Introduction
Colorectal cancer (CRC) is the third most common
cancer in men, the second most common cancer in women and the
fourth-leading cause of cancer-associated mortality worldwide
(1). CRC morbidity and mortality has
increased in recent years (2).
Although advancements in treatment have been made, the
identification of new prognostic biomarkers and improving the
understanding of the molecular mechanisms underlying CRC remains a
challenge (3). Therefore,
investigating the mechanisms underlying the occurrence and
development of CRC, and identifying novel diagnostic biomarkers and
effective therapeutics is of high importance (4,5).
Long non-coding RNAs (lncRNAs) are RNA transcripts
of >200 nucleotides in length that are located in the nucleus
and cytosol, and are often expressed in a disease-, tissue- or
developmental stage-specific manner (6). Previous studies have demonstrated that
lncRNAs serve important roles in transcriptional regulation, cell
growth, carcinogenesis and metastasis (7,8). Aberrant
expression of lncRNAs has been observed in CRC and may have an
oncogenic or tumor suppressive role in the cancer initiatome
(8,9).
Growth arrest specific 5 (GAS5) is a lncRNA that is associated with
cell proliferation, and serves an essential role in the growth
arrest of T-cells and non-transformed lymphocytes (10). Overexpression of GAS5 decreases the
rate of cell cycling, whereas downregulation of GAS5 inhibits
apoptosis and maintains faster cell cycle progression.
Mourtada-Maarabouni et al (11) demonstrated that GAS5 transcription
levels were significantly decreased in breast cancer samples
compared with adjacent healthy breast epithelial tissue. Inhibition
of cell growth and induction of apoptosis through GAS5
overexpression was independent of other stimuli in certain cell
lines (11). However, the role of
GAS5 in CRC remains to be completely elucidated.
To clarify the clinical significance of GAS5
expression in CRC, GAS5 expression in CRC tissues and cell lines
was investigated, and the association between GAS5 expression in
tumor tissue and patient outcome was analyzed. To further
understand the functional significance of GAS5, the effect of
altered GAS5 levels on the phenotype of CRC cells was examined. The
results of the present study demonstrated that GAS5 expression is
frequently decreased in CRC, indicating that GAS5 serves an
essential role in the suppression of CRC and is a predictor of poor
survival in patients with CRC. The present study demonstrates the
importance of developing lncRNA-directed diagnostic and therapeutic
agents.
Materials and methods
Tissue collection
A total of 53 CRC tissue samples and the adjacent
normal tissues were obtained from patients diagnosed with CRC
following histopathological evaluation between January 2010 and May
2010, according to the seventh edition of the American Joint
Committee on Cancer Staging Manual (12). The patients whose clinicopathological
data was incomplete or whose total RNA following extraction was
degraded were excluded. Patients underwent surgery at Peking
University People's Hospital (Beijing, China) and
clinicopathological information was recorded for all samples
(Table I). No local or systemic
treatment was given to patients prior to CRC tissue sample
excision. All specimens were immediately frozen in liquid nitrogen
and stored at −80°C until required for RNA extraction. The present
study was approved by the Research Ethics Committee of Peking
University (Beijing, China). Informed consent was obtained from all
patients.
 | Table I.Clinicopathological characteristics
of 53 patients with colorectal carcinoma. |
Table I.
Clinicopathological characteristics
of 53 patients with colorectal carcinoma.
| Clinicopathological
parameter | Number of patients
(%) |
|---|
| Gender |
|
Male | 35 (66.0) |
|
Female | 18 (34.0) |
| Age (years) |
|
<60 | 15 (34.0) |
|
>60 | 38 (66.0) |
| Tumor size
(cm) |
|
<2 | 23 (43.4) |
|
>2 | 30 (56.7) |
| Histological
differentiation |
|
Well | 2 (3.8) |
|
Moderate | 42 (79.2) |
|
Poor | 9 (17.0) |
| Depth of
invasion |
|
T1+T2 | 5 (9.4) |
|
T3+T4 | 48 (90.6) |
| TNM stage |
|
I+II | 25 (47.2) |
|
III+IV | 28 (52.8) |
| Lymphatic
metastasis |
|
Yes | 25 (47.2) |
| No | 28 (52.8) |
| Regional lymph
nodes |
|
pN0 | 28 (52.8) |
|
pN1 | 16 (30.2) |
|
pN2 | 8 (15.1) |
|
pNX | 1 (1.9) |
| Distant
metastasis |
|
Yes | 9 (17.0) |
| No | 44 (83.0) |
| Expression of
GAS5 |
|
High | 27 (50.9) |
| Low | 26 (49.1) |
Cell lines and culture conditions
A total of seven human CRC cell lines (SW480, SW620,
RKO, HCT116, HT-29, LoVo and LS174T) were purchased from the
American Type Culture Collection (Manassas, VA, USA). The wild-type
human colon mucosal epithelial cell line, NCM460, was purchased
from INCELL Corporation LLC (San Antonio, TX, USA). SW480 and SW620
cells were cultured in Leibovitz's L-15 medium, while the other
cell lines were cultured in RPMI-1640 medium, both supplemented
with 10% fetal bovine serum (FBS) (all Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), 100 IU/ml penicillin and 100
mg/ml streptomycin. Cells were incubated at 37°C with 5%
CO2.
