Introduction
Cyclin-dependent kinase (Cdk) 2 regulates the
initiation and progression of the S phase of the cell cycle, and
the regulation of Cdk2 involves cyclin binding and phosphorylation
(1). Monomeric Cdk2 is inactive, and
its activation requires binding to cyclin. Cdk2 associates with
cyclin E to drive G1/S phase transition, and it associates with
A-type cyclins to mediate DNA replication during S phase (2–5). The
Cdk2/cyclin complex is recognized by multiple kinases, which
phosphorylate the T14, Y15, and T160 amino acid residues in the
complex. Phosphorylation of T14 and Y15 induces inactivation of
Cdk2; activation of Cdk2 requires de-phosphorylation of both T14
and Y15 by Cdc25 and phosphorylation of T160 by CDK activating
kinase (CAK) (6,7). Cdk2 also mediates the cell cycle
inhibitory and tumor-suppressing activities of P21 Cip1 and P27
Kip1, respectively (2,8–10).
Human ribosomal protein S3 (hRpS3) is a component of
the small ribosomal subunit, which is involved in protein synthesis
(11–13). The functions of hRpS3 are not limited
to protein translation, and it performs multiple extra-ribosomal
activities, such as in DNA repair, cell death, inflammation, and
tumorigenesis (13–19). These extra-ribosomal functions of
hRpS3 are induced by its interactions many signaling molecules,
which result in different post-translation modifications that
confer different abilities to hRpS3 (13–19). hRpS3
is phosphorylated by IKKβ on Serine 209, which is important for
nuclear localization (7). In response
to DNA damage, hRpS3, phosphorylated by protein kinase C-δ (PKCδ)
at serine 6 (S6) and Threonine 221 (T221) residues, translocates
into the nucleus for repair (11,12,20).
Moreover, hRpS3 is also phosphorylated by extracellular
signal-regulated kinase 1 (ERK1), which is a MAP kinase that plays
an important role in the regulation of cell growth (11,21,22). ERK1
phosphorylates hRpS3 on Threonine 42 (T42) (11,22);
moreover, this phosphorylation is necessary for the nuclear
translocation of hRpS3 in response to DNA damage (21). Threonine 70 (T70) of hRpS3 is
phosphorylated by Akt, which promotes its nuclear translocation,
thereby preventing hRpS3-induced apoptosis by inhibiting its
interaction with E2F1 (13,20).
In this study, we showed that Cdk2 also interacts
with hRpS3. Moreover, using bioinformatics tools (NetPhos2.0,
www.cds.dtu.dk/services/NetPhos; KinasePhos,
kinasephos.mbc.nctu.edu.tw), we found
eight sites for putative Cdk2-mediated phosphorylation in hRpS3. We
also found that Cdk2 phosphorylates S6 and T221 of hRpS3. Cdk2 is
active in S phase, and our in vitro kinase assay revealed
that hRpS3 phosphorylation in cells arrested in the S-phase was
more than two folds that in asynchronous control cells. Cdk2
phosphorylates the non-ribosomal form of hRpS3, and this
phosphorylation is important for nuclear transport of hRpS3. We
also observed that knockdown of hRpS3 delays cell cycle
progression. Our findings demonstrate that hRpS3 is involved in
cell cycle regulation and that this function may be linked to DNA
damage repair.
Materials and methods
Cell lines
Human embryonic kidney cells (HEK293) were grown in
Dulbecco's Modified Eagle's medium (DMEM; Invitrogen Life
Technologies, Carlsbad, CA, USA) containing 10% fetal bovine serum
(FBS, Invitrogen Life Technologies) and 1% penicillin-streptomycin
solution (Sigma-Aldrich, St. Louis, MO, USA) at 37°C in a 5%
CO2 incubator.
Plasmid construction
Gene fragments corresponding to cDNA coding regions
of human RpS3 (Accession no. NM_001005) and Cdk2 (accession no.
NM_001798) were amplified using PCR. A Cdk2 fragment was inserted
into the EcoRI and SalI sites of pCMV Tag2C
(Stratagene, La Jolla, CA, USA), and hRpS3 fragments were inserted
into the BamHI and XhoI sites of pCMV Tag3A
(Stratagene). All constructs were confirmed by restriction enzyme
mapping and DNA sequencing.
pDNA transfection
HEK293 cells were seeded in 6-well plates at a
density of 1×105 cells per well and incubated for 24 h
before transfection. Recombinant plasmid DNAs were transiently
transfected into 80% confluent HEK293 cells using
Lipofectamine® 2000 reagent (Invitrogen Life
Technologies) according to the manufacturer's protocol. After 24 h
of incubation with the plasmids, the cells were collected and lysed
in lysis buffer (50 mM Tris-HCl, pH 8.0; 100 mM NaCl; 5 mM EDTA; 1
mM NaF; 1 mM Na3VO4; 1% Nonidet P-40; 10
µg/ml of PMSF; and protease inhibitor cocktail) for 40 min at 4°C.
After centrifugation, supernatants were collected and used in
immunoblot analysis and immunoprecipitation (IP).
GST pull-down assay
For GST pull-down assays, GST-Cdk2 was adsorbed onto
glutathione-Sepharose 4B beads (GE Healthcare, Bucks, UK).
His6-purified hRpS3 protein was then incubated with GST
or GST fusion protein in binding buffer (50 mM Tris-HCl, pH 7.5;
150 mM NaCl; 1 mM EDTA; 0.3 mM DTT; 0.1% NP-40; and protease
inhibitor cocktail). The binding reaction was performed for 3 h at
4°C; subsequently, the beads were washed and bound proteins were
subjected to SDS-PAGE analysis. Proteins were detected by
immunoblot analysis.
