Introduction
Hepatocellular carcinoma (HCC) is the third most
common cause of cancer-associated mortality worldwide and its
incidence rate is >20 per 100,000 individuals in East Asia
(1). Detecting HCC early is difficult
and the majority of patients are diagnosed at an advanced stage of
the disease (2). The prognosis of
patients suffering with HCC remains poor even following complete
tumor resection, and recurrence occurs in 70% of cases 5 years
after resection (3). Recurrence may
be partially predicted by evaluating the proliferation activity of
cells in the resected tumor, assessing microvascular invasion and
measuring serum levels of α-fetoprotein (4,5). However,
recurrent and metastatic cases of HCC are usually refractory to
chemotherapy and interventional therapy, and molecular targeting
therapy has limited efficacy in such cases (6). It is thus necessary to identify a novel
molecule able to predict poor HCC prognosis and to develop novel
targeting therapies to treat patients with HCC following tumor
resection.
The comprehensive profiling of molecules expressed
in human malignant tumors discloses their biology and pathogenesis.
Previous studies have performed proteomic analyses of HCC (7–11) and
identified candidate proteins for the early diagnosis (7,10) and
rapid progression of HCC (9,11). However, only a limited number of
patients were examined in these studies.
Liquid chromatography-tandem mass spectrometry
(LC-MS/MS) has the ability to profile a large number of proteins
and formalin-fixed, paraffin-embedded (FFPE) samples may be used
for analysis (12). This enables a
retrospective study to identify biomarkers for the prognosis of
patients with cancer and target molecules to treat HCC, as complete
clinical records are available.
The present study aimed to identify a novel
biomarker for the prognosis of HCC by investigating the protein
disulfide-isomerase A3 (PDIA3), which is highly expressed in HCC,
and to investigate the significance of any associations between
levels of PDIA3 expression and patient clinicopathological
factors.
Materials and methods
HCC cases
The FFPE tumor tissues of 86 cases of HCC were
procured from the archives of pathology at Nippon Medical School
Hospital (Tokyo, Japan). All patients underwent radical hepatic
resection between January 2007 and December 2010. Pathological
diagnoses were made following the criteria of the American Joint
Committee on Cancer/International Union Against Cancer
Tumor-Node-Metastasis classification (13). The present study was performed in
accordance with the principles embodied in the Declaration of
Helsinki 2013, and the Japanese Society of Pathology Ethics
Committee. Informed consent for the study was obtained from all
patients.
Comprehensive protein profiling by
LC-MS/MS using FFPE tissues
FFPE tissues from 11 HCC cases were used for
comprehensive protein profiling. Expressed proteins were profiled
in HCC and non-HCC tissues obtained from each case. Each section
(10 µm thick) was deparaffinized in xylene and rehydrated through a
graded alcohol series of 100, 90, 80 and 70%. Following staining
with hematoxylin, the HCC and non-HCC tissues were dissected under
a stereoscopic microscope. Protein was extracted from the tissues
by lysis buffer [6 M guanidine-hydrochloride, 40 mM Tris (pH 8.2)
and 65 mM dithiothreitol (DTT)] and protein concentration was
determined by the Bradford method (14). Extracted protein (10 µg) was reduced
in 45 mM DTT and 20 mM Tris (2-carboxyethyl) phosphine
hydrochloride and alkylated in 100 mM iodoacetamide. The protein
was further digested using proteomics-grade trypsin (Agilent
Technologies, Inc., Santa Clara, CA, USA) at 37°C for 24 h and the
digest was purified in a PepClean™ C-18 spin column. Digested
protein (2 µg) was injected into a peptide L-trap column (Chemicals
Evaluation and Research Institute, Tokyo, Japan) and further
separated through Advanced-nano ultra-high-performance liquid
chromatography (AMR, Inc., Tokyo, Japan) using a reverse-phase C-18
column (Zaplous column α, gel particles 3 µm diameter and 10 nm
pore size, 0.1×150 mm; AMR, Inc.). The protein solution was run
with the gradient concentration of acetonitrile from 5–35% in 0.1%
formic acid in acetonitrile at a flow rate of 500 nl/min for 2 h.