Total RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis
Total RNA was extracted from tissue samples/cultured
cells using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). cDNA was synthesized from RNA via reverse
transcription using the PrimeScript RT 5X Master Mix (cat no.
RR036A; Takara Biotechnology Co., Ltd., Dalian, China). qPCR was
performed using the SYBR® Green I Premix Ex Taq™ II
master mix (cat. no. RR820A; Takara Biotechnology Co., Ltd.)
according to the manufacturer's protocol. cDNA (50 ng) was used and
the thermocycling conditions were as follows: 30 sec at 95°C; then
95°C for 5 sec; and 60°C for 30 sec for 40 cycles. Results were
normalized to the expression of GAPDH. PCR primer sequences for
GAS5 and GAPDH were as follows: GAS5 forward,
5′-CTTCTGGGCTCAAGTGATCCT-3′ and reverse,
5′-TTGTGCCATGAGACTCCATCAG-3′; and GAPDH forward,
5′-GTCAACGGATTTGGTCTGTATT-3′ and reverse,
5′-AGTCTTCTGGGTGGCAGTGAT-3′. RT-qPCR and data collection was
performed using an ABI® 7500 Real-Time PCR System
running version 2.0 7500 software (Applied Biosystems; Thermo
fisher Scientific, Inc.). GAS5 expression was calculated and
subsequently normalized to the expression of GAPDH in SW480, SW620,
RKO, HCT116, HT-29, LoVo and LS174T and NCM460 cells using the
2−ΔΔCq method (13).
GAS5 overexpression, knockdown and
transfection
The full-length GAS5 sequence (National Center for
Biotechnology Information code, NR_002578) synthesized by PCR was
purchased and cloned into a pCDNA3.1(+) vector with NheI and
BamHI sites (both Invitrogen; Thermo fisher Scientific,
Inc.) to produce pCDNA-GAS5. An empty pCDNA3.1(+) vector was used
as the vehicle control. Small interfering RNAs (siRNAs) targeting
human GAS5 mRNA (si-h-GAS5) and the negative control siRNA (cat no.
siN05815122147) were purchased from Guangzhou RiboBio Co., Ltd.
(Guangzhou, China). The sequences of the anti-GAS5 siRNAs were as
follows: si-1, CTTGCCTGGACCAGCTTAA; si-2, GCAAGCCTAACTCAAGCCA;
si-3, GCAAAGGACTCAGAATTCA. Transfections with 50 nM pCDNA-GAS5,
empty vector, siRNA-h-GAS5 or NC siRNA were performed using
Lipofectamine® 2000 (Invitrogen; Thermo fisher
Scientific, Inc.) according to the manufacturer's protocol and
cells were harvested following a 72 h incubation at 37°C with 5%
CO2. For functional analysis of GAS5, pCDNA-GAS5 was
transfected into SW480 and HCT-116 cells, and siRNA-h-GAS5 was
transfected into RKO cells.
Analysis of apoptosis and cell cycle
progression
A total of 2×105 cells were seeded into
12-well plates 1 day prior to transfection and cells were harvested
72 h following transfection. Apoptotic cells were analyzed using
the Alexa FluorR® 488 Annexin V/Dead Cell Apoptosis kit
(Invitrogen; Thermo fisher Scientific, Inc.) according to the
manufacturer's protocol. To assay the number of cells in each stage
of the cell cycle, cells were harvested and subsequently stained
using the BD Cycletest™ Plus DNA Reagent kit (BD Biosciences,
Franklin Lakes, NJ, USA), according to the manufacturer's
instructions. Cells were then detected using flow cytometry and
data analyzed using FlowJo software (version 7; Tree Star, Inc.,
Ashland, OR, USA).
Cell proliferation and colony
formation assays
SW480, HCT-116 and RKO cells (2×105) were
seeded into 12-well plates day prior to transfection and cells were
harvested 72 h following transfection. Cell proliferation assays
were performed over the next 24–120 h using the Cell Counting Kit-8
(CCK8; Sigma-Aldrich; Merck Millipore, Darmstadt, Germany),
according to the manufacturer's instructions. For the colony
formation assay, SW480, HCT-116 and RKO cells were plated into
6-well plates at a density of 500 cells/well, and maintained in
media containing 10% FBS for 10 days at 37°C with 5%
CO2. Colonies were then fixed with methanol and stained
using 0.1% crystal violet (Sigma-Aldrich; Merck Millipore). Visible
colonies were manually counted. The colony assay was repeated three
times in duplicate.
