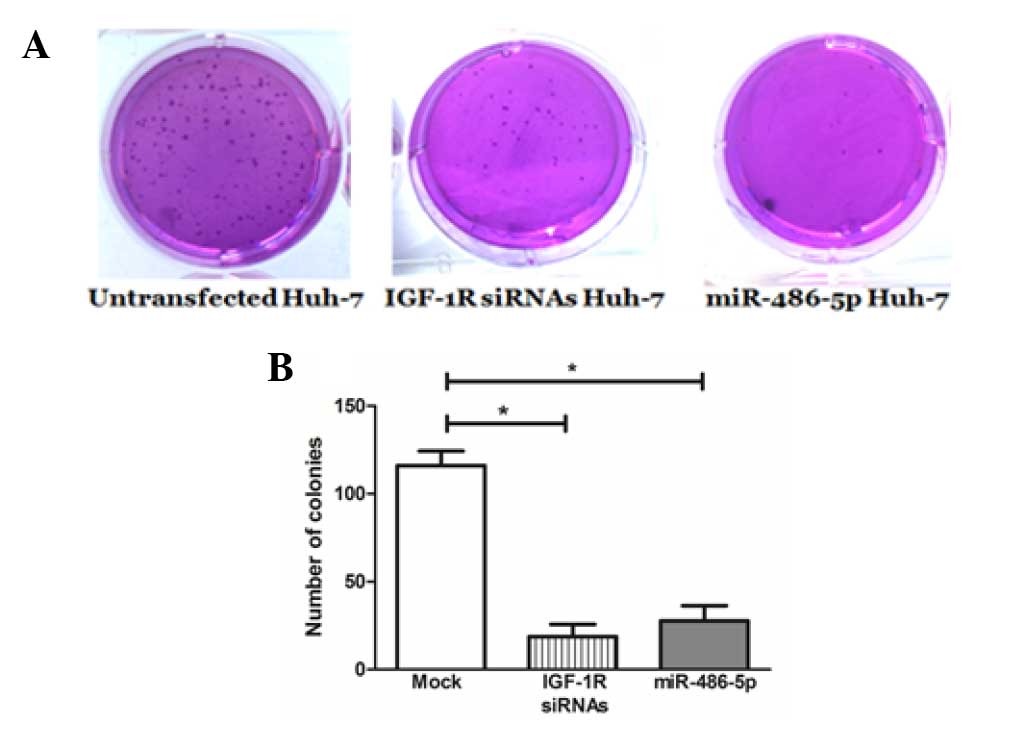
Impact of miR-486-5p on cellular
anchorage-independent growth in Huh-7 cells
miR-486-5p intended overexpression resulted in a
considerable inhibition of colony formation (26.7±2.4) in the Huh-7
cells compared with the mock cells (114.3±7.9), which was
comparable to the positive control knockdown cells that were
transfected with IGF-1R siRNAs (22.4±2.8) (Fig. 5).
Discussion
Deregulation of the IGF-axis is renowned to
contribute to each stage of HCC progression and treatment
resistance (1,26–28). On
the other hand, studies have identified few potential targets for
miR-486-5p, including the antiapoptotic factor olfactomedin-4
(29), Rho GTPase-activating protein
5 (30), the tumor suppressor
phosphatase and tensin homolog (31),
the proto-oncogene Pim-1 kinase (32)
and the chief gatekeeper of the IGF-axis IGF-1R, which was also
reported to be a validated target for miR-486-5p in lung cancer
(23) thus rendering it as a
potential candidate for investigation in HCC.
Studies investigating miR-486-5p in HCC are scarce,
and its expression profile has not previously been investigated in
HCV-induced HCC liver tissues. Therefore, the present study aimed
to screen for miR-486-5p in HCV-induced HCC liver tissues compared
with healthy liver tissues. Significant downregulation of
miR-486-5p was found in the HCV-induced HCC tissues, as well as in
Huh-7 cells (Fig. 2). This
corroborates a recent study performed on Chinese patients in which
miR-486-5p was also downregulated in HBV-induced HCC tissues
(33).
