Introduction
Lung adenocarcinoma is a type of non-small cell lung
cancer. It is frequently observed in females and smokers. Lung
adenocarcinoma usually originates in the bronchial epithelium, with
certain cases originating from the large bronchial mucous glands.
Its growth is generally slow, but occasionally hematogenous
metastasis occurs in the early phase. Lymph node metastasis occurs
late in the disease (1). To date,
although the histopathological progression of lung adenocarcinoma
has been thoroughly described, the molecular underpinnings are less
well understood (2).
Special adenine-thymine-rich sequence-binding
protein 1 (SATB1) is a nuclear matrix attachment region binding
protein which is located in the short arm of chromosome 3 and
includes 763 amino acids (3). SATB1
is involved in the development of thymocytes, maturation of T cells
and the formation of chromosome structure. Loss of SATB1 has been
demonstrated to lead to a change of at least 2% of genes in mice
(4). A study by Han et al
revealed that SATB1 plays a key role in the development of breast
carcinoma (5). A subsequent study
demonstrated strong expression of SATB1 in various tumor cells
(6–9).
There have been no studies of the SATB1 gene in lung
adenocarcinoma. In the present study, the lung adenocarcinoma cell
line A549 was cultured in vitro, and SATB1 siRNA was
constructed and transfected into A549 cells to silence the SATB1
gene. The changes in cell proliferation, invasion, migration and
apoptosis were observed.
Materials and methods
Ethics
The present study was approved and registered by the
ethics committee of the First Affiliated Hospital of Liaoning
Medical University, China, in January 2012. The related screening
and analysis of the resected samples was approved by the ethics
committee of Liaoning Medical University, and written consent forms
for the use of these samples were signed and participation in the
study was agreed upon by all subjects.
Sample collection
A total of 60 lung adenocarcinoma and 16 adjacent
normal tissue samples (at least 5 cm away from the edge of the
cancer tissue) were collected from the sample preservation center
at the First Affiliated Hospital of Liaoning Medical University.
These samples were all resected in the Department of Thoracic
Surgery between January 2012 and December 2013. The inclusion
criteria of the samples were: i) Patients had not received any
prior radiotherapy or chemotherapy treatment; ii) each patient had
received a medical examination including cranial computed
tomography (CT) scan, chest CT scan, abdominal CT scan and emission
computed tomography, from which the tumor-node-metastasis (TNM)
stage of the patient could be clearly defined; iii) patients had
received radical surgery with sufficient tissue samples prepared in
paraffin blocks for further testing; iv) patients who had at least
two concurrent primary tumors were excluded. All samples were fixed
in 10% formaldehyde and paraffin-embedded. The samples were
routinely and serially sectioned at a thickness of 5 µm, and then
immunohistochemically stained. The lung adenocarcinoma cases were
staged according to the TNM staging system stipulated in the
seventh edition of the American Joint Committee on Cancer (AJCC)
Cancer Staging Manual (2009) (10).
Immunohistochemistry
Sections were deparaffinized and hydrated in a
stepwise xylene and graded ethanol, washed with phosphate-buffered
saline (PBS), and recovered through microwave irradiation. A 3%
H2O2 solution was added and cultured for 10
min, and then washed with PBS. Goat blocking serum was supplied and
cultured under room temperature and the diluted primary antibodies
were applied (1:100). After storing overnight at 4°C, the sections
were washed in PBS, then secondary antibodies were added and the
sections were cultured at 37°C for 20 min. Freshly prepared
3,3′-diaminobenzidine chromogenic reagent was applied, and the
sections were cultured at 37°C for 5 to 10 min. Nuclei were then
stained with hematoxylin and eosin (HE).
Cell lines and culture conditions
The human lung adenocarcinoma cell line A549 was
obtained from Shanghai Biological Sciences Institute in China.
Cells were cultured in RPMI-1640 supplemented with 10% fetal bovine
serum (FBS), 10 U/l penicillin G and 100 mg/l streptomycin at 37°C
in a humidified atmosphere containing 5% CO2.
Preparation of target vector and
transfection
Based on the SATB1 cDNA sequence in Gene Bank, three
pairs of synthesized oligonucleotide were designed (Dalian
Biotechnologies, Dalian, China). The sequences used were as
follows: (Si-1) F:
5′-GATCCCCGGATTTGGAAGAGAGTGTCTTCAAGAGAGACACTCTCTTCCAAATCCTTTTTGGAAA-3′;
(Si-1) R:
5′-AGCTTTTCCAAAAAGGATTTGGAAGAGAGTGTCTCTCTTGAAGACACTCTCTTCCAAATCCGGG-3′;
(Si-2) F:
5′-GATCCCCGTCCACCTTGTCTTCTCTCTTCAAGAGAGAGAGAAGACAAGGTGGACTTTTTGGAAA-3′;
(Si-2) R:
5′-AGCTTTTCCAAAAAGCCACCTTGTCTTCTCTCTCTCTTGAAGAGAGAAGACAAGGTGGACGGG-3′;
(Si-N) (control group) F:
5′-GATCCGCGAGACCTCAGTATGTTACCTGTGAAGCCACAGATGGGGTAACATACTGAGGTCTCGCTTTTTTG-3′.