Statistical analysis
Statistical analysis was performed using SPSS
software (version 20.0; SPSS Inc., Chicago, IL, USA). Values are
presented as the mean ± standard deviation. Statistical differences
between groups were analyzed using the Student's t-test. The
association between GAS5 expression and the clinicopathological
features of CRC was analyzed using the Chi-squared test. The
difference in GAS5 expression between CRC tissue and adjacent
normal tissue was analyzed using a Student's t-test.
Survival analysis was performed using the Kaplan-Meier estimator.
The log-rank test was used to analyze differences between the high
and low GAS5 expression groups. A Cox proportional hazards analysis
was performed to evaluate the independent prognostic factors of
overall survival (OS) in patients with CRC. P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression of GAS5 is downregulated in
human CRC tissues and cell lines
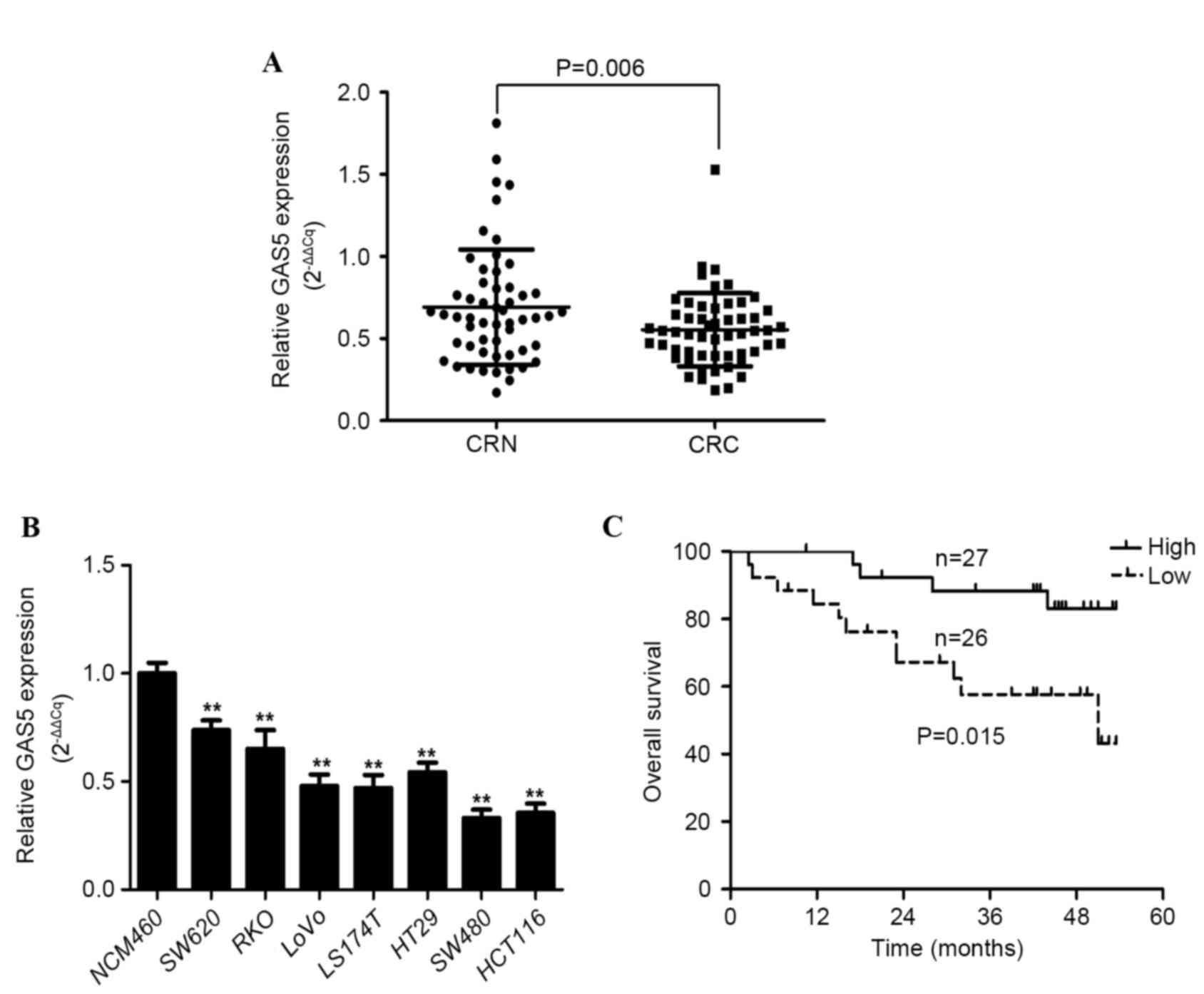
GAS5 expression was examined in CRC tissue samples
and adjacent histologically normal tissue samples from 53 patients
using RT-qPCR, with results normalized to GAPDH. GAS5 expression
was significantly decreased in CRC tissue samples compared with the
adjacent healthy tissue by a median relative expression difference
of 0.2568±0.6722 (P=0.006; Fig. 1A).