It is important to note that miR-486-5p has also
been found to be downregulated in numerous other cancer types,
including pancreatic cancer (34),
gastric cancer (29), osteosarcoma
(35), and lung cancer (23), however, the mechanism of miR-486-5p
downregulation in those types of cancer, including HCC, is largely
unknown. According to previous studies, miR-486-5p downregulation
in HCC tissues could be highly accredited to its chromosomal
location (8p11.21), which is one of the most typically occurring
genomic deletion regions, containing potential tumor suppressor
genes (29). Epigenetic silencing
through DNA methylation and/or histone deacetylation could also be
one of the reasons for miR-486-5p downregulation, particularly in
HCV-induced HCC tissues, as HCV core protein was recently reported
to directly induce DNA methylation in HCC tissues (36,37).
However, further studies are required to investigate the exact
mechanisms underlying miR-486-5p suppression in HCC.
To elucidate the molecular mechanism of miR-486-5p
in HCC, the IGF-axis was the main concern of the present study.
miR-486-5p was found to negatively regulate its validated target
IGF-1R in Huh-7 cells. Nevertheless, miR-486-5p was able to block
the IGF-signaling pathways downstream of IGF-1R, namely the
PI3K/AKT/mTOR, JAK/STAT and RAS/RAF/MAPK pathways, by the
downregulation of mTOR, STAT3 and c-Myc mRNA levels, respectively
(Fig. 2). This could be a direct or
indirect effect of miR-486-5p on the downstream mediators, as
bioinformatically, miR-486-5p was predicted to target the 3′-UTR of
STAT3. By contrast, miR-486-5p was recently found to target an
upstream regulator of PI3K/AKT/mTOR known as p85α, thus inhibiting
the activation of the PI3K/AKT/mTOR pathway and its interlocked
RAS/RAF/MAPK signaling cascade, and indirectly acting as a
repressor for mTOR and c-Myc transcription (33,38). It is
important to note that due to the limited efficacy of IGF-1R
inhibitors as a single therapy in HCC and the compensatory
activation of downstream signaling pathways resulting in drug
resistance (9–13), a combination of agents targeting
multiple molecules in IGF-axis, known as vertical blockade, were
previously evaluated in several preclinical studies, and
synergistic anti-cancer efficacy was demonstrated (39,40).
However, the current study nominates miR-486-5p as a single
endogenous biological molecule that could vertically and
horizontally block the IGF-signaling pathway, as miR-486-5p was not
only able to repress the downstream signaling cascades of IGF-1R,
but it could also repress the mRNA level of its other validated
target IGF-1, the main ligand of the IGF-axis (23) in Huh-7 cells (Youness et al,
unpublished data). This feature ranks miR-486-5p as a possible
therapeutic target for HCC, with an expected synergistic anti-tumor
effect and reduced resistance compared with any other IGF-1R
inhibitors. Further investigations are required to experimentally
validate the superiority of miR-486-5p over other IGF-1R inhibitors
in the treatment of HCC.
Due to the fact that miR-486-5p was found to have a
dual action and can function as a cancer-specific tumor suppressor,
as in gastric cancer (29) and
non-small cell lung cancer (23,30), or as
an aggressive oncomiR as in case of colorectal cancer (41) and glioblastoma (42), it was tempting to mechanistically
investigate the role for miR-486-5p as either a tumor suppressor
miRNA or an oncomiR in HCC. In the current study, miR-486-5p was
shown to act as a tumor suppressor miRNA in HCC as it markedly
reduced the Huh-7 cell viability, proliferation, migration and
clonogenicity in a similar pattern to that exhibited by IGF-1R
siRNAs (Figs. 3–5). These results are in line with those of a
recent study performed on QGY-7701 and QGY-7703 HCC cell lines,
which reported that miR-486-5p also repressed the migration and
induced apoptosis of those types of cells (33).
