Introduction
Laryngeal squamous cell carcinoma (LSCC) is the
second most common head and neck cancer in the world (1). The incidence of LSCC in China has
exhibited an increasing trend in the young population (2,3). LSCC is a
complex disease, and it is hypothesized that both genetic and
environmental factors play a role in it (4). Epigenetic modifications, including DNA
methylation, are known as the bridging factors of genetics and the
environment (5). However, the role of
epigenetic modifications in the pathogenesis and progression of
LSCC remains to be explained.
DNA methylation of CpG islands (CGIs) is a
significant regulatory mechanism of gene expression (6). Epigenetic silencing of tumor suppressor
genes by promoter hypermethylation is considered to be a
significant event in the progression of cancers including leukemia
(7), lung cancer (8) and prostate cancer (9). Gene-body methylation levels are also
demonstrated to be correlated with gene expression (10,11).
TGFB2 is a member of the transforming growth
factor β (TGFB) family, which encodes multifunctional peptides in
the regulation of cell proliferation, differentiation, adhesion and
migration (12). TGFB is able to
inhibit tumor growth at the early stages and promote tumor growth
at the late stages of disease (13).
A previous study has demonstrated that increased TGFB2
promoter methylation level is associated with prostate cancer
progression (14). BCL2L11
encodes a member of the BCL-2 (B cell lymphoma 2) protein family,
which acts as an apoptotic activator that is involved in a wide
variety of cellular activities (15).
BCL2L11 methylation has been noted to be associated with the
risk of hematological and epithelial cancers (16).
In the current study, the methylation levels of
TGFB2 and BCL2L11 were investigated in male laryngeal
cancer patients to determine whether these epigenetic markers are
associated with LSCC risk.
Materials and methods
Tissue samples
LSCC and adjacent tissues were collected from 90
male patients (mean age, 60.5±8.65 years) who had undergone
surgical treatment at the Department of Otolaryngology at Ningbo
Lihuili Hospital, Zhejiang, China. The 90 tumors consisted of 43
well-differentiated cases, 32 moderately differentiated cases and
15 poorly differentiated cases. Among the LSCC patients, there were
27 stage I, 14 stage II, 11 stage III and 38 stage IV patients. All
specimens were obtained freshly during the surgery and stored at
−80°C. Permission was obtained from the bioethics committee at
Ningbo Lihuili Hospital. All patients signed informed consent
forms.
DNA bisulfite treatment and
pyrosequencing analysis
Genomic DNA was isolated from tumor samples and then
quantified as previously described (11,17).
TGFB2 and BCL2L11 methylation levels were measured
using bisulfite pyrosequencing technology. DNA conversion was
performed using reagents provided in the Zymo EZ DNA
Methylation-Gold kit (Zymo Research, Orange, CA, USA). The
pyrosequencing procedures were performed as described previously
(11,17). The primer sequences of TGFB2
were 5′-GAGTAATTTTAAGTTGGGGAGAAGTTAGT-3′ for the forward primer,
5′-Biotin-ACTCCAAACCCCAACCCAACCA-3′ for the reverse primer, and
5′-CCAACCCAACCACAA-3′ for the sequencing primer. The primer
sequences of BCL2L11 were 5′-GAGGAAGTTGTTGGAGGAGAAT-3′ for
the forward primer, 5′-Biotin-ACCCTACCCATCCCTATAC-3′ for the
reverse primer, and 5′-ATCCCCAAACCCAAT-3′ for the sequencing
primer.
Statistical analyses
Statistical analyses were performed by PASW
Statistics 18.0 software (SPSS Inc., Chicago, IL, USA). Logarithmic
transformation was applied for the data deviated from normality.
Analysis of variance was used to evaluate the association between
the risk factors (age, smoking history, histological
differentiation and clinical stage) and relative methylation rate
difference. A paired sample t-test was applied to compare
TGFB2 and BCL2L11 methylation between the LSCC and
adjacent tissues. All P-values were adjusted by logistic
regression. A two-tailed P<0.05 was considered to indicate a
statistically significant difference. All figures were drawn using
GraphPad Prism 6 software (GraphPad, San Diego, CA, USA).