An amaZon ETD Ion-Trap Mass Spectrometer (Bruker Corporation,
Billerica, MA, USA) was used to analyze eluted peptides. Data of
the 10 most intense peaks of each full MS scan were acquired. All
MS/MS spectral data were analyzed by MASCOT v2.3.01 (Matrix Science
Ltd., London, UK). The following parameter settings were used:
Trypsin cleavage; allowance of ≤2 missed cleavage peptides;
variable modifications of methionine oxidation; peptide mass
tolerance; fixed modifications of cysteine carbamidomethylation;
and fragment MS/MS tolerance ± 0.5 Da. Amounts of identified
proteins were expressed as normalized spectral abundance factor
(15). Relative expression levels of
identified proteins in HCC tissues relative to those in non-HCC
tissues were expressed as ratios of spectral counts (Rsc) in the
base 2 logarithmic scale (16). An
Rsc >1 indicated that the protein was upregulated in HCC tissue,
while an Rsc <-1 indicated that the protein was downregulated in
the HCC tissues compared with the non-HCC tissues. An Rsc between
−1 and 1 indicated that levels of protein expression were similar
in HCC and non-HCC tissue.
Bioinformatic analysis
Functional properties of identified proteins were
analyzed by the Database Annotation, Visualization and Integrated
Discovery (v6.7) using the Kyoto Encyclopedia of Gene and Genomes
(KEGG) database (17). Relative
abundance (%) of each functional category was calculated by
dividing the number of proteins in each category by the total
number of identified proteins.
Reverse-transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from FFPE tissues using the
RNeasy FFPE kit (Qiagen, Inc., Valencia, CA, USA). Expression of
PDIA3 mRNA and 18S rRNA as a reference was determined by the 7500
Fast Real-Time PCR system (Thermo Fisher Scientific, Inc., Waltham,
MA, USA) using cDNA, which was synthesized from 20 ng total RNA by
SuperScript® III using the SuperScript® VILO™
cDNA Synthesis kit (Thermo Fisher Scientific, Inc.). TaqMan primers
and probes were purchased from Thermo Fisher Scientific, Inc. These
consisted of PDIA3 (Hs04194196) and 18S rRNA (Hs03928990), and the
details of the primers and probes are available at the
manufacturer's website (http://www.thermofisher.com/jp/ja/home.html). qPCR was
subsequently performed, beginning with denaturation at 95°C for 10
min, followed by 40 cycles of amplification at 95°C for 15 sec and
extension at 60°C for 1 min. The level of expression of PDIA3 mRNA
was calculated as the ratio of PDIA3 mRNA to 18S rRNA, and PDIA3
expression in HCC tissue was further calculated as relative
increase, compared with that in non-HCC tissue using the
2−ΔΔCq method (18). The
expression levels were measured in triplicate.
Immunostaining and scoring
Sections 3-µm thick were used for immunostaining.
Following deparaffinization, sections were treated in
Histofine® Antigen Activation Liquid (pH 9.0; Nichirei
Biosciences, Inc., Tokyo, Japan) at 121°C for 15 min. Endogenous
peroxidase was blocked in 0.3% hydrogen peroxide and methanol for
30 min. Sections were then incubated with antibodies for PDIA3
(catalog no. ab13506; dilution, 1:150; Abcam, Tokyo, Japan) and
Ki-67 (MIB1; catalog no. M7240; dilution, 1:100; Dako Japan Co.,
Ltd., Tokyo, Japan) in phosphate-buffered saline containing 1%
bovine serum albumin (Sigma-Aldrich Japan K.K., Tokyo, Japan) for
16 h at 4°C. The sections were further incubated with the Histofine
Simple Stain™ MAX-PO (R; Nichirei Biosciences, Inc.) for 30 min,
and peroxidase activity was visualized by 3,3′-diaminobenzidine.
The sections were then counterstained with hematoxylin.
The intensity and proportion of
stained tumor cells were semi-quantitated
Cytoplasmic staining was considered a positive
reaction. If no tumor cell staining occurred, intensity and
proportion were scored as 0. Tumor cell staining intensity was
categorized into 3 grades: 1, weak; 2, moderate; and 3, strong. The
proportional score was divided into 3 grades: 1, <10%; 2,
10–50%; and 3, >50%. The total score was calculated as the sum
of the intensity and proportional scores. Two investigators
evaluated the score in a blind manner. HCC tissue with a mean total
score ≥5 was classified as having high expression and HCC tissue
with a mean total score <5 was classified as having low
expression. The Ki-67 index was calculated as the percentage of
Ki-67-positive cells in 1,000 tumor cells in the areas of the
highest nuclear labeling (so-called ‘hot spots’), which was
determined using a microscope (magnification, ×40).