Co-IP
HEK293 cells were seeded 5×105
cells/60-mm2 dish and cultured 80–90% confluency before
transfection. Lysates were collected from cells transiently
transfected with different sets of plasmid DNAs and incubated for 3
h with described antibodies at 4°C with gentle rotation. Antibodies
and bound proteins were incubated with protein A/G-Sepharose beads
(Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) at 4°C for 12
h. Samples were precipitated with centrifugation, washed with PBS,
and mixed with gel loading buffer. Immunoprecipitated samples were
resolved on SDS-PAGE gel and subjected to immunoblot analysis.
Immunoblot analysis
Proteins resolved on 8–12% SDS-PAGE gel were
transferred onto polyvinylidene difluoride (PVDF) membranes (Pall
Corporation, East Hill, NY, USA). The membranes were blocked with
5% non-fat dried milk in TBS-T (TBS with 0.05% Tween-20) for 20 min
at room temperature. The membrane was incubated with primary
antibodies against anti-hRpS3, p-Ser hRpS3, p-Thr hRpS3 (Cell
Signaling Technology Inc., MA, USA), anti-Cdk2, Cdk1, cyclin B1,
cyclin E1 (Santa Cruz Biotechnology). After appropriate washing,
the membranes were incubated with horseradish peroxidase
(HRP)-conjugated secondary antibodies (Santa Cruz Biotechnology).
Protein bands were detected using ECL Western blotting detection
reagents (Thermo Fisher Scientific, Waltham, MA, USA).
Cell cycle synchronization
To arrest cells the in the S-phase, cells
transfected for 24 h were treated with 3 mM thymidine (an inhibitor
of DNA synthesis) for 18 h (23,24). Then,
the cells were washed with medium and incubated in fresh medium for
2 h. To collect G2/M phase cells, the cells were treated with 0.1
µg/ml nocodazole (a mitotic inhibitor) for 12 h (23,24). An
aliquot of cells was stained with propidium iodide (PI;
Sigma-Aldrich) and analyzed by fluorescence-activated cell sorting
(FACS).
FACS analysis
Cells were trypsinized and centrifuged for 10 min.
The collected cells were fixed with 70% ethanol for 30 min,
followed by centrifugation. After washing with ice-cold PBS, the
cell pellets were resuspended in 0.5 ml PBS containing 50 µg/ml PI
and 100 µg/ml RNase (Sigma-Aldrich). After 30 min of incubation in
the dark, cell complex was estimated using a minimum of 10,000
cells per sample and was analyzed using a flow cytometer.
Fluorescence emitted from the PI-DNA was analyzed using Cell Quest
Alias software (BD Biosciences, Rockville, MD, USA).
In vitro Cdk2 kinase assay
After incubation of cell lysates with Cdk2-specific
monoclonal antibody (5 µg), samples were incubated with 50 µl
protein A/G-Sepharose beads at 4°C for 12 h. Immune complexes were
washed with washing buffer (1:1 mixture of lysis buffer and PBS)
and kinase reaction buffer (20 mM Tris-HCl, pH 7.4; 15 mM
MgCl2; and 1 mM DTT). After washing, samples were added
to a mixture of 1 µg substrate (histone H1 or hRpS3), 200 µM ATP,
and 5 µCi [32P-γ] ATP in 30 µl of reaction buffer.
Reactions were carried out for 25 min at 30°C and terminated by
addition of SDS-PAGE sample buffer. Samples were subjected to
SDS-PAGE, and phosphorylation was detected by autoradiography.
hRpS3 siRNA
hRpS3 siRNA oligonucleotides targeting the sequences
5′-GGGUUCCUAGUACUGCAATT-3′ and 5′-UUGCAGUACUAGGAACCCCTT-3′ were
purchased from Santa Cruz Biotechnology. Cells were transfected
with hRpS3 siRNAs (5 nM each) using Lipofectamine® 2000
(Invitrogen Life Technologies). Following 24 h of transfection, the
level of hRpS3 was determined using anti-hRpS3 antibody (Cell
Signaling Technology Inc., Danvers, MA, USA). A scrambled siRNA
[green fluorescent protein (GFP) siRNA,
5′-GGGCACAAGCUGGAGUACAACUACA-3′] was used as the control.
Statistical analysis
All experiments were performed at least three times
for statistical analysis. To determine the significance of the
differences between samples, we performed the Student's
t-test using Microsoft Excel. Date were expressed as mean ±
standard errors, and P<0.05 (paired two-tailed t-test,
P<0.01, P<0.001) was considered to indicate statistical
significance.