Results
In the present study, we assessed the TGFB2
and BCL2L11 methylation levels in LSCC and their adjacent
non-neoplastic tissues using bisulfite pyrosequencing technology.
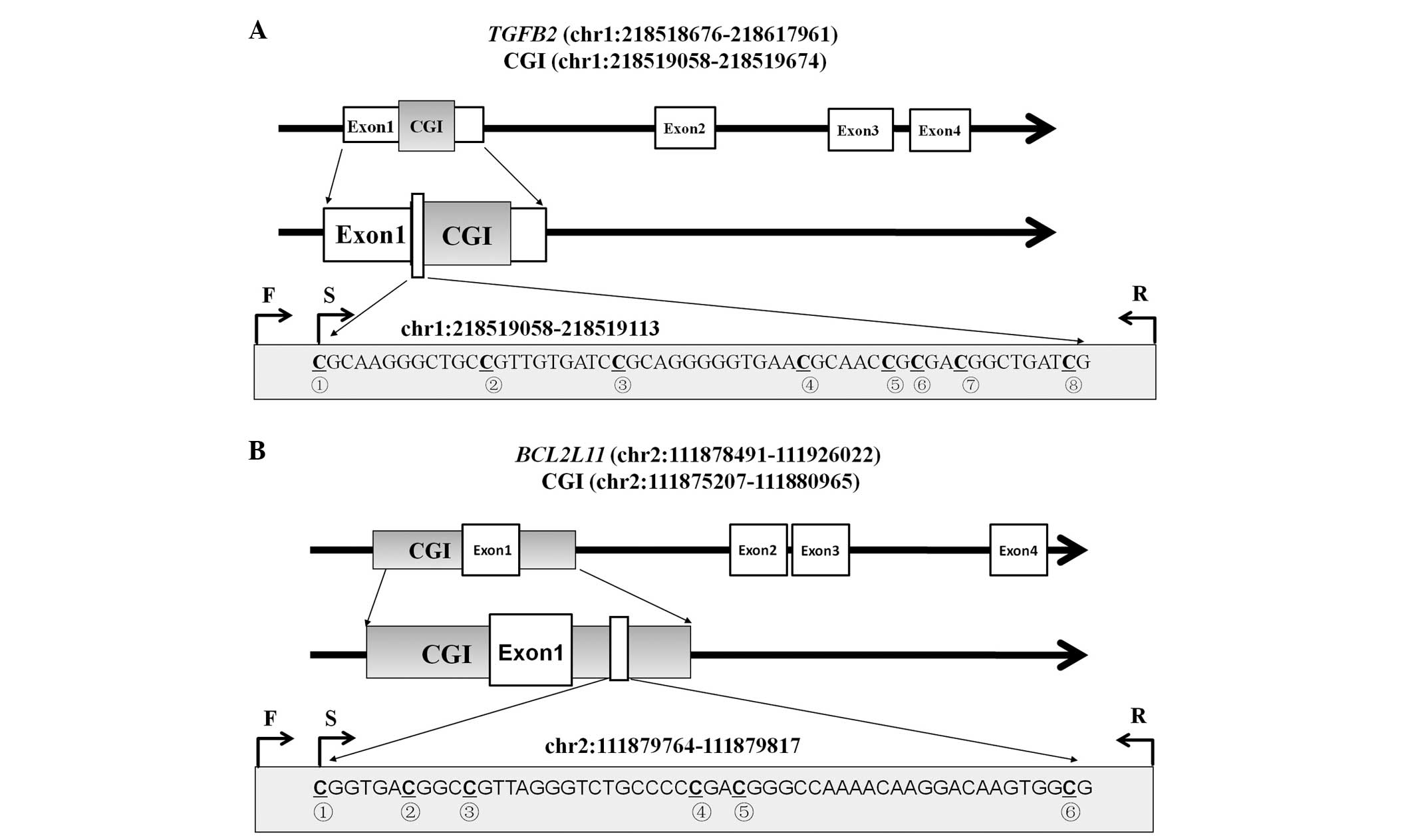
As shown in Fig. 1, the tested
fragments were located in the CGIs of the TGFB2 promoter
(chr1:218519058-218519113) and BCL2L11 gene-body
(chr2:111879764-111879817). A total of eight and six CpG
dinucleotides were measured for TGFB2 and BCL2L11,
respectively. The mean methylation levels of the tested CpG sites
were used to compare their differences between the LSCC tumor and
the adjacent normal tissues.