Terminal deoxynucleotidyl transferase
dUTP nick-end labeling (TUNEL) assay
Apoptotic cell death of HCC was determined by TUNEL
assay using an Apoptag® Peroxidase In Situ
Apoptosis Detection kit (EMD Millipore, Billerica, MA, USA).
Nuclear staining was considered a positive result. The TUNEL index
was calculated as the percentage of TUNEL-positive cells in 1,000
carcinoma cells in the areas of highest nuclear labeling under a
microscope (magnification ×40).
Statistical analysis
All data are presented as the mean ± standard error.
The data of two groups were compared by the Mann-Whitney U-test.
Clinicopathological parameters were analyzed by the χ2
test and Fisher's exact test. Cumulative survival rate was
calculated using the Kaplan-Meier method and the significance of
differences in survival rate were analyzed by the log-rank test.
P<0.05 was considered to indicate a statistically significant
difference. All statistical analyses were performed using GraphPad
Prism v5.0 (GraphPad Software, Inc., La Jolla, CA, USA).
Results
Comprehensive profiling of
proteins
A total of 378 proteins were identified from the
FFPE tissues, 295 from the HCC tissues and 270 proteins in the
non-HCC tissues. A total of 187 proteins were identified in HCC and
non-HCC. In total, 142 proteins were upregulated (Rsc >1) in the
HCC tissues compared with the non-HCC tissues and 60 proteins were
downregulated (Rsc <-1; Fig. 1A).
Overall, 176 proteins were equally expressed in the HCC and non-HCC
tissues, and housekeeping gene products, including β-actin and
histone H4, were equally expressed.
The functional properties of the identified proteins
were analyzed using the KEGG database. Among the upregulated
proteins, the most abundant functional category was antigen
processing and presentation (Fig.
1B), and 11 proteins out of 142 upregulated proteins (7.7%)
were classified within this category (Table I). None of the protein among the
downregulated proteins (0/60, 0%) and equally expressed proteins
(0/176, 0%) was classified in this functional category. It was thus
speculated that the upregulation of proteins involved in antigen
processing and presentation was a characteristic feature of HCC.
Among 11 proteins in the antigen processing and presentation
category, the clinicopathological significance of PDIA3 in HCC is
unknown, therefore PDIA3 expression at the mRNA level was
investigated.
 | Table I.Upregulated proteins in the antigen
processing and presentation category. |
Table I.
Upregulated proteins in the antigen
processing and presentation category.
|
|
|
| Spectral
counting |
|
|---|
|
|
|
|
|
|
|---|
| ID | Protein | AAa | HCC | Non-HCC | Rsc |
|---|
| HSP71 | Heat shock 70 kDa
protein 1A/1B | 641 | 14 | 0 | 3.8 |
| HSP76 | Heat shock 70 kDa
protein 6 | 643 | 12 | 0 | 3.6 |
| 1A01 | HLA class I
histocompatibility antigen, A-1 α chain | 365 | 6 | 0 | 2.72 |
| HS71L | Heat shock 70 kDa
protein 1-like | 64 | 5 | 0 | 2.51 |
| HS90B | Heat shock protein
HSP 90-β | 724 | 5 | 0 | 2.51 |
| GRP78 | 78 kDa
glucose-regulated protein | 654 | 8 | 1 | 2.23 |
| HS90A | Heat shock protein
90-α | 732 | 5 | 1 | 1.66 |
| HSP7C | Heat shock cognate
71 kDa protein | 646 | 10 | 3 | 1.59 |
| PDIA3 | Protein
disulfide-isomerase A3 | 505 | 6 | 2 | 1.34 |
| HLAE | HLA class I
histocompatibility antigen, α chain E | 358 | 1 | 0 | 1.03 |
| PSME2 | Proteasome
activator complex subunit 2 | 239 | 1 | 0 | 1.03 |
RT-qPCR analysis of PDIA3 (data not
shown)
The expression of PDIA3 in HCC tissues was verified
by TaqMan probes in 11 cases of HCC and non-HCC. The relative
expression of PDIA3 mRNA was significantly elevated in the HCC
tissues (3.43±2.93) compared with the non-HCC tissues (1.20±0.81;
P<0.05). Protein profiling and quantitation of mRNA confirmed
the upregulation of PDIA3 in HCC. The clinicopathological
significance of increased PDIA3 expression was subsequently
examined in all cases of HCC.