The median ratio between relative GAS5 expression in cancerous
tissue compared with normal tissue was 0.532 (Fig. 1A). GAS5 expression was downregulated
in 35/53 (66%) CRC tissue samples compared with the corresponding
adjacent healthy tissue. In addition, the relative expression level
of GAS5 in the CRC cell lines (SW480, SW620, RKO, LOVO, LS174T,
HT-29 and HCT116) was significantly decreased compared with the
normal colorectal mucosa cell line (NCM460) (P<0.001, P=0.003,
P=0.004, P<0.001, P<0.001, P=0.003 and P<0.001,
respectively; Fig. 1B). These results
indicate that abnormal GAS5 expression is associated with CRC
tumorigenesis and pathogenesis.
Association between GAS5 expression
and clinicopathological features in patients with CRC
The clinicopathological characteristics of 53
patients with colorectal carcinoma are presented in Table I. According to the median ratio of
GAS5 mRNA expression in CRC tissue compared with adjacent healthy
tissue (0.532), the 53 patients with CRC were classified into two
groups, high GAS5 expression (n=27; relative GAS5 expression
≥0.532) and low GAS5 expression (n=26; relative GAS5 expression
<0.532) (Table II). Follow-ups
demonstrated that the low GAS5 expression group exhibited a
significantly increased tumor size [odds ratio (OR), 0.176; 95% CI,
0.053–0.586; P=0.003) and more advanced tumor-node-metastasis (TNM)
stage (OR, 0.261; 95% CI, 0.083–0.819; P=0.019) (Table II). No significant differences were
observed in the distribution of gender, age, histological
differentiation, depth of invasion, lymphatic metastasis, regional
lymph node status or presence of distant metastasis between
patients in the high and low GAS5 mRNA expression groups (Tables I and II).
 | Table II.Correlation between GAS5 mRNA
expression and clinicopathological characteristics in patients with
colorectal carcinoma. |
Table II.
Correlation between GAS5 mRNA
expression and clinicopathological characteristics in patients with
colorectal carcinoma.
|
| GAS5 mRNA
expression group |
|
|---|
|
|
|
|
|---|
| Clinicopathological
parameter | High (number of
patients) | Low (number of
patients) | Chi-squared test
P-value |
|---|
| Sex |
|
|
|
|
Male | 15 | 20 | 0.101 |
|
Female | 12 | 6 |
|
| Age |
|
|
|
|
<60 | 7 | 8 | 0.696 |
|
>60 | 20 | 18 |
|
| Tumor size
(cm) |
|
|
|
|
<2 | 17 | 6 | 0.003a |
|
>2 | 10 | 20 |
|
| Histological
differentiation |
|
|
|
|
Well | 0 | 2 | 0.492 |
|
Moderate | 22 | 20 |
|
|
Poor | 5 | 4 |
|
| Depth of
invasion |
|
|
|
|
T1+T2 | 4 | 1 | 0.370 |
|
T3+T4 | 23 | 25 |
|
| TNM stage |
|
|
|
|
I+II | 17 | 8 | 0.019a |
|
III+IV | 10 | 18 |
|
| Lymphatic
metastasis |
|
|
|
|
Yes | 14 | 11 | 0.487 |
| No | 13 | 15 |
|
| Regional lymph
nodes |
|
|
|
|
pN0 | 13 | 15 | 0.771 |
|
pN1 | 8 | 8 |
|
|
pN2 | 4 | 4 |
|
|
pNX | 1 | 0 |
|
| Distant
metastasis |
|
|
|
|
Yes | 3 | 6 | 0.427 |
| No | 24 | 20 |
|
Decreased GAS5 mRNA expression is a
predictor of poor prognosis in patients with CRC
The correlation between GAS5 expression and the
outcome of patients with CRC following a colectomy was examined. OS
curves of patients were plotted according to high or low GAS5
expression status using the Kaplan-Meier estimator. As shown in
Fig. 1C, patients expressing low
levels of GAS5 mRNA had a significantly shorter median survival
time (37.8±3.8 months) compared with patients expressing high
levels of GAS5 (49.2±2.1 months) (P=0.015). These results suggest
that downregulated GAS5 expression is significantly associated with
poor OS in patients with CRC.