Oligonucleotide was annealed and ligated with pRNAT-U6.1/Neo-siRNA
using T4 DNA ligase. The three constructed recombinant plasmids
SATB1-siRNA-1, SATB1-siRNA-2 and SATB1-siRNA-N were verified by
sequencing and restriction endonuclease digestion. The A549 cells
were seeded (2×105 cells/well) in six-well plates.
Following incubation for 24 h, the cells were transfected with
SATB1-siRNA-1, SATB1-siRNA-2 and SATB1-siRNA-N plasmid in
serum-free medium using Lipofectamine 2000 reagent (Invitrogen,
Carlsbad, CA, USA) according to the manufacturer's
instructions.
Western blot analysis of SATB1
Forty-eight hours after transfection, cells were
washed three times with ice-cold PBS, and then centrifuged at 4°C
for 30 min (13,000 × g). The supernatant was collected and the
bicinchoninic acid method was used to determine the protein
concentration. A 10% polyacrylamide gel was prepared to load
protein samples, and 5% nonfat dry milk was added to block the
non-specific antigen. The primary antibody (1:250; rabbit
anti-human SATB1 polyclonal antibody) and the secondary antibody
(1:500; goat anti-rabbit antibody) were applied. Each sample was
also probed with β-actin antibody as a loading control.
MTT assay
Twenty-four hours after transfection, cells from the
four groups were loaded on a 96-well plate at 1×103
cells/well and cultured with RPMI-1640 medium with 10% FBS, at time
points of 24, 48 and 72 h. The medium was removed from each well,
and 20 µl 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium
bromide (MTT; 5 mg/ml in PBS) was added in the absence of light.
Formazan crystals were produced over a 4-h incubation period. Then
the supernatant was removed, and 150 µl dimethyl sulfoxide was
added to each well. The dark blue MTT crystals were dissolved by
agitating the plates at room temperature for 10 min, and the
absorbance was then measured on a Bio-Rad microplate reader
(Bio-Rad, Hercules, CA, USA) using a test wavelength of 490 nm and
a reference wavelength of 570 nm.
Transwell assay
The invasion of A549 cells was assayed using
modified Transwell chambers. The polycarbonate filter (pore size, 8
µm) which separates the upper and lower compartments of the chamber
was coated with 50 µg reconstituted basement membrane (Matrigel, BD
Biosciences, Bedford, MA, USA). Thirty-six hours after
transfection, the full medium was replaced with serum-free culture
medium. Eight hours later, it was digested to a suspension at a
density of 1×104/ml. Cells were seeded into the
Transwell chamber. The chamber was placed into a 24-well culture
plate with 500 µl RPMI-1640 medium containing 15% serum added
outside of the chamber, and 200 µl cell suspension was added to the
chamber. After 48 h of incubation at 37°C, cells on the upper
surface of the filter that had not invaded through the Matrigel
were removed completely with cotton swabs. Cells that had invaded
remained on the filter. Cells on the polycarbonate filter were
fixed with HE. The number of invasive cells was counted under a
microscope (magnification, ×200).
Scratch assay
The migration ability of A549 cells was performed in
a 24-well culture plate. Following transfection, a horizontal wound
(scratch) was made on the cells with a tiny spear. Cell migration
was observed at 24, 48 and 72 h after transfection, and the scratch
spaces were analyzed.
Flow cytometry
Forty-eight hours after transfection, the cells were
washed with PBS twice, and then digested and centrifuged. Then
apoptosis detection was processed according to the instructions of
the kit (BD Pharmingen™ Annexin V-PE Apoptosis Detection Kit I; BD
Biosciences): Cells were resuspended with 1X binding buffer
(1×106/ml). A total of 100 µl was drawn, and 5 µl PE
Annexin V and 5 µl 7-AAD were added. Cells were cultured in a
rotary system at room temperature for 15 min, then 400 µl 1X
binding buffer was added, and the cells were analyzed within one
hour.