Cases of HCC and immunostaining of
PDIA3
Immunostaining was performed in 86 HCC cases. Of
these, 51 patients were men and 35 were women, with a mean age of
68 years (range, 34–87). Hepatitis B surface antigen (HBsAg) was
positive in 26 cases and hepatitis C virus (HCV) was positive in 51
cases. A total of 6 cases were positive for HBsAg and HCV. Local
recurrence was noted in 47 cases and 6 patients exhibited
metastasis to the lungs, brain and lymph nodes. Early recurrence or
metastasis within 6 months of surgery was reported in 13 cases
(15%).
Representative immunostaining for PDIA3 in the HCC
and non-HCC tissues is presented in Fig.
2A. The HCC tissues demonstrated potent staining for PDIA3,
while the non-HCC tissues exhibited weak staining (Fig. 2B). At a higher magnification,
cytoplasm of the non-HCC cells exhibited a vaguely positive
reaction (Fig. 2C), while the
cytoplasm of HCC cells exhibited a clear positive reaction
(Fig. 2D). Staining intensity varied
among the HCC tissues; 56 cases (65%) were classified as high
expression (Fig. 2E) and 30 cases
(35%) were classified as low expression (Fig. 2F).
Association between PDIA3 expression
and clinicopathological factors
Associations between PDIA3 expression and patient
clinicopathological factors were examined in 86 cases of HCC
(Table II). No significant
correlations were observed between expression levels of PDIA3 and
any patient characteristics. However, low PDIA3 was associated with
poorly differentiated HCC and a smaller tumor size, though these
associations were not significant (P>0.05). Local recurrence in
the residual liver occurred in 29 cases (52%) of HCC with high
expression and in 18 cases (60%) of low expression. Distant
metastasis occurred in 5 cases (9%) with high expression and in 1
case (3%) of low expression.
 | Table II.Associations between
clinicopathological factors and PDIA3 expression in hepatocellular