In conclusion, the potential role of miR-486-5p in
vertically and horizontally blocking the IGF-axis, represented by
repressing the expression levels of IGF-1R and its downstream
mediators mTOR, STAT3 and c-Myc in Huh-7 cells, was investigated in
this study. Mechanistically, miR-486-5p was found to act as a
potential tumor suppressor miRNA in HCC. Further studies are
required to determine if miR-486-5p could be used as a potential
therapeutic target for the treatment of HCC.
Glossary
Abbreviations
Abbreviations:
|
HCC
|
hepatocellular carcinoma
|
|
IGF-1R
|
insulin-like growth factor 1
receptor
|
|
BrdU
|
bromodeoxyuridine
|
|
STAT3
|
signal transducer and activator of
transcription 3
|
|
mTOR
|
mammalian target of rapamcyin
|
|
MTT
|
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
|
References
|
1
|
Pollak M: The insulin and insulin-like
growth factor receptor family in neoplasia: An update. Nat Rev
Cancer. 12:159–169. 2012.PubMed/NCBI
|
|
2
|
Cornellà H, Alsinet C and Villanueva A:
Molecular pathogenesis of hepatocellular carcinoma. Alcohol Clin
Exp Res. 35:821–825. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cervello M, McCubrey JA, Cusimano A,
Lampiasi N, Azzolina A and Montalto G: Targeted therapy for
hepatocellular carcinoma: Novel agents on the horizon. Oncotarget.
3:236–260. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chang AY and Wang M: In-vitro growth
inhibition of chemotherapy and molecular targeted agents in
hepatocellular carcinoma. Anticancer Drugs. 24:251–259. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cohen BD, Baker DA, Soderstrom C,
Tkalcevic G, Rossi AM, Miller PE, Tengowski MW, Wang F, Gualberto
A, Beebe JS and Moyer JD: Combination therapy enhances the
inhibition of tumor growth with the fully human anti-type 1
insulin-like growth factor receptor monoclonal antibody CP-751,871.
Clin Cancer Res. 11:2063–2073. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lin RX, Wang ZY, Zhang N, Tuo CW, Liang
QD, Sun YN and Wang SQ: Inhibition of hepatocellular carcinoma
growth by antisense oligonucleotides to type I insulin-like growth
factor receptor in vitro and in an orthotopic model. Hepatol Res.
37:366–375. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rodon J, DeSantos V, Ferry RJ Jr and
Kurzrock R: Early drug development of inhibitors of the
insulin-like growth factor-I receptor pathway: Lessons from the
first clinical trials. Mol Cancer Ther. 7:2575–2588. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen KF, Yeh PY, Yeh KH, Lu YS, Huang SY
and Cheng AL: Down-regulation of phospho-Akt is a major molecular
determinant of bortezomib-induced apoptosis in hepatocellular
carcinoma cells. Cancer Res. 68:6698–6707. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
van Malenstein H, Dekervel J, Verslype C,
Van Cutsem E, Windmolders P, Nevens F and van Pelt J: Long-term
exposure to sorafenib of liver cancer cells induces resistance with
epithelial-to-mesenchymal transition, increased invasion and risk
of rebound growth. Cancer Lett. 329:74–83. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tai WT, Cheng AL, Shiau CW, Liu CY, Ko CH,
Lin MW, Chen PJ and Chen KF: Dovitinib induces apoptosis and
overcomes sorafenib resistance in hepatocellular carcinoma through
SHP-1-mediated inhibition of STAT3. Mol Cancer Ther. 11:452–463.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gedaly R, Angulo P, Hundley J, Daily MF,
Chen C, Koch A and Evers BM: PI-103 and sorafenib inhibit
hepatocellular carcinoma cell proliferation by blocking
Ras/Raf/MAPK and PI3K/AKT/mTOR pathways. Anticancer Res.