Results
Cdk2 physically interacts with
hRpS3
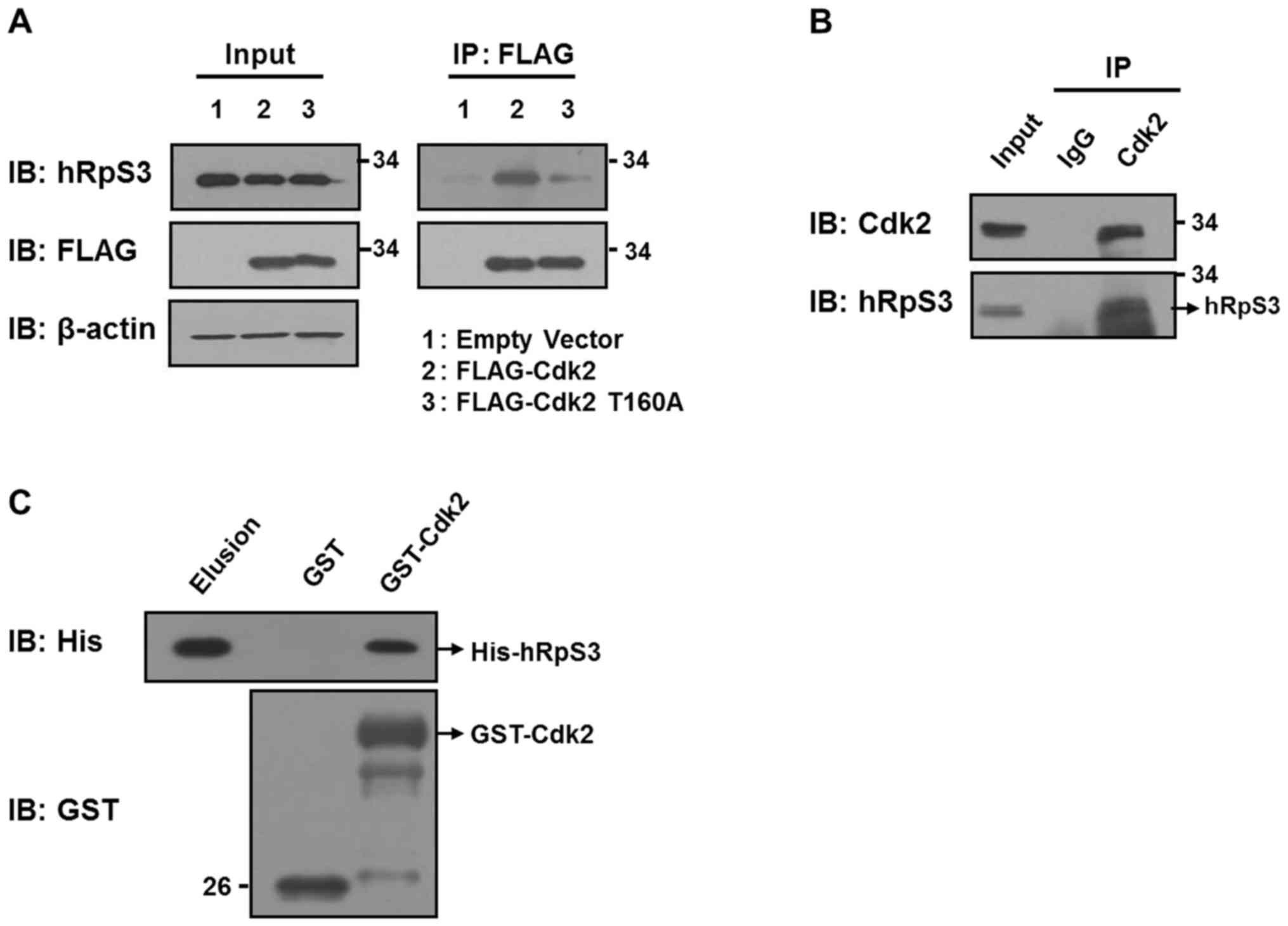
To determine whether hRpS3 interacts with Cdk2,
co-IP was conducted using HEK293 cells transfected with FLAG empty
vector or with FLAG-tagged Cdk2 (FLAG-Cdk2) and c-Myc-tagged hRpS3.
After 24 h incubation, cells were lysed and immunoprecipitated with
anti-RpS3 antibody. The co-IP results indicated that Cdk2
interacted with ectopically expressed hRpS3 (Fig. 1A). A similar co-IP assay with
antibodies against endogenous Cdk2 and hRpS3 also revealed that
Cdk2 interacted with hRpS3 (Fig. 1B).
Rabbit IgG was used as a control. To determine whether hRpS3
interacts directly with Cdk2, we conducted a GST pull-down assay
(Fig. 1C). His-hRpS3 and GST-Cdk2
were generated and purified before conducting the assay. Purified
His-hRpS3 interacted with GST-Cdk2 but not with GST alone,
indicating that Cdk2 interacts directly with hRpS3. Next, we
investigated whether the interaction between Cdk2 and hRpS3
required Cdk2 activation. For this, we prepared FLAG-Cdk2 and a
mutant of Cdk2 with modifications at T160 (T160A; Cdk2
phosphorylated at T160 is the active form), and transfected these
into HEK293 cells (Fig. 1A; lanes 2
and 3). Co-IP revealed that substitution of threonine with alanine
inhibited Cdk2 phosphorylation at the site. These results show that
hRpS3 interacts only with the active form of Cdk2 (Fig. 1A; lanes 2 and 3).
hRpS3 was phosphorylated by Cdk2
during the S-phase
It is already known that the highest activity of
Cdk2 is observed in the S-phase (1).