The Cox proportional hazards model demonstrated that
the level of GAS5 mRNA expression [hazard ratio (HR), 0.236; 95%
confidence interval (CI), 0.067–0.827; P=0.024], TNM stage (HR,
0.164; 95% CI, 0.032–0.754; P=0.010) and distant metastasis status
(HR, 0.089; 95% CI, 0.025–0.317; P<0.001) were significantly
associated with the OS rate of patients with CRC, and may be used
as independent prognostic factors (Table III). These results indicate that a
low GAS5 expression level is an independent risk factor for CRC and
a predictor of poor prognosis.
 | Table III.Cox proportional hazards model of
variables associated with overall survival in patients with
colorectal carcinoma (n=53). |
Table III.
Cox proportional hazards model of
variables associated with overall survival in patients with
colorectal carcinoma (n=53).
|
| Multivariate
analysis |
|---|
|
|
|
|---|
| Variables | HR (95% CI) | P-value |
|---|
| Age (<60 vs. ≥60
years old) | 0.996
(0.954–1.954) | 0.996 |
| Sex (males vs.
females) | 0.805
(0.231–2.231) | 0.805 |
| Tumor size (<2
vs. ≥2 cm) | 0.489
(0.193–2.193) | 0.484 |
| Histological
differentiation | 0.366
(0.100–1.100) | 0.129 |
| (well and moderate
differentiation vs. poor differentiation) |
| TNM stage (I+II vs.
III+IV) | 0.164
(0.032–0.032) | 0.010a |
| Depth of invasion
(T1+T2 vs. T3+T4) | 2.258
(0.202–25.202) | 0.508 |
| Lymphatic
metastasis (present vs. absent) | 0.506
(0.159–1.159) | 0.249 |
| Distant metastasis
(present vs. absent) | 0.089
(0.025–0.025) | 0.000a |
| GAS5 mRNA
expression (high vs. low) | 0.236
(0.067–0.067) | 0.024a |
GAS5 decreases CRC cell growth and
colony formation, and induces G0/G1 cell cycle arrest and
apoptosis
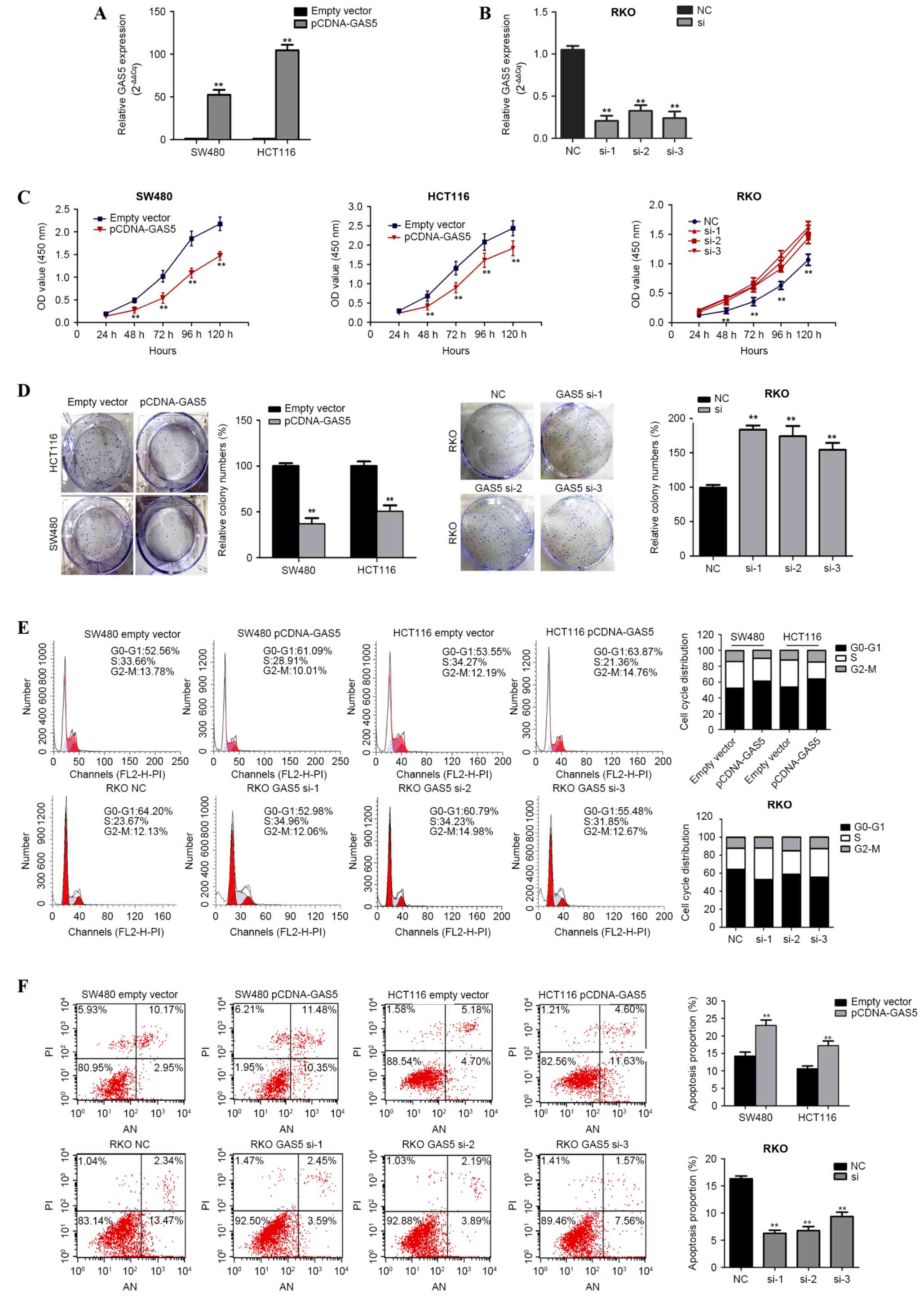
The role of GAS5 in CRC pathology was functionally
analyzed in vitro through the overexpression or knockdown of
GAS5 in CRC cell lines. SW480 and HCT116 cells were transfected
with pCDNA3.1-GAS5 in order to induce GAS5 overexpression, and RKO
cells were transfected with siRNA-h-GAS5 in order to knockdown
GAS5. GAS5 mRNA overexpression and knockdown was confirmed using
RT-qPCR (Fig. 2A and B). CCK-8
analysis demonstrated that GAS5 overexpression significantly
repressed the rate of cell proliferation in the SW480 and HCT116
cell lines, whereas knockdown of GAS5 increased proliferation in
siRNA-h-GAS5-transfected RKO cells (Fig.
2C). Furthermore, a colony formation assay revealed that
clonogenic survival was significantly decreased in SW480 and HCT116
cells transfected with pCDNA3.1-GAS5 compared with the negative