carcinoma. |
Table II.
Associations between
clinicopathological factors and PDIA3 expression in hepatocellular
carcinoma.
|
|
| PDIA3
expression |
|
|---|
|
|
|
|
|
|---|
| Characteristic | n | High (n=56) | Low (n=30) | P-value |
|---|
| Gender |
|
|
| 0.819 |
|
Male | 51 | 34 | 17 |
|
|
Female | 35 | 22 | 13 |
|
| Age, years |
|
|
| 0.482 |
|
<65 | 31 | 22 | 9 |
|
|
≥65 | 55 | 34 | 21 |
|
| HBsAg |
|
|
| 0.806 |
|
Positive | 26 | 16 | 10 |
|
|
Negative | 60 | 40 | 20 |
|
| HCV |
|
|
| 0.819 |
|
Positive | 51 | 34 | 17 |
|
|
Negative | 35 | 22 | 13 |
|
| Cirrhosis |
|
|
| 0.251 |
|
Yes | 51 | 36 | 15 |
|
| No | 35 | 20 | 15 |
|
| Preoperative AFP,
ng/ml |
|
|
| 1 |
|
<20 | 44 | 29 | 15 |
|
|
≥20 | 42 | 27 | 15 |
|
| Preoperative DCP,
mAU/ml |
|
|
| 0.645 |
|
<40 | 32 | 22 | 10 |
|
|
≥40 | 54 | 34 | 20 |
|
| Tumor size, cm |
|
|
| 0.143 |
|
<5 | 59 | 35 | 24 |
|
| ≥5 | 27 | 21 | 6 |
|
| Tumor number |
|
|
| 0.24 |
| 1 | 54 | 32 | 20 |
|
|
<1 | 32 | 24 | 8 |
|
| Vascular
invasion |
|
|
| 0.815 |
|
Positive | 31 | 21 | 10 |
|
|
Negative | 55 | 35 | 20 |
|
| UICC stage |
|
|
| 0.287 |
| I | 35 | 19 | 16 |
|
| II | 44 | 31 | 13 |
|
|
III | 6 | 5 | 1 |
|
| IV | 1 | 1 | 0 |
|
|
Differentiation |
|
|
| 0.066 |
|
Well | 17 | 15 | 2 |
|
|
Moderate | 54 | 31 | 23 |
|
|
Poor | 15 | 10 | 5 |
|
| Local
recurrence |
|
|
| 0.503 |
|
Yes | 47 | 29 | 18 |
|
| No | 39 | 27 | 12 |
|
| Distal
metastasis |
|
|
| 0.66 |
|
Yes | 6 | 5 | 1 |
|
| No | 80 | 51 | 29 |
|
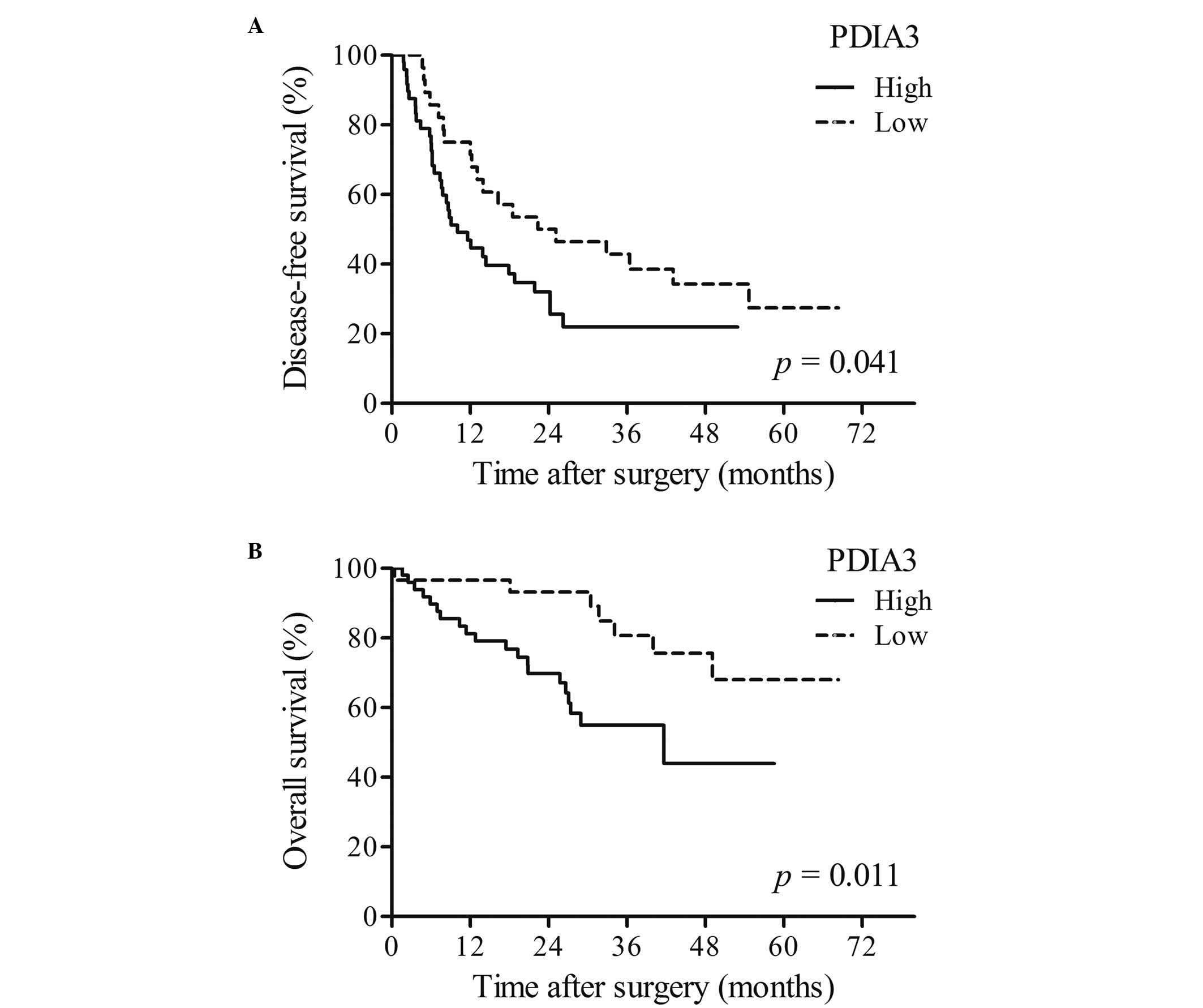
Disease-free survival (DFS) and
overall survival time (OS) of patients with HCC
The DFS and OS times significantly differed between
HCC patients with high and low expression of PDIA3. OS and DFS
times were significantly shorter in patients with high PDIA3
expression (P<0.05; Fig. 3A)
compared with that of patients with low PDIA3 expression
(P<0.05; Fig. 3B).