Statistical analysis
The images were analyzed by Quantity One software
(Bio-Rad, Berkeley, CA, USA). All laboratory data are presented as
the means ± standard deviation. The χ2 test and single
factor analysis of variance were performed with SPSS 17.0 software
(SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
Patient characteristics
A total of 60 samples were selected successfully, 27
of which were resected from males and 33 from females, with an
average age of 54 years (range, 37 to 75). Among the samples, 20
cases were well differentiated, 23 cases were moderately
differentiated, and 17 cases were poorly differentiated. Forty-four
cases were stage I or II, and 16 cases were stage III (Table I).
 | Table I.Correlation between positive
expression of SATB1 protein in lung adenocarcinoma tissues and
patient characteristics. |
Table I.
Correlation between positive
expression of SATB1 protein in lung adenocarcinoma tissues and
patient characteristics.
|
|
| SATB1 |
|
|
|---|
|
|
|
|
|
|
|---|
| Characteristic | n (60) | − | + | ++ | χ2 | P-value |
|---|
| Gender |
|
| Male | 27 | 10 | 5 | 12 | 2.184 | 0.336 |
|
Female | 33 | 14 | 10 | 9 |
|
|
| Age |
|
| ≤60 | 24 | 10 | 6 | 8 | 0.060 | 0.971 |
|
>60 | 36 | 14 | 9 | 13 |
|
|
| Degree of
differentiation |
|
| Well | 20 | 14 | 3 | 3 |
|
|
|
Moderate | 23 | 7 | 7 | 9 |
|
|
| Poor | 17 | 3 | 5 | 9 | 12.315 | 0.015 |
| TNM stage |
|
| Stages I
and II | 44 | 20 | 13 | 11 |
|
|
| Stage
III | 16 | 4 | 2 | 10 | 7.305 | 0.026 |
SATB1 expression in lung
adenocarcinoma tissues and adjacent normal tissues
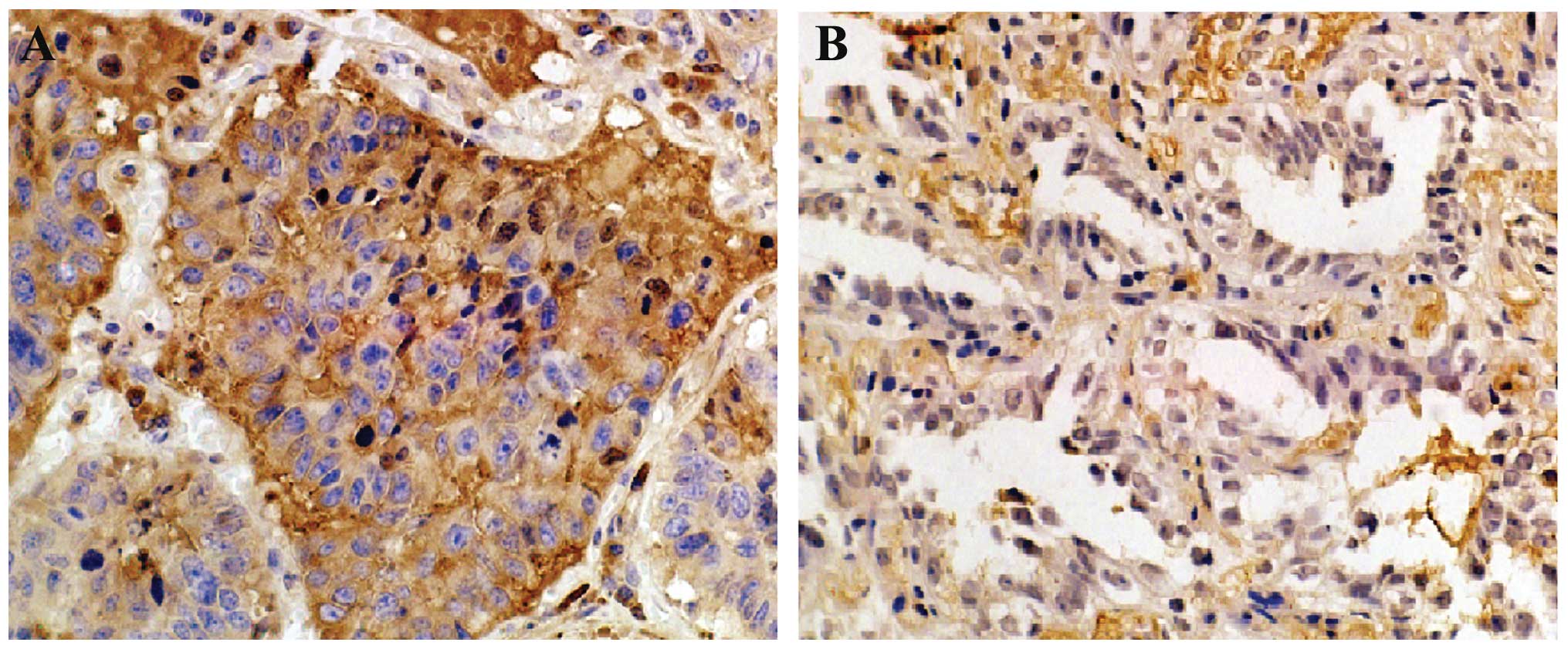
Immunohistochemistry revealed that SATB1 protein was
mainly expressed in the cytoplasm and the nuclei, and exhibited