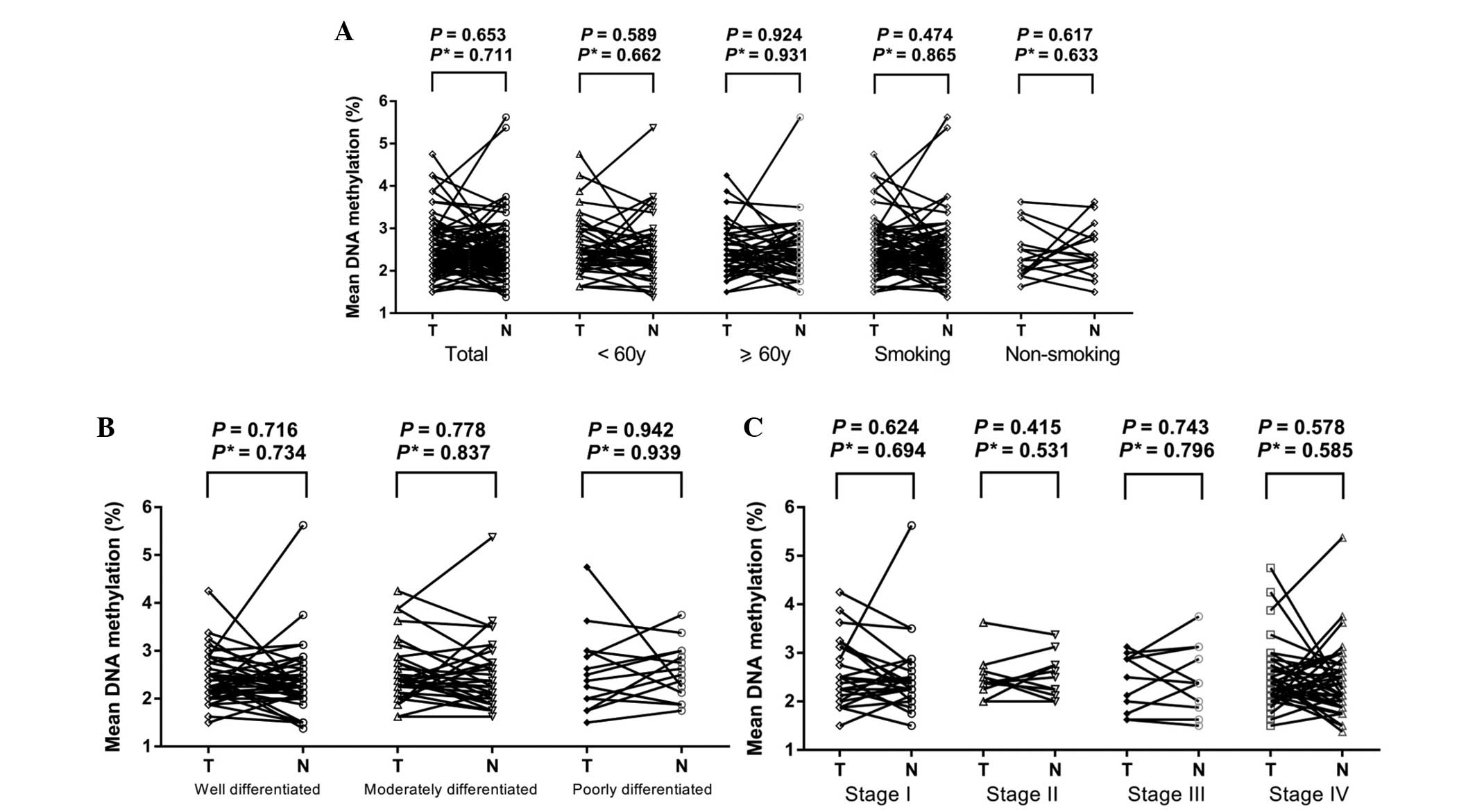
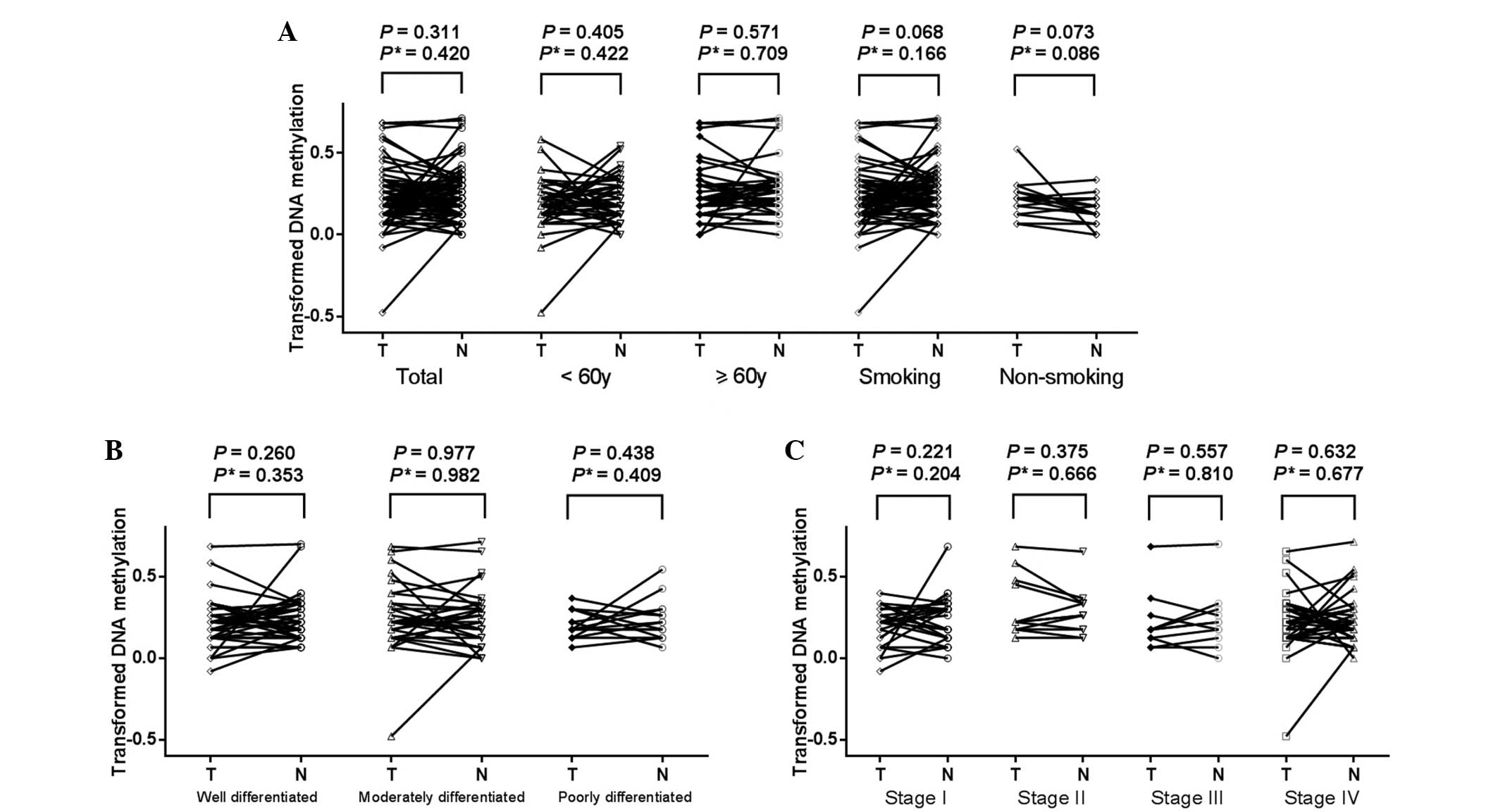
Our results revealed that there were no
statistically significant differences of the methylation levels of
TGFB2 promoter (Fig. 2,
P=0.653) and BCL2L11 gene-body (Fig. 3, P=0.311) between LSCC tissues and the
paired normal tissues. Further tests demonstrated no change in the
results when adjusted for age, smoking history, histological
differentiation and clinical stage (all adjusted P>0.05).