Ki-67 index and TUNEL indices
Proliferation activity and apoptotic cell death were
examined in 24 cases of HCC. Representative immunostaining of Ki-67
and TUNEL in HCC tissues with high and low expression of PDIA3 was
performed (Fig. 4A). The Ki-67 index
in the HCC tissues with high expression was significantly greater
than those in the HCC tissues with low PDIA3 expression (P<0.05;
Fig. 4B), indicating increased cell
proliferation in HCC tissues with high expression of PDIA3. By
contrast, the TUNEL index in the HCC tissues with high expression
was significantly lower than those in the HCC tissues with low
expression (P<0.05; Fig. 4C),
indicating decreased cell apoptosis in HCC tissues with high PDIA3
expression.
Discussion
In the current study, 378 proteins were identified
from HCC and non-HCC tissues by LC-MS/MS. Proteins involved in
antigen processing and presentation were the most abundant among
the upregulated proteins, and proteins in this category were not
detected among the downregulated and equally expressed proteins.
These upregulated proteins, including PDIA3, are usually present in
the endoplasmic reticulum (ER) (19)
and the enrichment of ER proteins in HCC has been reported in a
previous protein profiling study (7).
It is therefore plausible that the upregulation of ER proteins is a
characteristic feature of HCC.
Among the proteins classified in the category of
antigen processing and presentation, the clinicopathological
significance of increased PDIA3 expression in HCC was previously
unknown. The current study therefore examined PDIA3 expression in
HCC and investigated its association with various patient
clinicopathological features. The DFS and OS times of HCC patients
with high expression of PDIA3 were significantly shorter than those
of patients with low expression, thus PDIA3 expression may be
considered as a potential biomarker for the poor prognosis of
HCC.
PDIA3, also known as ERp57 or glucose-regulated
protein 58 kDa, is a thiol-oxidoreductase chaperone belonging to
the PDI family (20). The molecule is
involved in multiple cellular functions, including the folding of
newly synthesized proteins and assembly of major histocompatibility
complex I. Previous studies have reported the upregulation of PDIA3
protein and mRNA levels in HCC (7,10), which
is consistent with the results of the current study. However, in
previous proteomic studies, PDIA3 was not identified as a candidate
biomarker indicating early recurrence and metastasis of HCC
(9,11). This may be partially due to patient
selection as these studies were conducted with patients in which
recurrence and metastasis occurred within 6 months of the
treatment, while only 15% of patients in the current study
experienced recurrence and metastasis within this same time frame.
Therefore, PDIA3 may not be a suitable marker for patients with
rapidly progressing HCC.
The expression of PDIA3 has been observed in various
types of human cancer, including ovarian, mammary, uterine,
pulmonary and gastric cancer (21,22).
Increased PDIA3 expression was associated with a poor prognosis in
adenocarcinoma of the uterine cervix (23). However, its downregulation was
associated with poor outcomes in squamous cell carcinoma of the
uterine cervix (24), and loss of
expression of PDIA3 is correlated with more aggressive forms of
gastric cancer (25). Therefore,
although PDIA3 is expressed in various types of cancer, the
pathogenetic role of PDIA3 may vary among various types of human
cancer, depending on the primary organs affected and the histology
of the cancer.
In the present study, the shorter DFS and OS times
observed in HCC patients with high expression of PDIA3 appeared to
be associated with an increase in proliferation and reduction in
apoptotic cell death of cancer cells. In addition to its function
as a chaperone, PDIA3 is involved in the ER stress signaling
pathway, otherwise known as the ‘unfolded protein response’
(20). It has been demonstrated that
PDIA3 protects cells from ER-stress-induced apoptosis and that
silencing PDIA3 induces apoptosis (26). However, the direct signaling pathway
from PDIA3 to cell proliferation is not well known. Increased
proliferation of HCC may, in part, be due to the interaction of
PDIA3 with the mammalian target of rapamycin (mTOR) (27). The mTOR pathway serves an important
role in the carcinogenesis of HCC (28,29) and
increased expression of PDIA3 may induce cell proliferation through
enhanced interaction with mTOR. Further studies are required to
determine the precise molecular mechanism of cell proliferation and
interaction between PDIA3 and mTOR in order to establish an
effective targeting therapy.