yellow or brown colored staining. Its expression was significantly
higher in the poorly differentiated cells when compared with the
well-differentiated cells. The expression of SATB1 in the lung
adenocarcinoma tissues was not correlated with patient age or
gender (Table I). A statistical
difference was noted in SATB1 protein levels among the lung
adenocarcinoma tissues with different degrees of differentiation
(χ2=12.315, P<0.001), which were negatively
correlated with the expression of SATB1 (Fig. 1). A statistical difference was
observed in the expression of SATB1 among the TNM stages in the
lung adenocarcinoma cases (χ2=7.305, P=0.026), and the
expression was positively correlated with TNM stage. There were 60%
(36/60) of cases with positive expression of SATB1 protein among
the lung adenocarcinoma tissues while the rate of positive
expression was significantly low in the adjacent normal tissues
(18.75%, 3/16, P<0.001, Table
II).
 | Table II.Correlation between positive
expression of SATB1 protein in lung adenocarcinoma tissues and
adjacent normal lung tissues. |
Table II.
Correlation between positive
expression of SATB1 protein in lung adenocarcinoma tissues and
adjacent normal lung tissues.
|
|
|
| SATB1 |
|
|
|---|
|
|
|
|
|
|
|
|---|
| Tissue | n | − | + | ++ | χ2 | P-value |
|---|
| Tumor tissues | 60 | 24 | 15 | 21 |
|
|
| Adjacent normal | 16 | 13 | 2 | 1 | 8.904 | 0.012 |
Fluorescence microscopy
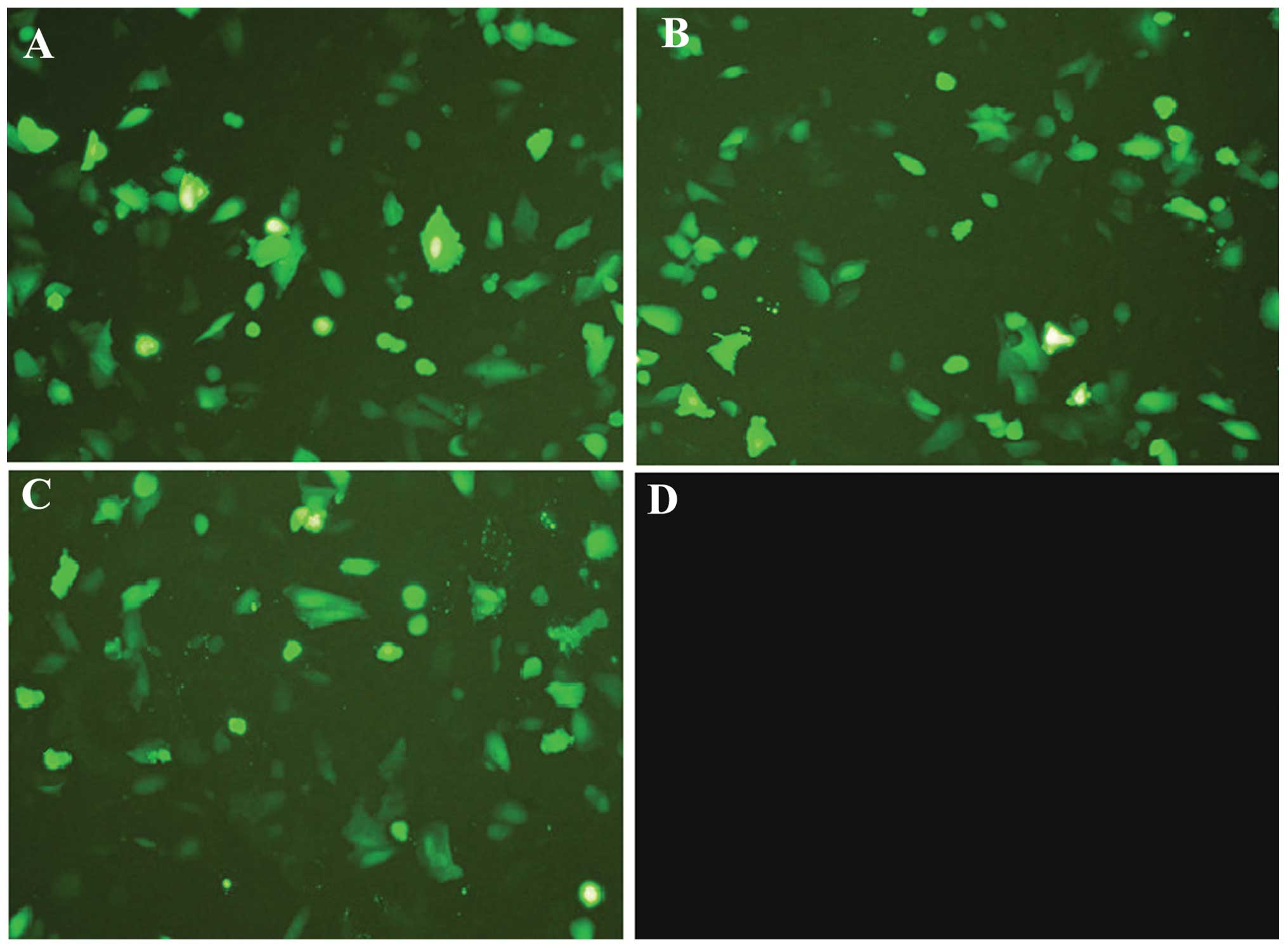
After the A549 cells were transfected with
SATB1-siRNA-1, SATB1-siRNA-2 or SATB1-siRNA-N, green fluorescence
was observed in the cytoplasm. As shown in Fig. 2, under fluorescence microscopy, the
transfection efficiency of the three groups (SATB1-siRNA-1,