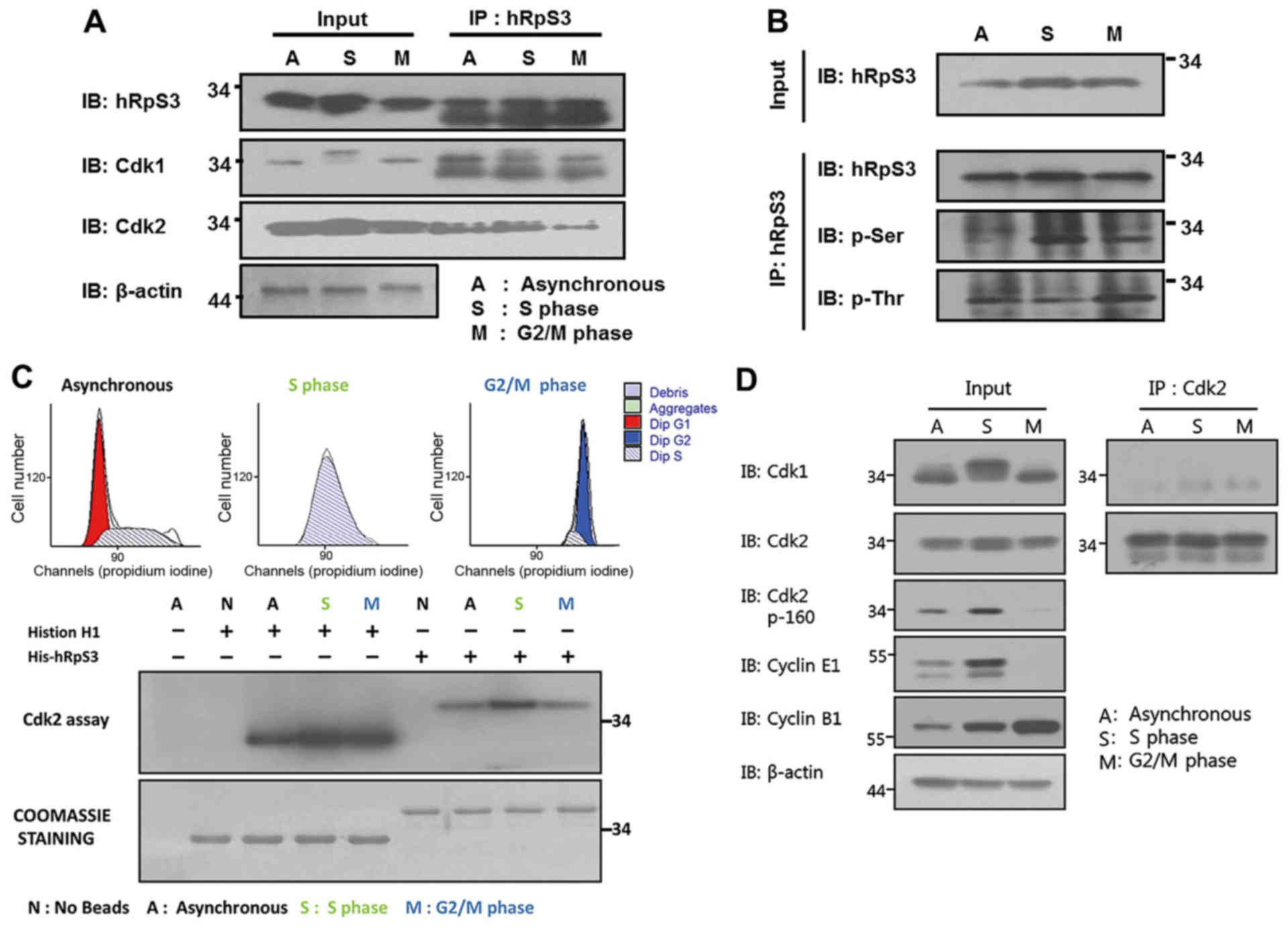
Therefore, we investigated the association between hRpS3
phosphorylation and Cdk2 activity. Co-IP of asynchronous and
synchronous cells arrested at the S and G2/M phases was conducted
using anti-hRpS3 antibody (Fig. 2A).
Consistent with the results in Fig.
1A, endogenous hRpS3 was found to interact with Cdk2. Moreover,
Fig. 2A shows that this interaction
was higher in the S-phase than in the G2/M phase. This result
indicates that hRpS3 interacts with the active form of Cdk2 in the
S-phase.
Cdk2 phosphorylates its substrate at the
serine/threonine residues. Therefore, the possibility of hRpS3
being a substrate of Cdk2 was examined. We have previously
demonstrated that Cdk1 interacts and phosphorylates hRpS3, and this
phosphorylation increased in the G2/M phase (21). Thus, we conducted a co-IP assay using
asynchronous and synchronous cells (arrested at the S and G2/M
phases). Whole cell lysates were subjected to immunoprecipitation
with anti-hRpS3 antibody. Following immunoblot analysis using
anti-hRpS3, hRpS3 was detected in both input and
immunoprecipitation samples (Fig.
2B). Phospho-serine (p-Ser) levels in the immunoprecipitated
sample were higher in the cells arrested at the S-phase than in the
cells arrested at the G2/M phase. In contrast, an intense band of
phospho-threonine (p-Thr) was detected in cells in the G2/M phase.
Since Cdk2 is active in the S-phase, these results indicate that
Cdk2 phosphorylates the serine of hRpS3 in the S-phase.
Fig. 1. A shows that
hRpS3 binds with the active form of Cdk2. To demonstrate the
ability of Cdk2 to phosphorylate hRpS3, we conducted an in
vitro kinase assay (Fig. 2C).
Cells were synchronized at the G2/M- and S-phases using nocodazole
and thymidine, respectively. Cell synchronization was confirmed by
FACS analysis (Fig. 2C, upper
histogram). The in vitro kinase assay was conducted using
hRpS3 and histone H1. Cdk2 was isolated from the synchronized
samples by immunoprecipitation with anti-Cdk2 antibody. histone H1,
a known substrate of Cdk2, was used as a positive control.
Autoradiography revealed phosphorylation of both histone H1 and
hRpS3 by Cdk2. Moreover, phosphorylation of hRpS3 by Cdk2 increased
in the S-phase.
Cdk1 and Cdk2 are closely related
proteins with high similarity
Thus, there might be a cross-reaction among the
antibodies used for co-IP. To exclude this possibility, we tested
the specificity of anti-Cdk1 and anti-Cdk2 antibodies by
immunoprecipitation. The first panel in Fig. 2D shows that the antibody against Cdk1
specifically recognized Cdk1 in input, but it did not detect Cdk1
or Cdk2 in the immunoprecipitation assay. The same result was
obtained for the anti-Cdk2 antibody (Fig.
2D, second panel). Thus, the result for the in vitro
Cdk1/2 kinase assay was specific for the indicated protein.