Moreover, there was no difference in the TGFB2 promoter or
BCL2L11 gene-body methylation levels between tumor tissues
and paired normal tissues in the breakdown analyses by age, smoking
history, histological differentiation or clinical stage of the
patients (Figs. 2 and 3, all P>0.05, data not shown).
Discussion
The aim of the present study was to determine
whether DNA methylation of the TGFB2 promoter and
BCL2L11 gene-body was associated with LSCC risk. Through a
series of statistical analyses of these two genes, no clear
correlation with laryngeal cancer was observed. Further subgroup
tests by age, smoking history, histological differentiation and
clinical stage also produced similar results.
Epigenetic modifications are central to
tumorigenesis (18). In cancer cells,
global hypomethylation is accompanied by hypermethylation of
certain gene promoter CGIs (6). DNA
methylation of the gene promoter region leads to gene silencing by
blocking the binding of transcription factors or methyl-binding
proteins (6), while a positive
correlation has been observed in a variety of human tissue types
between gene-body methylation and gene expression (10,19). As
for LSCC, aberrant DNA methylation of tumor suppressor genes,
including MGMT, CHD5, p16, p14, PTEN,
MYCT1 and FHIT, has been reported (4,20–23). DNA methylation levels of the promoter
and gene-body are relatively dynamic (24). However, there are no previous studies
on gene-body methylation in LSCC.
TGFB2 promoter hypermethylation was first
identified as a potential epigenetic marker of prostate cancer in
whole genome methylation profiling studies (25). A previous study revealed that the gene
suppression of TGFB2 was not associated with DNA methylation
but with chromatin remodeling in breast cancer tissue samples
(26). In the present study, we did
not observe any association between TGFB2 methylation and
LSCC, suggesting that there were other genetic or epigenetic
modifications in LSCC.
The apoptosis-related gene BCL2LL1 has been
noted to be hypermethylated in colon cancer (27), hematological and epithelial cancers
(16). In addition, low
BCL2L11 expression in chronic myeloid leukemia was ascribed
to BCL2LL1 hypermethylation at the gene promoter (28). Due to the limitation of not
identifying a suitable primer set to assess the promoter CGI of the
BCL2LL1 gene, we selected the CGI in the gene-body region as
an alternative. Although our study did not identify any significant
association with BCL2L11 gene-body methylation, the
contribution of other CGIs to LSCC risk remains to be explored.
In conclusion, our study suggests a lack of
contribution of the TGFB2 promoter and BCL2L11
gene-body methylation to LSCC in male patients. However, this study
was designed as a pilot study, and further investigations are
required to confirm our findings.