30:4951–4958. 2010.PubMed/NCBI
|
|
12
|
O'Reilly KE, Rojo F, She QB, Solit D,
Mills GB, Smith D, Lane H, Hofmann F, Hicklin DJ, Ludwig DL, et al:
mTOR inhibition induces upstream receptor tyrosine kinase signaling
and activates Akt. Cancer Res. 66:1500–1548. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zitzmann K, Rüden Jv, Brand S, Göke B,
Lichtl J, Spöttl G and Auernhammer CJ: Compensatory activation of
Akt in response to mTOR and Raf inhibitors-a rationale for
dual-targeted therapy approaches in neuroendocrine tumor disease.
Cancer Lett. 295:100–109. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pillai RS, Bhattacharyya SN and Filipowicz
W: Repression of protein synthesis by miRNAs: How many mechanisms?
Trends Cell Biol. 17:118–126. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lan FF, Wang H, Chen YC, Chan CY, Ng SS,
Li K, Xie D, He ML, Lin MC and Kung HF: Hsa-let-7g inhibits
proliferation of hepatocellular carcinoma cells by downregulation
of c-Myc and upregulation of p16(INK4A). Int J Cancer. 128:319–331.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
He L, He X, Lim LP, de Stanchina E, Xuan
Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al: A microRNA
component of the p53 tumour suppressor network. Nature.
447:1130–1134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
El Tayebi HM, Hosny KA, Esmat G, Breuhahn
K and Abdelaziz AI: miR-615-5p is restrictedly expressed in
cirrhotic and cancerous liver tissues and its overexpression
alleviates the tumorigenic effects in hepatocellular carcinoma.
FEBS Lett. 586:3309–3316. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Law PT, Ching AK, Chan AW, Wong QW, Wong
CK, To KF and Wong N: MiR-145 modulates multiple components of the
insulin-like growth factor pathway in hepatocellular carcinoma.
Carcinogenesis. 33:1134–1341. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li D, Liu X, Lin L, Hou J, Li N, Wang C,
Wang P, Zhang Q, Zhang P, Zhou W, et al: MicroRNA-99a inhibits
hepatocellular carcinoma growth and correlates with prognosis of
patients with hepatocellular carcinoma. J Biol Chem.
286:36677–36685. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Peng Y, Dai Y, Hitchcock C, Yang X, Kassis
ES, Liu L, Luo Z, Sun HL, Cui R, Wei H, et al: Insulin growth
factor signaling is regulated by microRNA-486, an underexpressed
microRNA in lung cancer. Proc Natl Acad Sci USA. 110:15043–15048.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mazzaferro V, Regalia E, Doci R, Andreola
S, Pulvirenti A, Bozzetti F, Montalto F, Ammatuna M, Morabito A and
Gennari L: Liver transplantation for the treatment of small
hepatocellular carcinomas in patients with cirrhosis. N Engl J Med.
334:693–699. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
El Tayebi HM, Omar K, Hegy S, El Maghrabi
M, El Brolosy M, Hosny KA, Esmat G and Abdelaziz AI: Repression of
miR-17-5p with elevated expression of E2F-1 and c-MYC in
non-metastatic hepatocellular carcinoma and enhancement of cell
growth upon reversing this expression pattern. Biochem Biophys Res
Commun. 434:421–427. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
De Meyts P: Insulin and its receptor:
Structure, function and evolution. Bioessays. 26:1351–1362. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Durzyńska J: IGF axis and other factors in
HPV-related and HPV-unrelated carcinogenesis (review). Oncol Rep.