Cdk2 phosphorylates hRpS3 at specific
residues
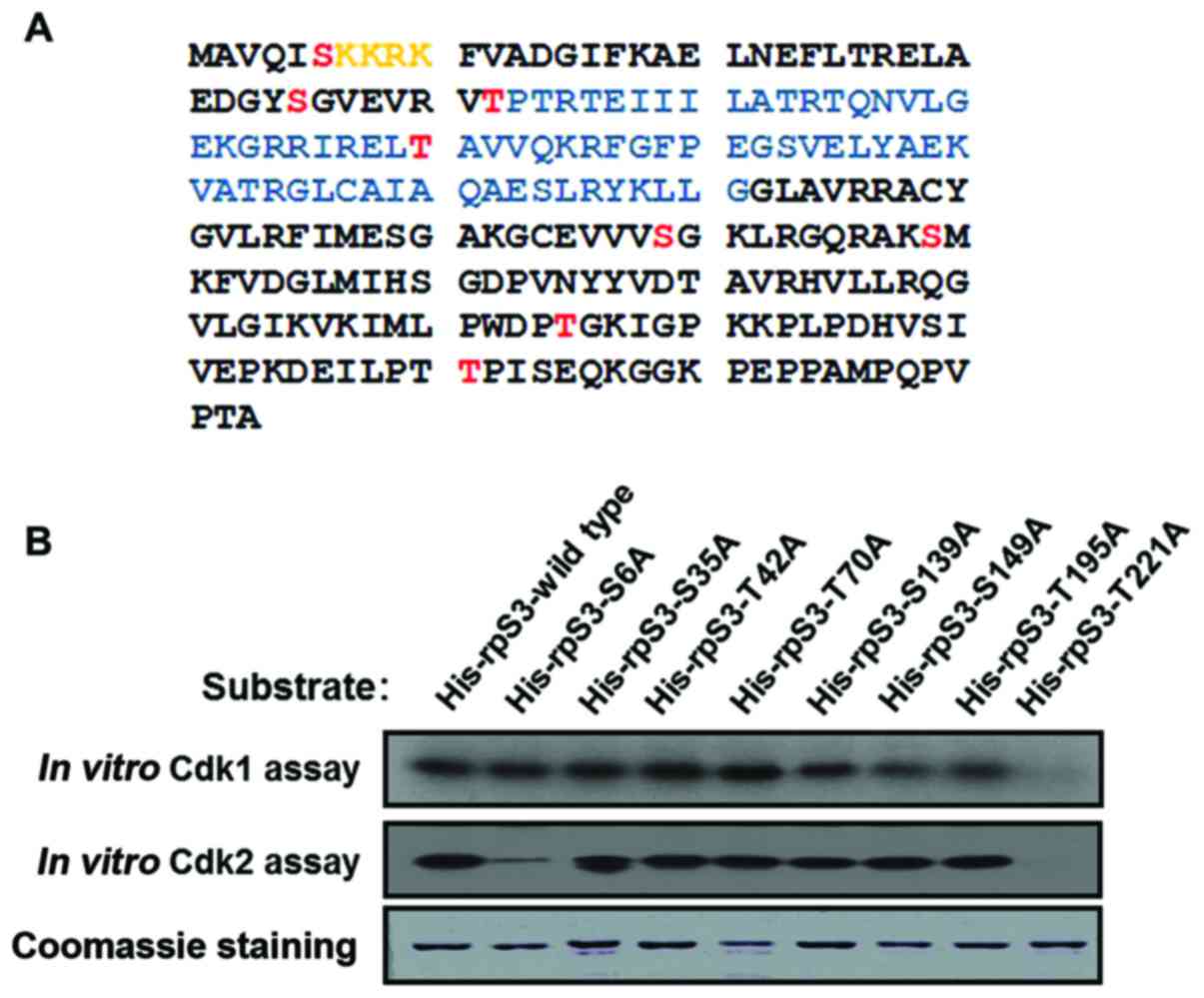
Based on the analysis of hRpS3 amino acid sequence
using bioinformatics tools (NetPhos2.0, www.cds.dtu.dk/services/NetPhos; KinasePhos,
kinasephos.mbc.nctu.edu.tw), eight
possible sites for phosphorylation by Cdk2 were identified
(Fig. 3A) (18). To observe which amino acid is
phosphorylated by Cdk2, hRpS3 constructs containing a
serine/threonine-to-alanine substitution were created and used as
the substrate in the in vitro kinase assay. As shown in
Fig. 3B, phosphorylation was
significantly lower in the hRpS3-S6A and -T221A mutants than in the
wild type. This result suggests that Cdk2 phosphorylates the S6 and
T221 residues of hRpS3. Coomassie blue staining in the lowest panel
was used as a loading control.
hRpS3 knockdown induced
phosphorylation of Cdk2 and P53
The physiological effect of interaction between Cdk2
and hRpS3 on the cell cycle was observed by knockdown of hRpS3. For
hRpS3 knockdown, HEK293 cells were transfected with varying
concentrations siRpS3. Cells transfected with siGFP were used as
control. As shown in Fig. 4A, hRpS3
knockdown was successful in all transfected cells. Whole cell
lysates were subjected to immunoblotting for the detection of
phosphor-Cdk2 (p-Cdk2) (T14, Y15; inactive form), phospho-P53
(p-P53), and P53. The expression of p-Cdk2 (T14, Y15) and p-P53
increased with increasing concentration of siRpS3. Phosphorylation
of P53 induced production of p21, an inhibitor of Cdk2 activation
(9). Therefore, increased
phosphorylation of P53 and Cdk2 (T14, Y15) is indicative of delayed
cell cycle progression. This result suggests that hRpS3 is involved
in cell cycle regulation by inducing phosphorylation of P53.