Acknowledgements
The study was supported by grants from Zhejiang
Provincial Natural Science Foundation of China (no. LY14H160003),
the Scientific Innovation Team Project of Ningbo (no. 2012B2019),
Ningbo Social Development Key Research Project (no. 2012C5015),
Ningbo Natural Science Foundation (no. 2012A610208, 2012A610217 and
2013A610217), the Medical and Health Research Project of Zhejiang
Province (no. 2012ZDA042) and the Medical and Health Training
Project of Zhejiang Province (no. 2014PYA 017).
References
|
1
|
Morshed K, Polz-Dacewicz M, Szymański M
and Polz D: Short-fragment PCR assay for highly sensitive
broad-spectrum detection of human papillomaviruses in laryngeal
squamous cell carcinoma and normal mucosa: clinico-pathological
evaluation. Eur Arch Otorhinolaryngology. 265(Suppl 1): S89–S96.
2008. View Article : Google Scholar
|
|
2
|
Chen K, Song F, He M, Li H, Qian B, Zhang
W, Wei Q and Hao X: Trends in head and neck cancer incidence in
Tianjin, China, between 1981 and 2002. Head Neck. 31:175–182. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cattaruzza MS, Maisonneuve P and Boyle P:
Epidemiology of laryngeal cancer. Eur J Cancer B Oral Oncol.
32B:293–305. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang M, Li W, Liu YY, Fu S, Qiu GB, Sun KL
and Fu WN: Promoter hypermethylation-induced transcriptional
down-regulation of the gene MYCT1 in laryngeal squamous cell
carcinoma. BMC Cancer. 12:2192012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Toporcov TN, Znaor A, Zhang ZF, Yu GP,
Winn DM, Wei Q, Vilensky M, Vaughan T, Thomson P, Talamini R, et
al: Risk factors for head and neck cancer in young adults: a pooled
analysis in the INHANCE consortium. Int J Epidemiol. 44:169–185.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Laird PW: Cancer epigenetics. Hum Mol
Genet. 14:R65–R76. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jiang D, Hong Q, Shen Y, Xu Y, Zhu H, Li
Y, Xu C, Ouyang G and Duan S: The diagnostic value of DNA
methylation in leukemia: a systematic review and meta-analysis.
PloS One. 9:e968222014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huang T, Chen X, Hong Q, Deng Z, Ma H, Xin
Y, Fang Y, Ye H, Wang R, Zhang C, et al: Meta-analyses of gene
methylation and smoking behavior in non-small cell lung cancer
patients. Sci Rep. 5:88972015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jiang D, Shen Y, Dai D, Xu Y, Xu C, Zhu H,
Huang T and Duan S: Meta-analyses of methylation markers for
prostate cancer. Tumour Biol. 35:10449–10455. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Aran D, Toperoff G, Rosenberg M and
Hellman A: Replication timing-related and gene body-specific
methylation of active human genes. Hum Mol Genet. 20:670–680. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu L, Zheng D, Wang L, Jiang D, Liu H, Xu
L, Liao Q, Zhang L, Liu P, Shi X, et al: GCK gene-body
hypomethylation is associated with the risk of coronary heart
disease. Biomed Res Int. 2014:1517232014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sporn MB and Roberts AB: Transforming
growth factor-beta: recent progress and new challenges. J Cell
Biol. 119:1017–1021. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bierie B and Moses HL: Tumour
microenvironment: TGFbeta: the molecular Jekyll and Hyde of cancer.
Nat Rev Cancer. 6:506–520. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu L, Kron KJ, Pethe VV, Demetrashvili N,
Nesbitt ME, Trachtenberg J, Ozcelik H, Fleshner NE, Briollais L,
van der Kwast TH and Bapat B: Association of tissue promoter
methylation levels of APC, TGFβ2, HOXD3 and RASSF1A with prostate
cancer progression. Int J Cancer. 129:2454–2462. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Leo E, Mancini M, Aluigi M, Castagnetti F,
Martinelli G, Barbieri E and Santucci MA: DNA hypermethylation
promotes the low expression of pro-apoptotic BCL2L11 associated
with BCR-ABL1 fusion gene of chronic myeloid leukaemia. Br J
Haematol. 159:373–376. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dunwell T, Hesson L, Rauch TA, Wang L,
Clark RE, Dallol A, Gentle D, Catchpoole D, Maher ER, Pfeifer GP
and Latif F: A genome-wide screen identifies frequently methylated
genes in haematological and epithelial cancers. Mol Cancer.