SATB1-siRNA-2 and SATB1-siRNA-N) was satisfactory with all
exceeding 75%. It was observed that certain cells treated with
SATB1-siRNA were less confluent or became smaller and orbicular
compared with the control. Consistently, there were fewer cells in
the SATB1-siRNA-1 and SATB1-siRNA-2 transfected cells compared with
the SATB1-siRNA-N transfected and control cells cultured for 72 h
after transfection. The SATB1-siRNA treatment decreased the number
of A549 cells, implying that SATB1 is involved in cell cycle
progression and cell survival.
Western blot analysis of SATB1
In order to assess whether SATB1-siRNA effectively
silenced SATB1 expression in A549 cells, protein was extracted from
the A549 cells. Cell lysates were analyzed for SATB1 protein
expression using western blot analysis with SATB1 antibodies. As
shown in Fig. 3, following
transfection with SATB1-siRNA-1 and SATB1-siRNA-2, a significant
decrease in SATB1 protein expression was observed. SATB1 protein
levels were noted to be significantly downregulated following
transfection with SATB1-siRNA-1 and SATB1-siRNA-2 compared with the
untransfected cells or those transfected with SATB1-siRNA-N.
Cell proliferation assay
A549 cell proliferation was analyzed using the MTT
assay. As shown in Fig. 4, compared
with the A549 cells transfected with SATB1-siRNA-N, the
proliferation of the A549 cells transfected with SATB1-siRNA-1 and
SATB1-siRNA-2 was significantly reduced to 72.50% and 70.00%
(P<0.05), 65.85% and 69.51% (P<0.05), and 58.68% and 57.02%
(P<0.01) at 24, 48 and 72 h, respectively. No significant
difference was observed between the untransfected A549 cells and
those transfected with SATB1-siRNA-N (P>0.05).