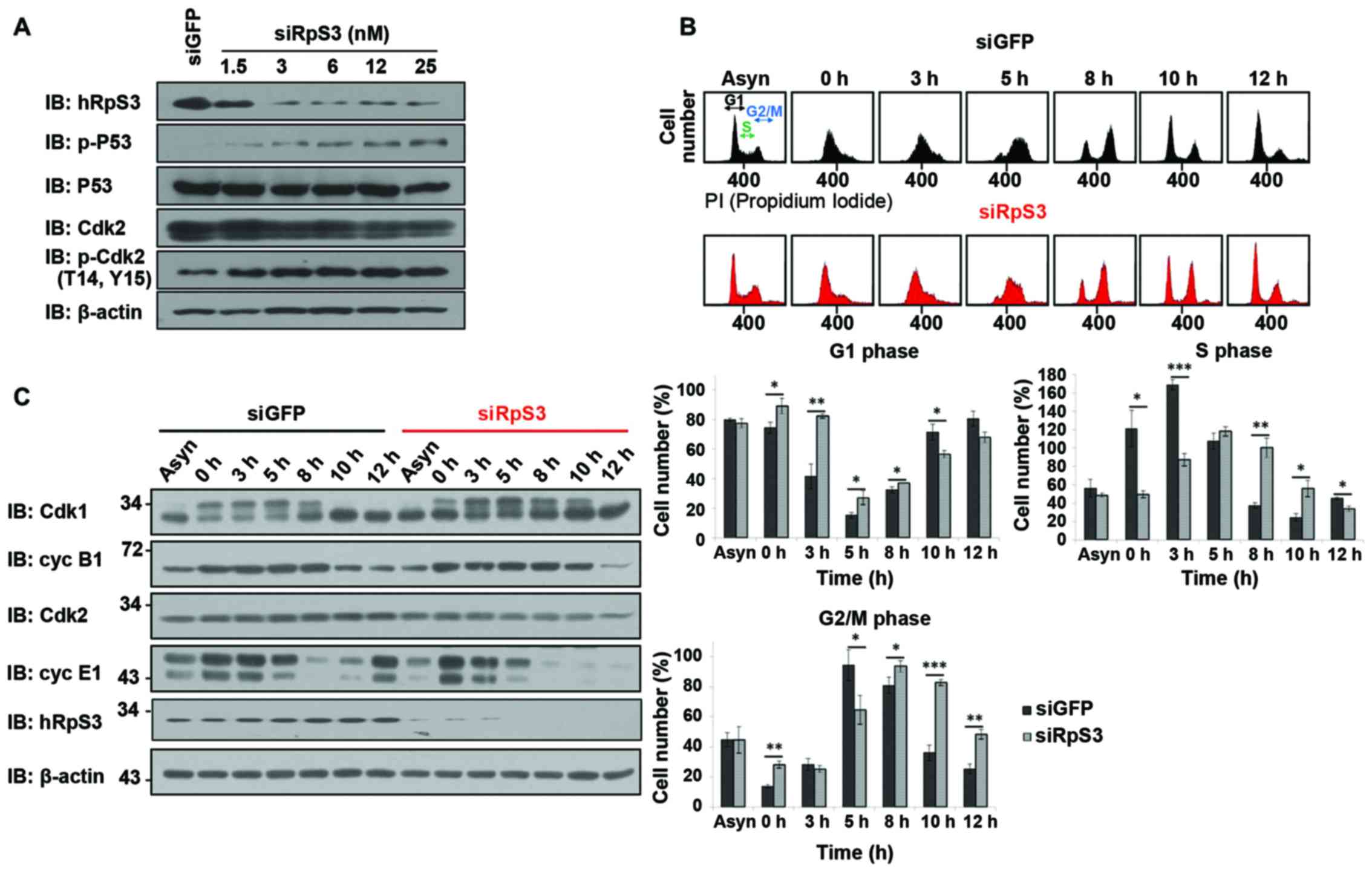 | Figure 4.Cell cycle delay induced by knockdown
of hRpS3. (A) hRpS3 knockdown induces phosphorylation of Cdk2 and
P53. Cells were either treated with siGFP as a control or with
various concentrations of siRpS3 (1.5, 3, 6, 12 and 25 nM). Whole
cell lysates were subjected to immunoblotting with antibodies
against hRpS3, Cdk2, p-Cdk2 (T14, Y15), p-P53. β-Actin was used as
loading control. (B and C) HEK293 cells were transfected with siGFP
(control) or siRpS3. Cells were arrested in the G1 phase by
incubation with aphidicoline for 15 h, released into fresh medium,
and harvested at indicated times. (B) Each harvested sample was
subjected to flow cytometry. Harvested cells were stained with
propidium iodide for FACS analysis. The percentage of siGFP- and
siRpS3-treated cells in the indicated cell cycle phase is shown as
mean ± standard error (of three independent experiments). P-value
was calculated using a paired t-test, *P<0.05, **P<0.01,
***P<0.001. (C) Lysates were immunoblotted with antibodies
against Cdk1, Cdk2, cyclin B1, cyclin E1, and hRpS3. |
Cell cycle delay was induced by
knockdown of hRpS3
The effect of hRpS3 knockdown was examined in
synchronous and asynchronous cells by FACS and immunoblotting. To
synchronize cells at the late G1 phase, they were treated with
aphidicolin (Aph). After 15 h, Aph was released into fresh medium,
and cells were harvested at different release times such that they
would synchronize and enter the S-phase in 2–4 h (22). Cells enter the G2-phase after 8 h of
releasement.
We observed a slight difference in cell cycle
progression in siRpS3-transfected cells and siGFP-transfected
control cells (Fig. 4B). In the hRpS3
knockdown cells, the S and G2/M phases were delayed compared with
that in siGFP-transfected control cells. siGFP-transfected cells
entered the S-phase at 0–3 h and the G2/M phase at 5 h after
release of Aph into fresh media. In contrast, the
siRpS3-transfected cells were still in the G1-phase at 0–3 h, and
entered the G2/M phase at 5 h after Aph release. Notably, at 5 h
after Aph release, the number of siGFP-transfected cells that
entered the G2/M phase was considerably higher than that of
siRpS3-transfected cells. Thus, the hRpS3 knockdown cells showed
delayed cell cycle progression in each phase compared with the
siGFP-transfected control cells. These results suggest that hRpS3
is required for cell cycle progression.