9:442010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tang L, Ye H, Hong Q, Wang L, Wang Q, Wang
H, Xu L, Bu S, Zhang L, Cheng J, et al: Elevated CpG island
methylation of GCK gene predicts the risk of type 2 diabetes in
Chinese males. Gene. 547:329–333. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Torrisani J, Hanoun N, Laurell H, Lopez F,
Maoret JJ, Souque A, Susini C, Cordelier P and Buscail L:
Identification of an upstream promoter of the human somatostatin
receptor, hSSTR2, which is controlled by epigenetic modifications.
Endocrinology. 149:3137–3147. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ball MP, Li JB, Gao Y, Lee JH, LeProust
EM, Park IH, Xie B, Daley GQ and Church GM: Targeted and
genome-scale strategies reveal gene-body methylation signatures in
human cells. Nat Biotechnol. 27:361–368. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang J, Zhu XB, He LX, Gu ZW, Jin MZ and
Ji WY: Clinical significance of epigenetic silencing and
re-expression of O6-methylguanine-DNA methyltransferase using
epigenetic agents in laryngeal carcinoma. Oncol Lett. 9:35–42.
2015.PubMed/NCBI
|
|
21
|
Kis A, Tatár TZ, Gáll T, Boda R, Tar I,
Major T, Redl P, Gergely L and Szarka K: Frequency of genetic and
epigenetic alterations of p14ARF and p16INK4A in head and neck
cancer in a Hungarian population. Pathol Oncol Res. 20:923–929.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang J, Chen H, Fu S, Xu ZM, Sun KL and Fu
WN: The involvement of CHD5 hypermethylation in laryngeal squamous
cell carcinoma. Oral Oncol. 47:601–608. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yin D and Dong M: Methylation of promoter
and expression of FHIT gene in laryngeal squamous cell carcinoma.
Lin Chuang Er Bi Yan Hou Ke Za Zhi. 19:253–255, 258. 2005.(In
Chinese). PubMed/NCBI
|
|
24
|
Suzuki MM and Bird A: DNA methylation
landscapes: provocative insights from epigenomics. Nat Rev Genet.
9:465–476. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kron K, Pethe V, Briollais L, Sadikovic B,
Ozcelik H, Sunderji A, Venkateswaran V, Pinthus J, Fleshner N, van
der Kwast T and Bapat B: Discovery of novel hypermethylated genes
in prostate cancer using genomic CpG island microarrays. PloS One.
4:e48302009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hinshelwood RA, Huschtscha LI, Melki J,
Stirzaker C, Abdipranoto A, Vissel B, Ravasi T, Wells CA, Hume DA,
Reddel RR and Clark SJ: Concordant epigenetic silencing of
transforming growth factor-beta signaling pathway genes occurs
early in breast carcinogenesis. Cancer Res. 67:11517–11527. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cho Y, Turner ND, Davidson LA, Chapkin RS,
Carroll RJ and Lupton JR: Colon cancer cell apoptosis is induced by
combined exposure to the n-3 fatty acid docosahexaenoic acid and
butyrate through promoter methylation. Exp Biol Med (Maywood).
239:302–310. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
San Jose-Eneriz E, Agirre X,
Jimenez-Velasco A, Cordeu L, Martín V, Arqueros V, Gárate L,
Fresquet V, Cervantes F, Martínez-Climent JA, et al: Epigenetic
down-regulation of BIM expression is associated with reduced
optimal responses to imatinib treatment in chronic myeloid
leukaemia. Eur J Cancer. 45:1877–1889. 2009. View Article : Google Scholar : PubMed/NCBI
|