control group (P<0.001; Fig. 2D);
however, the opposite phenomenon was observed in
siRNA-h-GAS5-transfected RKO cells (P=0.001; Fig. 2D).
The inhibitory effect of GAS5 on cell cycle
progression and apoptosis was examined using flow cytometry.
Compared with their respective controls, upregulation of GAS5
expression resulted in the accumulation of G0/G1 cells in the SW480
(52.83±3.16% vs. 61.58±3.47%; P=0.032) and HCT116 (53.88±3.82% vs.
63.95±2.97%; P=0.022) cell lines, whereas downregulation of GAS5
expression reduced the number of G0/G1 cells in the
siRNA-h-GAS5-transfected RKO cells (66.22±1.24% vs. 56.42±2.30%;
P=0.020 vs. the control) (Fig. 2E).
In addition, the rate of apoptosis was significantly increased
following ectopic expression of GAS5 in SW480 (13.37±0.54% vs.
21.89±0.85%) and HTC116 (9.92±0.56% vs. 16.33±0.85%) cells (both
P<0.001; Fig. 2F), but decreased
in siRNA-h-GAS5-transfected RKO cells (16.19±0.32% vs. 7.08±1.02%);
(P<0.001 vs. the control group; Fig.
2F). These results suggest that GAS5 inhibits CRC cell growth
and colony formation, and induces G0/G1 arrest and apoptosis.
Discussion
The cancer transcriptome is more complex than was
previously expected (14,15). Although initially thought to be
spurious transcriptional noise, lncRNAs are now known to
participate in the regulation of cellular development, cell growth
and the development of human disease, including cancer (16–19). A
number of lncRNAs serve important regulatory roles in chromosome
modification (20), transcription in
the nucleus and post-transcriptional processing in the cytoplasm
(21). Accumulating evidence has
demonstrated that lncRNA dysregulation affects epigenetic
regulation and induces cell growth, resulting in progressive and
uncontrolled tumor growth (8,20,22–26). The
lncRNA GAS5 is non-coding, hosts multiple small nucleolar (sno) RNA
sequences in its introns and contains 12 exons (27,28). GAS5
was initially identified during screening for potential tumor
suppressor genes (29) and is a
stress-inducible gene, which is differentially expressed in healthy
and tumor tissues/cell lines (30).
In addition, GAS5 has been demonstrated to be involved in the
regulation of the cell cycle (31)
and to function as a tumor suppressor in human T cells, and breast
and prostate cancer cell lines by inducing apoptosis (10,11,32,33).
Furthermore, reduced expression of GAS5 and/or its snoRNAs has been
observed in head and neck squamous cell carcinomas, and gastric and
cervical cancer (34–36), indicating that it serves an important
role in tumorigenesis.