Further, to elucidate the mechanism through which
hRpS3 regulates cell cycle progression, we examined the expression
of cell cycle-related proteins using immunoblotting (Fig. 4C). In the siGFP-transfected cells, the
level of cyclin B1 (binds to Cdk1 and involved in regulating G2/M
transition) significantly decreased after 10 h of Aph releasement
(Fig. 4C). Additionally, in hRpS3
knockdown cells, the level of cyclin B1 significantly decreased
after release for 12 h. These results indicate that hRpS3 knockdown
induced delay of G2/M progression. The level of cyclin E1 (binds to
Cdk2 and required for progression to the S-phase) in
siGFP-transfected and siRpS3-transfected cells decreased after 8 h
of releasement. The level of cyclin E1 in siGFP-transfected cells
increased significantly after 12 h releasement, whereas it was not
detected in siRpS3-transfected cells after 8 h of release. This
expression pattern of cyclin E1 showed that hRpS3 knockdown delayed
the progression to S-phase. These findings suggest that hRpS3 is
regulates cell cycle progression modulating the expression of cell
cycle-related proteins (cyclins B and E1).
Cell cycle progression is mainly regulated by CDKs.
CDK activity increased or decreased as the cell cycle progressed.
Among the CDKs, the Cdk1/cyclin B complex is known to control G2/M
transition, while Cdk2/cyclin E1/A complexes regulate G1/S and S/G2
transitions (9–11,21,25). We
found that the levels of Cdk2/cyclin E1 and Cdk1/cyclin B1 were
decreased in hRpS3 knockdown cells. Thus, hRpS3 knockdown delayed
cell cycle progression by downregulating the proteins involved in
cell cycle regulation.
Discussion
The cell cycle is regulated by the CDK family of
proteins (26). CDKs are the
catalytic subunits of a large family of heterodimeric
serine/threonine protein kinases that regulate cell cycle
progression (27). The catalytic
activity of CDKs requires the binding of a regulatory subunit,
cyclin, which is synthesized and degraded during each cycle
(28). The catalytic activity of Cdk1
requires the binding of the cyclin B1 (27). When Cdk1/cyclin B1 activity is maximal
during G2-M phase, this complex phosphorylates a number of proteins
that regulate several cellular events (29). Cdk2 plays a role in G1/S transition
and initiation of DNA synthesis in the S-phase by forming complexes
with cyclins E and A (1,5–7).
DNA damage repair and cell cycle progression are
tightly regulated. As a ribosomal protein, hRpS3, is also known to
perform other extra-ribosomal functions, such as in DNA repair, via
its N-glycosylase activity and by releasing 8-oxoG from a DNA
lesion (30,31). Furthermore, hRpS3 binds and stimulates
the activity of uracil-DNA glycosylase (UNG), as well as the base
excision repair (BER) enzymes, hOGG1 and APE/Ref-1 (32,33).
Previous reports have shown that hRpS3 can be translocated into the
nucleus for regulation of cell cycle progression, similar to some
other ribosomal proteins such as ribosomal protein L6 and ribosomal
protein S13, which are involved in the regulation of cyclin E and
the promotion of cell growth (34,35).
Therefore, different functions of hRpS3 are believed to result from
the post-translational modifications that result from its
interaction with different molecules.
Previously, we reported that hRpS3 physically
interacts with and is phosphorylated by Cdk1, especially, in the
G2/M phase. This phosphorylation occurs at T221 and is important
for the nuclear translocation of hRpS3 (21). Here, we studied the interaction
between hRpS3 and Cdk2 (Fig. 1).
Interestingly, the interaction between hRpS3 and Cdk2 was increased
in the S-phase. Previous observations prompted us to determine
whether hRpS3 is a substrate of Cdk2. We conducted a Cdk2 kinase
assay in asynchronous and synchronous cells (arrested in the S and
G2/M phases) using hRpS3 and purified histone H1 as substrates
(Fig. 2C) and observed that hRpS3
phosphorylation by Cdk2 occurs in the S-phase. These results
revealed that hRpS3 interacts with Cdk2 and is phosphorylated at a
serine residue by Cdk2. Subsequently, based on a previous report
and the hRpS3 amino acid sequence analysis performed in this study,
eight phosphorylation sites were identified (Fig. 3A). The serine/threonine residues in
these putative phosphorylation sites were substituted by alanine
and a Cdk1/2 kinase assay was performed (Fig. 3B). S6 and T221 of hRpS3 were
phosphorylated by Cdk2, and T221 was phosphorylated by Cdk1.
Knockdown of hRpS3 induced phosphorylation of P53,
which functions upstream of Cdk1/2, resulting in a delay in cell
cycle progression and cell cycle arrest (Fig. 4A). Furthermore, we observed that
knockdown of hRpS3 induced delay in cell cycle progression and
downregulation of cyclins E1 and B1 (Fig.
4B and C). Our results show that the hRpS3 participates in cell
cycle regulation by modulating the expression of cell cycle-related
proteins.
In summary, we have shown that hRpS3 is involved in
the Cdk2-mediated regulation of cell cycle progression. Our
findings indicate a possible link between DNA damage repair and
cell cycle progression. Further studies are required to determine
other phosphorylation sites of hRpS3 that are linked to cell cycle
progression and to elucidate the relationship between the
mechanisms involved in DNA damage repair and cell cycle
control.