32:2295–2306. 2014.PubMed/NCBI
|
|
28
|
Shaw RJ, Lamia KA, Vasquez D, Koo SH,
Bardeesy N, Depinho RA, Montminy M and Cantley LC: The kinase LKB1
mediates glucose homeostasis in liver and therapeutic effects of
metformin. Science. 310:1642–1646. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Oh HK, Tan AL, Das K, Ooi CH, Deng NT, Tan
IB, Beillard E, Lee J, Ramnarayanan K, Rha SY, et al: Genomic loss
of miR-486 regulates tumor progression and the OLFM4 antiapoptotic
factor in gastric cancer. Clin Cancer Res. 17:2657–2667. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang J, Tian X, Han R, Zhang X, Wang X,
Shen H, Xue L, Liu Y, Yan X, Shen J, et al: Downregulation of
miR-486-5p contributes to tumor progression and metastasis by
targeting protumorigenic ARHGAP5 in lung cancer. Oncogene.
33:1181–1189. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Small EM, O'Rourke JR, Moresi V,
Sutherland LB, McAnally J, Gerard RD, Richardson JA and Olson EN:
Regulation of PI3-kinase/Akt signaling by muscle-enriched
microRNA-486. Proc Natl Acad Sci USA. 107:4218–4223. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pang W, Tian X, Bai F, Han R, Wang J, Shen
H, Zhang X, Liu Y, Yan X, Jiang F and Xing L: Pim-1 kinase is a
target of miR-486-5p and eukaryotic translation initiation factor
4E, and plays a critical role in lung cancer. Mol Cancer.
13:2402014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang XP, Hou J, Shen XY, Huang CY, Zhang
XH, Xie YA and Luo XL: MicroRNA-486-5p, which is downregulated in
hepatocellular carcinoma, suppresses tumor growth by targeting
PIK3R1. FEBS J. 282:579–594. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ali S, Saleh H, Sethi S, Sarkar FH and
Philip PA: MicroRNA profiling of diagnostic needle aspirates from
patients with pancreatic cancer. Br J Cancer. 107:1354–1360. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Namløs HM, Meza-Zepeda LA, Barøy T,
Østensen IH, Kresse SH, Kuijjer ML, Serra M, Bürger H,
Cleton-Jansen AM and Myklebost O: Modulation of the osteosarcoma
expression phenotype by microRNAs. PLoS One. 7:e480862012.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lim JS, Park SH and Jang KL: Hepatitis C
virus Core protein overcomes stress-induced premature senescence by
down-regulating p16 expression via DNA methylation. Cancer Lett.
321:154–161. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dong Y and Wang A: Aberrant DNA
methylation in hepatocellular carcinoma tumor suppression (Review).
Oncol Lett. 8:963–968. 2014.PubMed/NCBI
|
|
38
|
Pourdehnad M, Truitt ML, Siddiqi IN,
Ducker GS, Shokat KM and Ruggero D: Myc and mTOR converge on a
common node in protein synthesis control that confers synthetic
lethality in Myc-driven cancers. Proc Natl Acad Sci USA.
110:11988–11993. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mazzoletti M, Bortolin F, Brunelli L,
Pastorelli R, Di Giandomenico S, Erba E, Ubezio P and Broggini M:
Combination of PI3K/mTOR inhibitors: Antitumor activity and
molecular correlates. Cancer Res. 71:4573–4584. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Floc'h N, Kinkade CW, Kobayashi T, Aytes
A, Lefebvre C, Mitrofanova A, Cardiff RD, Califano A, Shen MM and
Abate-Shen C: Dual targeting of the Akt/mTOR signaling pathway
inhibits castration-resistant prostate cancer in a genetically
engineered mouse model. Cancer Res. 72:4483–4493. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ragusa M, Majorana A, Statello L, Maugeri
M, Salito L, Barbagallo D, Guglielmino MR, Duro LR, Angelica R,
Caltabiano R, et al: Specific alterations of microRNA transcriptome
and global network structure in colorectal carcinoma after
cetuximab treatment. Mol Cancer Ther. 9:3396–3409. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Song L, Lin C, Gong H, Wang C, Liu L, Wu
J, Tao S, Hu B, Cheng SY, Li M and Li J: miR-486 sustains NF-kB
activity by disrupting multiple NF-kB-negative feedback loops. Cell
Res. 23:274–289. 2013. View Article : Google Scholar : PubMed/NCBI
|