In conclusion, the present study performed
comprehensive protein profiling of HCC by LC-MS/MS using FFPE
tissues and determined that expression was upregulated in HCC.
Increased PDIA3 expression predicted poor prognosis and was
associated with an increase in cell proliferation and a reduction
of apoptosis. To the best of our knowledge, the current study is
the first to demonstrate the significance of PDIA3 in the prognosis
of HCC, and may enable an effective targeting strategy to be
developed to treat patients with refractory and metastatic HCC.
Acknowledgements
The authors would like to thank Mr. Kiyoshi Teduka,
Mr. Takenori Fujii, Ms. Yoko Kawmoto, Ms. Kiyoko Kawahar and Ms.
Taeko Suzuki for technical assistance (Department of Integrated
Diagnostic Pathology, Nippon Medical School, Tokyo, Japan). The
present study was supported by grants-in-aid for Clinical Rebiopsy
Bank Project for Comprehensive Cancer Therapy Development to
Professor Zenya Naito from the Ministry of Education, Culture,
Sport, Science, and Technology, Japan (S1311022).
Glossary
Abbreviations
Abbreviations:
|
HCC
|
hepatocellular carcinoma
|
|
PDIA3
|
protein disulfide-isomerase A3
|
|
AFP
|
alpha fetoprotein
|
|
LC-MS/MS
|
liquid chromatography-tandem mass
spectrometry
|
|
FFPE
|
formalin-fixed paraffin-embedded
|
|
NSAF
|
normalized spectral abundance
factor
|
|
Rsc
|
ratios of spectral counts
|
|
KEGG
|
Kyoto Encyclopedia of Gene and
Genomes
|
|
TUNEL
|
terminal deoxynucleotidyl transferase
dUTP nick-end labeling
|
|
DFS
|
disease-free survival
|
|
OS
|
overall survival
|
|
ER
|
endoplasmic reticulum
|
|
mTOR
|
mammalian target of rapamycin
|
References
|
1
|
Mittal S and El-Serag HB: Epidemiology of
hepatocellular carcinoma: Consider the population. J Clin
Gastroenterol. 47:(Suppl). S2–S6. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Roxburgh P and Evans TR: Systemic therapy
of hepatocellular carcinoma: Are we making progress? Adv Ther.
25:1089–1104. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
de Lope CR, Tremosini S, Forner A, Reig M
and Bruix J: Management of HCC. J Hepatol. 56:(Suppl 1). S75–S87.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bruix J and Sherman M: Practice Guidelines
Committee, American Association for the Study of Liver Diseases:
Management of hepatocellular carcinoma. Hepatology. 42:1208–1236.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mann CD, Neal CP, Garcea G, Manson MM,
Dennison AR and Berry DP: Prognostic molecular markers in
hepatocellular carcinoma: A systematic review. Eur J Cancer.
43:979–992. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Poon RT: Prevention of recurrence after
resection of hepatocellular carcinoma: A daunting challenge.
Hepatology. 54:757–759. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chignard N, Shang S, Wang H, Marrero J,
Bréchot C, Hanash S and Beretta L: Cleavage of endoplasmic
reticulum proteins in hepatocellular carcinoma: Detection of
generated fragments in patient sera. Gastroenterology.
130:2010–2022. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Corona G, De Lorenzo E, Elia C, Simula MP,
Avellini C, Baccarani U, Lupo F, Tiribelli C, Colombatti A and
Toffoli G: Differential proteomic analysis of hepatocellular
carcinoma. Int J Oncol. 36:93–99. 2010.PubMed/NCBI
|
|
9
|
Huang X, Zeng Y, Xing X, Zeng J, Gao Y,
Cai Z, Xu B, Liu X, Huang A and Liu J: Quantitative proteomics
analysis of early recurrence/metastasis of huge hepatocellular
carcinoma following radical resection. Proteome Sci. 12:222014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Teramoto R, Minagawa H, Honda M, Miyazaki
K, Tabuse Y, Kamijo K, Ueda T and Kaneko S: Protein expression
profile characteristic to hepatocellular carcinoma revealed by
2D-DIGE with supervised learning. Biochim Biophys Acta.