However, the underlying mechanisms behind the
effects of GAS5 in CRC remain unclear. In the present study, the
clinical and prognostic significance of GAS5 in 53 patients with
CRC was investigated. Analysis of GAS5 mRNA expression using
RT-qPCR demonstrated that GAS5 was significantly downregulated in
CRC tissue samples compared with adjacent normal tissue. Decreased
GAS5 expression was also identified in several CRC cell lines
compared with a wild-type colorectal mucosa cell line. In addition,
the present study revealed that decreased GAS5 expression was
associated with increased tumor size and an increased TNM stage.
Furthermore, downregulated GAS5 expression was associated with poor
prognosis in patients with CRC. Ectopic expression of GAS5 in
multiple CRC cell lines resulted in an increase in apoptosis, a
reduction in the rate of proliferation and inhibition of cell cycle
progression. Conversely, downregulation of GAS5 inhibited
apoptosis, and increased proliferation and cell cycle
progression.
In conclusion, the results of the present study
indicate that GAS5 negatively regulates the survival of CRC cells,
and functions as a tumor suppressor by regulating cell growth and
apoptosis, which is consistent with the results of previous studies
performed in lymphoid cells and other epithelial cell lines
(10,11,37,38).
However, the mechanisms underlying the effects of GAS5 in CRC
remain to be completely elucidated. The results of the present
study suggest that GAS5 has a complex role in CRC development.
Dysregulation of GAS5 may be an important diagnostic and prognostic
marker in patients with CRC. An improved understanding of the
GAS5-mediated pathogenesis and development of CRC may facilitate
the development of lncRNA-directed cancer therapeutics.
Acknowledgements
The present study was supported by the National
Natural Science Foundation (grant nos. 81372290 and 81372291), the
National High Technology Research and Development Program of China
(grant no. 2015AA020110), the Specialized Research Fund for the
Doctoral Program of Higher Education (grant no. 20130001120064),
and the Peking University People's Hospital Research and
Development Fund (grant no. RDC2013-17).
References
|
1
|
Stewart BW and Wild CP: World Cancer
Report 2014International Agency for Research on Cancer. Lyon: 2014,
View Article : Google Scholar
|
|
2
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Papadopoulos V, Tsapakidis K, Del Riobo
Galdo NA, Papandreou CN, Del Galdo F, Anthoney A, Sakellaridis N,
Dimas K and Kamposioras K: The prognostic significance of the
hedgehog signaling pathway in colorectal cancer. Clin Colorectal
Cancer. 15:116–127. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang B, Shen ZL, Jiang KW, Zhao G, Wang
CY, Yan YC, Yang Y, Zhang JZ, Shen C, Gao ZD, et al: MicroRNA-217
functions as a prognosis predictor and inhibits colorectal cancer
cell proliferation and invasion via an AEG-1 dependent mechanism.
BMC Cancer. 15:4372015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
De Roock W, De Vriendt V, Normanno N,
Ciardiello F and Tejpar S: KRAS, BRAF, PIK3CA, and PTEN mutations:
Implications for targeted therapies in metastatic colorectal
cancer. Lancet Oncol. 12:594–603. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang X, Gejman R, Mahta A, Zhong Y, Rice
KA, Zhou Y, Cheunsuchon P, Louis DN and Klibanski A: Maternally
expressed gene 3, an imprinted noncoding RNA gene, is associated
with meningioma pathogenesis and progression. Cancer Res.
70:2350–2358. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gutschner T and Diederichs S: The
hallmarks of cancer: A long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mourtada-Maarabouni M, Hedge VL, Kirkham
L, Farzaneh F and Williams GT: Growth arrest in human T-cells is
controlled by the non-coding RNA growth-arrest-specific transcript
5 (GAS5). J Cell Sci. 121:939–946. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mourtada-Maarabouni M, Pickard MR, Hedge
VL, Farzaneh F and Williams GT: GAS5, a non-protein-coding RNA,
controls apoptosis and is downregulated in breast cancer. Oncogene.
28:195–208. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gibb EA, Vucic EA, Enfield KS, Stewart GL,
Lonergan KM, Kennett JY, Becker-Santos DD, MacAulay CE, Lam S,
Brown CJ and Lam WL: Human cancer long non-coding RNA
transcriptomes. PLoS One. 6:e259152011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Park JY, Lee JE, Park JB, Yoo H, Lee SH
and Kim JH: Roles of Long Non-Coding RNAs on tumorigenesis and
glioma development. Brain Tumor Res Treat. 2:1–6. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Michalik KM, You X, Manavski Y,
Doddaballapur A, Zörnig M, Braun T, John D, Ponomareva Y, Chen W,
Uchida S, et al: Long noncoding RNA MALAT1 regulates endothelial
cell function and vessel growth. Circ Res. 114:1389–1397. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang EB, Yin DD, Sun M, Kong R, Liu XH,
You LH, Han L, Xia R, Wang KM, Yang JS, et al: P53-regulated long
non-coding RNA TUG1 affects cell proliferation in human non-small
cell lung cancer, partly through epigenetically regulating HOXB7
expression. Cell Death Dis. 5:e12432014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tripathi V, Ellis JD, Shen Z, Song DY, Pan
Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al: The
nuclear-retained noncoding RNA MALAT1 regulates alternative
splicing by modulating SR splicing factor phosphorylation. Mol
Cell. 39:925–938. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nie Y, Liu X, Qu S, Song E, Zou H and Gong
C: Long non-coding RNA HOTAIR is an independent prognostic marker
for nasopharyngeal carcinoma progression and survival. Cancer Sci.