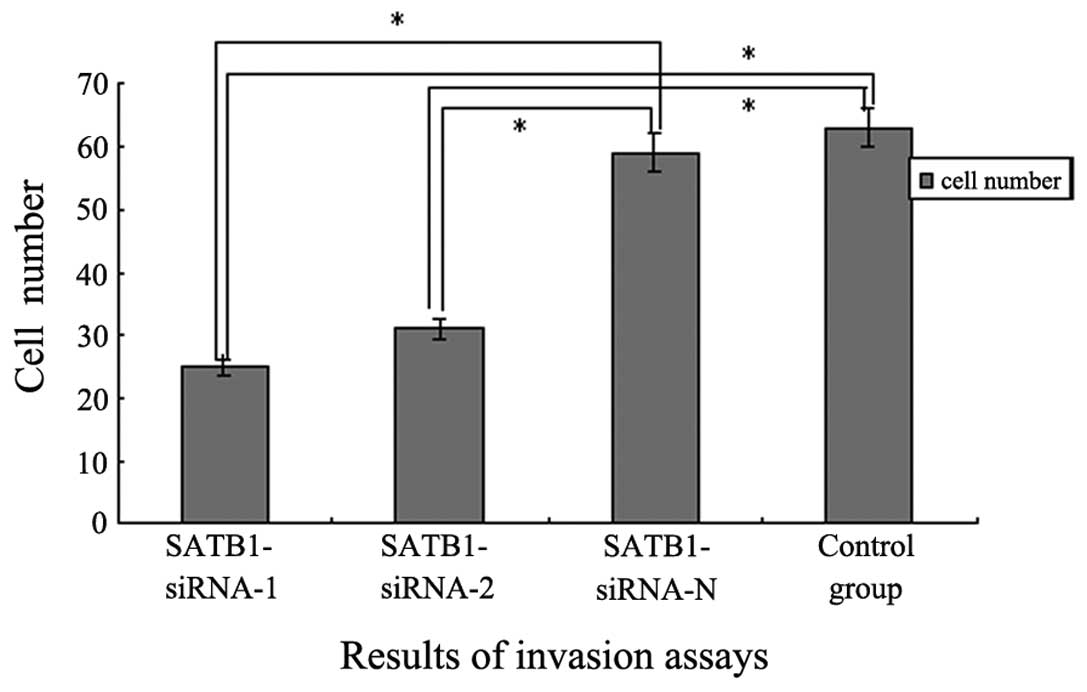
Cell invasion and migration assay
To investigate the role of SATB1-siRNA in lung
adenocarcinoma cell invasion, Transwell invasion assays were
performed. Untransfected A549 cells and cells transfected with
SATB1-siRNA-1, SATB1-siRNA-2 and SATB1-siRNA-N were incubated for
48 h on Matrigel-coated filters which were then stained with HE and
analyzed using a microscope. The number of A549 cells transfected
with SATB1-siRNA-1 and SATB1-siRNA-2 observed on the filter
significantly decreased compared with the number of untransfected
and SATB1-siRNA-N transfected cells (P<0.05). Furthermore, no
significant difference in cell number was observed between the
SATB1-siRNA-N group and the control group (P>0.05; Fig. 5). These data suggested that silencing
SATB1 using transient transfection with SATB1-siRNA may inhibit
lung adenocarcinoma cell invasion in vitro.
The migration ability test of transfected cells was
performed 72 h after transfection. The transmembrane cell number in
the SATB1-siRNA-1 and SATB1-siRNA-2 groups was only half of the
number in the SATB1-siRNA-N and blank control groups. This
indicated that the migration ability was inhibited significantly in
the SATB1-siRNA-1 and SATB1-siRNA-2 groups. The scratch width
measure results supported the results of the above-mentioned test:
the reducing space in SATB1-siRNA-1 and SATB1-siRNA-2 from 0 to 72
h following transfection was far below that measured from the
SATB1-siRNA-N and blank control groups (data shown in Table III).
 | Table III.Width of scratch in each time period
for each group of cells (mean ± standard deviation, n=5). |
Table III.
Width of scratch in each time period
for each group of cells (mean ± standard deviation, n=5).
| Time | SATB1-siRNA-1 | SATB1-siRNA-2 | SATB1-siRNA-N | Blank control |
|---|
| 0 h (mm) | 34.56±1.29 | 33.06±2.31 | 34.26±1.36 | 34.76±1.85 |
| 24 h (mm) | 27.77±2.17 | 26.48±1.68 | 26.12±1.16 | 25.79±1.11 |
| 48 h (mm) |
23.61±1.25a |
23.61±2.30a | 14.95±1.13 | 13.85±1.69 |
| 72 h (mm) |
16.35±1.56a |
17.89±1.95a | 7.82±1.23 | 6.59±2.36 |
Apoptosis rate increases following
transfection
Flow cytometric analysis revealed that the apoptosis
rate significantly increased (P<0.01) in the SATB1-siRNA-1 and
SATB1-siRNA-2 transfected cells compared with the SATB1-siRNA-N
transfected cells; there was no significant difference in the
apoptotic rate between cells transfected with SATB1-siRNA-N and the
blank control (P>0.05; Table
IV).
 | Table IV.Analysis of rate of apoptosis for
each cell group. |
Table IV.