1784:764–772. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yokoo H, Kondo T, Okano T, Nakanishi K,
Sakamoto M, Kosuge T, Todo S and Hirohashi S: Protein expression
associated with early intrahepatic recurrence of hepatocellular
carcinoma after curative surgery. Cancer Sci. 98:665–673. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gustafsson OJ, Arentz G and Hoffmann P:
Proteomic developments in the analysis of formalin-fixed tissue.
Biochim Biophys Acta. 1854:559–580. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bradford MM: A rapid and sensitive method
for the quantitation of microgram quantities of protein utilizing
the principle of protein-dye binding. Anal Biochem. 72:248–254.
1976. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zybailov B, Coleman MK, Florens L and
Washburn MP: Correlation of relative abundance ratios derived from
peptide ion chromatograms and spectrum counting for quantitative
proteomic analysis using stable isotope labeling. Anal Chem.
77:6218–6224. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Old WM, Meyer-Arendt K, Aveline-Wolf L,
Pierce KG, Mendoza A, Sevinsky JR, Resing KA and Ahn NG: Comparison
of label-free methods for quantifying human proteins by shotgun
proteomics. Mol Cell Proteomics. 4:1487–1502. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
da W Huang, Sherman BT, Stephens R,
Baseler MW, Lane HC and Lempicki RA: DAVID gene ID conversion tool.
Bioinformation. 2:428–430. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Paulsson K and Wang P: Chaperones and
folding of MHC class I molecules in the endoplasmic reticulum.
Biochim Biophys Acta. 1641:1–12. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Turano C, Gaucci E, Grillo C and
Chichiarelli S: ERp57/GRP58: A protein with multiple functions.
Cell Mol Biol Lett. 16:539–563. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Celli CM and Jaiswal AK: Role of GRP58 in
mitomycin C-induced DNA cross-linking. Cancer Res. 63:6016–6025.
2003.PubMed/NCBI
|
|
22
|
Chay D, Cho H, Lim BJ, Kang ES, Oh YJ,
Choi SM, Kim BW, Kim YT and Kim JH: ER-60 (PDIA3) is highly
expressed in a newly established serous ovarian cancer cell line,
YDOV-139. Int J Oncol. 37:399–412. 2010.PubMed/NCBI
|
|
23
|
Liao CJ, Wu TI, Huang YH, Chang TC, Wang
CS, Tsai MM, Lai CH, Liang Y, Jung SM and Lin KH: Glucose-regulated
protein 58 modulates cell invasiveness and serves as a prognostic
marker for cervical cancer. Cancer Sci. 102:2255–2263. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chung H, Cho H, Perry C, Song J, Ylaya K,
Lee H and Kim JH: Downregulation of ERp57 expression is associated
with poor prognosis in early-stage cervical cancer. Biomarkers.
18:573–579. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Leys CM, Nomura S, LaFleur BJ, Ferrone S,
Kaminishi M, Montgomery E and Goldenring JR: Expression and
prognostic significance of prothymosin-alpha and ERp57 in human
gastric cancer. Surgery. 141:41–50. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Corazzari M, Lovat PE, Armstrong JL, Fimia
GM, Hill DS, Birch-Machin M, Redfern CP and Piacentini M: Targeting
homeostatic mechanisms of endoplasmic reticulum stress to increase
susceptibility of cancer cells to fenretinide-induced apoptosis:
The role of stress proteins ERdj5 and ERp57. Br J Cancer.
96:1062–1071. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ramírez-Rangel I, Bracho-Valdés I,
Vazquez-Macías A, Carretero-Ortega J, Reyes-Cruz G and
Vázquez-Prado J: Regulation of mTORC1 complex assembly and
signaling by GRp58/ERp57. Mol Cell Biol. 31:1657–1671. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Matter MS, Decaens T, Andersen JB and
Thorgeirsson SS: Targeting the mTOR pathway in hepatocellular
carcinoma: Current state and future trends. J Hepatol. 60:855–865.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chang Z, Shi G, Jin J, Guo H, Guo X, Luo
F, Song Y and Jia X: Dual PI3K/mTOR inhibitor NVP-BEZ235-induced
apoptosis of hepatocellular carcinoma cell lines is enhanced by
inhibitors of autophagy. Int J Mol Med. 31:1449–1456.
2013.PubMed/NCBI
|