104:458–464. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhou Y, Zhang X and Klibanski A: MEG3
noncoding RNA: A tumor suppressor. J Mol Endocrinol. 48:R45–R53.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Niinuma T, Suzuki H, Nojima M, Nosho K,
Yamamoto H, Takamaru H, Yamamoto E, Maruyama R, Nobuoka T, Miyazaki
Y, et al: Upregulation of miR-196a and HOTAIR drive malignant
character in gastrointestinal stromal tumors. Cancer Res.
72:1126–1136. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kotake Y, Nakagawa T, Kitagawa K, Suzuki
S, Liu N, Kitagawa M and Xiong Y: Long non-coding RNA ANRIL is
required for the PRC2 recruitment to and silencing of p15(INK4B)
tumor suppressor gene. Oncogene. 30:1956–1962. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Khalil AM, Guttman M, Huarte M, Garber M,
Raj A, Morales Rivea D, Thomas K, Presser A, Bernstein BE, van
Oudenaarden A, et al: Many human large intergenic noncoding RNAs
associate with chromatin-modifying complexes and affect gene
expression. Proc Natl Acad Sci USA. 106:11667–11672. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Smith CM and Steitz JA: Classification of
gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member
of the 5′-terminal oligopyrimidine gene family reveals common
features of snoRNA host genes. Mol Cell Biol. 18:6897–6909. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Muller AJ, Chatterjee S, Teresky A and
Levine AJ: The gas5 gene is disrupted by a frameshift mutation
within its longest open reading frame in several inbred mouse
strains and maps to murine chromosome 1. Mamm Genome. 9:773–774.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Schneider C, King RM and Philipson L:
Genes specifically expressed at growth arrest of mammalian cells.
Cell. 54:787–793. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Renganathan A, Kresoja-Rakic J, Echeverry
N, Ziltener G, Vrugt B, Opitz I, Stahel RA and Felley-Bosco E: GAS5
long non-coding RNA in malignant pleural mesothelioma. Mol Cancer.
13:1192014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lodygin D, Tarasov V, Epanchintsev A,
Berking C, Knyazeva T, Körner H, Knyazev P, Diebold J and Hermeking
H: Inactivation of miR-34a by aberrant CpG methylation in multiple
types of cancer. Cell cycle. 7:2591–2600. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pickard MR, Mourtada-Maarabouni M and
Williams GT: Long non-coding RNA GAS5 regulates apoptosis in
prostate cancer cell lines. Biochim Biophys Acta. 1832:1613–1623.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yacqub-Usman K, Pickard MR and Williams
GT: Reciprocal regulation of GAS5 lncRNA levels and mTOR inhibitor
action in prostate cancer cells. Prostate. 75:693–705. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sun M, Jin FY, Xia R, Kong R, Li JH, Xu
TP, Liu YW, Zhang EB, Liu XH and De W: Decreased expression of long
noncoding RNA GAS5 indicates a poor prognosis and promotes cell
proliferation in gastric cancer. BMC Cancer. 14:3192014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cao S, Liu W, Li F, Zhao W and Qin C:
Decreased expression of lncRNA GAS5 predicts a poor prognosis in
cervical cancer. Int J Clin Exp Pathol. 7:6776–6783.
2014.PubMed/NCBI
|
|
36
|
Gee HE, Buffa FM, Camps C, Ramachandran A,
Leek R, Taylor M, Patil M, Sheldon H, Betts G, Homer J, et al: The
small-nucleolar RNAs commonly used for microRNA normalisation
correlate with tumour pathology and prognosis. Br J Cancer.
104:1168–1177. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mourtada-Maarabouni M, Hasan AM, Farzaneh
F and Williams GT: Inhibition of human T-cell proliferation by
mammalian target of rapamycin (mTOR) antagonists requires noncoding
RNA growth-arrest-specific transcript 5 (GAS5). Mol Pharmacol.
78:19–28. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kino T, Hurt DE, Ichijo T, Nader N and
Chrousos GP: Noncoding RNA gas5 is a growth arrest- and
starvation-associated repressor of the glucocorticoid receptor. Sci
Signal. 3:ra82010. View Article : Google Scholar : PubMed/NCBI
|