Analysis of rate of apoptosis for
each cell group.
| Group | Apoptosis rate
(%) |
|---|
| SATB1-siRNA-1 |
21.36±1.19a |
| SATB1-siRNA-2 |
20.45±1.85a |
| SATB1-siRNA-N | 6.12±1.36 |
| Blank control | 5.35±1.65 |
Discussion
Lung adenocarcinoma, in which hematogenous
metastasis usually occurs in the early stage, is a type of
non-small-cell lung cancer. Although radiotherapy and chemotherapy
have produced modest benefits in certain patients, they have a
tendency to relapse and become resistant to numerous drugs
following traditional therapies (11). Therefore early detection and providing
a reliable therapeutic target are crucial. The identification of
siRNAs may well be one of the transforming events in biology in the
past decade (12). This technology
has become an essential tool in the studies of gene function,
carcinoma and viral disease therapy (13,14). In
the present study, siRNAs targeting SATB1-SATB1-siRNA-1 and
SATB1-siRNA-2 were constructed successfully. Our results revealed
that transfection with SATB1-siRNA-1 or SATB1-siRNA-2 into the lung
adenocarcinoma cell line A549 could inhibit cell proliferation and
invasion significantly. In addition, SATB1-siRNA could induce the
apoptosis of lung adenocarcinoma cells in vitro.
SATB1, which was originally characterized as a
regulator in T cell differentiation, was noted to be overexpressed
in metastatic breast cancer cell lines and in human tissue
specimens from the advanced stages of breast carcinoma with
metastasis (15). There are few
studies reporting on the expression of SATB1 in lung cancers, but
the role of SATB1 is controversial: a study in squamous cell lung
cancer and non-small-cell lung cancers revealed that SATB1
expression was lost, and the loss of SATB1 predicted poor prognosis
in squamous cell carcinomas (16).
Furthermore, Huang et al demonstrated that the expression of
SATB1 was much higher in SCLC tissues with or without metastasis
than in normal lung tissues (17).
In the present study, we noted that SATB1 was highly
expressed in lung adenocarcinoma tissues compared with adjacent
normal tissue. Significantly, the expression of SATB1 was observed
to be closely correlated with the cancer cell differentiation
degree and TNM stage, which suggested that SATB1 might contribute
to invasion and metastasis in lung adenocarcinoma. Based on this,
we constructed SATB1-siRNA and silenced SATB1 in A549 cells. Our
results revealed that SATB1-siRNA could induce lung adenocarcinoma
cell apoptosis following transfection of SATB1-siRNA into lung
adenocarcinoma cells. Metastasis is the final step in solid tumor
progression and the most common cause of mortality in cancer
patients. Controlling the proliferation, invasion and migration is
likely to improve the survival rate of cancer patients (18). A previous study demonstrated that
SATB1 plays an significant role in the process of proliferation,
invasion and migration (19).
Consistent with that previous study, the present results also
revealed that the proliferation, invasion and migration ability of
lung adenocarcinoma cells declined notably following transfection
with SATB1-siRNA. These results suggest that SATB1 may be an ideal
target for the treatment of lung adenocarcinoma. As for the
mechanisms of SATB1 influencing proliferation, invasion and
migration in lung adenocarcinoma cells, there has been little
research to date. However, numerous studies into the roles of SATB1
in breast and other cancers have been conducted. Cai et al
observed that SATB1 could induce a change in the expression of more
than 1000 genes (20). Some of these
genes are associated with cancer invasion and metastasis, including
MMP2, MMP9 and CTGF (19,21). We intend to further study the detailed
mechanisms in lung adenocarcinoma cells.
In summary, our study revealed that SATB1 was highly
expressed in lung adenocarcinoma tissues compared with adjacent
normal tissues. The expression of SATB1 was closely correlated with
cancer cell differentiation and TNM stage. The transfection vector
system we constructed significantly downregulated the SATB1 level
in lung adenocarcinoma cells and it also inhibited cell
proliferation, invasion and migration, and induced cell apoptosis.
SATB1 could regulate the invasion and migration of lung
adenocarcinoma cells, which may provide essential clues for more
effective targeting of lung adenocarcinoma and other cancers with
aberrant SATB1 activation.
Acknowledgements
This study was supported by the Education Department
of Liaoning Province (series number L2012300) and the Science and
Technology Department of Liaoning Province (series number
2013022038 and 2013022046).
References
|
1
|
Yatabe Y, Borczuk AC and Powell CA: Do all
lung adenocarcinomas follow a stepwise progression? Lung Cancer.
74:7–11. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kadara H, Kabbout M and Wistuba II:
Pulmonary adenocarcinoma: a renewed entity in 2011. Respirology.
17:50–65. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xu L, Deng HX, Xia JH, Yang Y, Fan CH,
Hung WY and Siddque T: Assignment of SATB1 to human chromosome band
3p23 by in situ hybridization. Cytogenet Cell Genet. 77:205–206.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Beyer M, Thabet Y, Müller RU, Sadlon T,
Classen S, Lahl K, Basu S, Zhou X, Bailey-Bucktrout SL, Krebs W, et
al: Repression of the genome organizer SATB1 in regulatory T cells
is required for suppressive function and inhibition of effector
differentiation. Nat Immunol. 12:898–907. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Han HJ, Russo J, Kohwi Y and
Kohwi-Shigematsu T: SATB1 reprogrammes gene expression to promote
breast tumour growth and metastasis. Nature. 452:187–193. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang Y, Tian X, Ji H, Guan X, Xu W, Dong
B, Zhao M, Wei M, Ye C, Sun Y, et al: Expression of SATB1 promotes
the growth and metastasis of colorectal cancer. PLoS One.
9:e1004132014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Han B, Luan L, Xu Z and Wu B: Expression
and biological roles of SATB1 in human bladder cancer. Tumour Biol.
34:2943–2949. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shukla S, Sharma H, Abbas A, MacLennan GT,
Fu P, Danielpour D and Gupta S: Upregulation of SATB1 is associated
with prostate cancer aggressiveness and disease progression. PLoS
One. 8:e535272013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zuk K, Peczek L, Stec-Michalska K, Medrek
M and Nawrot B: SATB1 expression in gastric mucosa in relation to
Helicobacter pylori infection and family history of gastric cancer.
Adv Med Sci. 57:237–243. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Groome PA, Bolejack V, Crowley JJ, Kennedy
C, Krasnik M, Sobin LH and Goldstraw P: IASLC International Staging
Committee; Cancer Research and Biostatistics; Observers to the
Committee; Participating Institutions: The IASLC Lung Cancer
Staging Project: Validation of the proposals for revision of the T,
N, and M descriptors and consequent stage groupings in the
forthcoming (seventh) edition of the TNM classification of
malignant tumours. J Thorac Oncol. 2:694–705. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Notsuda H, Sakurada A, Endo C, Okada Y,
Horii A, Shima H and Kondo T: p190A RhoGAP is involved in EGFR
pathways and promotes proliferation, invasion and migration in lung
adenocarcinoma cells. Int J Oncol. 43:1569–1577. 2013.PubMed/NCBI
|
|
12
|
Keaney J, Campbell M and Humphries P: From
RNA interference technology to effective therapy: how far have we
come and how far to go? Ther Deliv. 2:1395–1406. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xia L, Guan W, Wang D, Zhang YS, Zeng LL,
Li ZP, Wang G and Yang ZZ: Killing effect of Ad5/F35-APE1 siRNA
recombinant adenovirus in combination with hematoporphrphyrin
derivative-mediated photodynamic therapy on human nonsmall cell
lung cancer. Biomed Res Int. 2013:9579132013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yi X, Zhao G, Zhang H, Guan D, Meng R,
Zhang Y, Yang Q, Jia H, Dou K, Liu C, et al: MITF-siRNA formulation
is a safe and effective therapy for human melasma. Mol Ther.
19:362–371. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hanker LC, Karn T, Mavrova-Risteska L,
Ruckhäberle E, Gaetje R, Holtrich U, Kaufmann M, Rody A and
Wiegratz I: SATB1 gene expression and breast cancer prognosis.
Breast. 20:309–313. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Selinger CI, Cooper WA, Al-Sohaily S,
Mladenova DN, Pangon L, Kennedy CW, McCaughan BC, Stirzaker C and
Kohonen-Corish MR: Loss of special AT-rich binding protein 1
expression is a marker of poor survival in lung cancer. J Thorac
Oncol. 6:1179–1189. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang B, Zhou H, Wang X and Liu Z:
Silencing SATB1 with siRNA inhibits the proliferation and invasion
of small cell lung cancer cells. Cancer Cell Int. 13:82013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bankert RB, Balu-Iyer SV, Odunsi K, Shultz
LD, Kelleher RJ Jr, Barnas JL, Simpson-Abelson M, Parsons R and
Yokota SJ: Humanized mouse model of ovarian cancer recapitulates
patient solid tumor progression, ascites formation, and metastasis.
PLoS One. 6:e244202011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fang XF, Hou ZB, Dai XZ, Chen C, Ge J,
Shen H, Li XF, Yu LK and Yuan Y: Special AT-rich sequence-binding
protein 1 promotes cell growth and metastasis in colorectal cancer.
World J Gastroenterol. 19:2331–2339. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cai S, Han HJ and Kohwi-Shigematsu T:
Tissue-specific nuclear architecture and gene expression regulated
by SATB1. Nat Genet. 34:42–51. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mir R, Pradhan SJ and Galande S: Chromatin
organizer SATB1 as a novel molecular target for cancer therapy.
Curr Drug Targets. 13:1603–1615. 2012. View Article : Google Scholar : PubMed/NCBI
|