Acknowledgements
This study was supported by a grant of the Korea
Health Technology R&D Project through the Korea Health Industry
Development Institute (KHIDI), funded by the Ministry of Health
& Welfare, Republic of Korea (grant no. HI15C1540) and the
National Research Foundation of Korea (NRF) grant funded by the
Korean government (MSIP) (no. NRF-2015R1C1A2A01053623).
Glossary
Abbreviations
Abbreviations:
|
hRpS3
|
human ribosomal protein S3
|
|
CDK
|
cyclin-dependent kinase
|
|
Aph
|
aphidicolin
|
|
GFP
|
green fluorescent protein
|
|
p-P53
|
phospho-P53
|
|
p-Cdk2
|
phosphor-Cdk2
|
|
NLS
|
nucleotide localization signal
|
|
KH
|
K homologue domain
|
|
IP
|
immunoprecipitation
|
References
|
1
|
Gu Y, Rosenblatt J and Morgan DO: Cell
cycle regulation of CDK2 activity by phosphorylation of Thr160 and
Tyr15. EMBO J. 11:3995–4005. 1992.PubMed/NCBI
|
|
2
|
Aleem E, Kiyokawa H and Kaldis P:
Cdc2-cyclin E complexes regulate the G1/S phase transition. Nat
Cell Biol. 7:831–836. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Koff A, Giordano A, Desai D, Yamashita K,
Harper JW, Elledge S, Nishimoto T, Morgan DO, Franza BR and Roberts
JM: Formation and activation of a cyclin E-cdk2 complex during the
G1 phase of the human cell cycle. Science. 257:1689–1694. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dulić V, Lee E and Reed SI: Association of
human cyclin E with a periodic G1-S phase protein kinase. Science.
257:1958–1961. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lew DJ, Dulić V and Reed SI: Isolation of
three novel human cyclins by rescue of G1 cyclin (Cln) function in
yeast. Cell. 66:1197–1206. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Coulonval K, Bockstaele L, Paternot S and
Roger PP: Phosphorylations of cyclin-dependent kinase 2 revisited
using two-dimensional gel electrophoresis. J Biol Chem.
278:52052–52060. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wan F, Weaver A, Gao X, Bern M, Hardwidge
PR and Lenardo MJ: IKKβ phosphorylation regulates RPS3 nuclear
translocation and NF-κB function during infection with
Escherichia coli strain O157:H7. Nat Immunol. 12:335–343.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Morgan DO: Cyclin-dependent kinases:
Engines, clocks, and microprocessors. Annu Rev Cell Dev Biol.
13:261–291. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Adams PD, Sellers WR, Sharma SK, Wu AD,
Nalin CM and Kaelin WG Jr: Identification of a cyclin-cdk2
recognition motif present in substrates and p21-like
cyclin-dependent kinase inhibitors. Mol Cell Biol. 16:6623–6633.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Martin A, Odajima J, Hunt SL, Dubus P,
Ortega S, Malumbres M and Barbacid M: Cdk2 is dispensable for cell
cycle inhibition and tumor suppression mediated by p27(Kip1) and
p21(Cip1). Cancer Cell. 7:591–598. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim SH, Lee JY and Kim J: Characterization
of a wide range base-damage-endonuclease activity of mammalian
rpS3. Biochem Biophys Res Commun. 328:962–967. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hegde V, Kelley MR, Xu Y, Mian IS and
Deutsch WA: Conversion of the bifunctional 8-oxoguanine/beta-delta
apurinic/apyrimidinic DNA repair activities of Drosophila
ribosomal protein S3 into the human S3 monofunctional
beta-elimination catalyst through a single amino acid change. J
Biol Chem. 276:27591–27596. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gao X and Hardwidge PR: Ribosomal protein
S3: A multifunctional target of attaching/effacing bacterial
pathogens. Front Microbiol. 2:1372011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Warner J and McIntosh KB: How common are
extraribosomal functions of ribosomal protein? Mol Cell. 34:3–11.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zimmermann RA: The double life of
ribosomal proteins. Cell. 15:130–132. 2003. View Article : Google Scholar
|
|
16
|
Naora H: Involvement of ribosomal proteins
in regulating cell growth and apoptosis: Translational modulation
or recruitment for extraribosomal activity? Immunol Cell Biol.
77:197–205. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kim J, Chubatsu LS, Admon A, Stahl J,
Fellous R and Linn S: Implication of mammalian ribosomal protein S3
in the processing of DNA damage. J Biol Chem. 270:13620–13629.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kim TS, Kim HD and Kim J:
PKCdelta-dependent functional switch of rpS3 between translation
and DNA repair. J Biochim Biophys Acta. 1793:395–405, 1793. 2009.
View Article : Google Scholar
|
|
19
|
Kim TS, Kim HD, Shin HS and Kim J:
Phosphorylation status of nuclear ribosomal protein S3 is
reciprocally regulated by protein kinase C{delta} and protein
phosphatase 2A. J Biol Chem. 284:21201–21208. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee SB, Kwon IS, Park J, Lee KH, Ahn Y,
Lee C, Kim J, Choi SY, Cho SW and Ahn JY: Ribosomal protein S3, a
new substrate of Akt, serves as a signal mediator between neuronal
apoptosis and DNA repair. J Bio Chem. 285:29457–29468. 2010.
View Article : Google Scholar
|
|
21
|
Yoon IS, Chung JH, Hahm SH, Park MJ, Lee
YR, Ko SI, Kang LW, Kim TS, Kim J and Han YS: Ribosomal protein S3
is phosphorylated by Cdk1/cdc2 during G2/M phase. BMB Rep.
8:529–534. 2011. View Article : Google Scholar
|
|
22
|
Spandari S, Sala F and Pedrali-Noy G:
Aphidicolin: A specific inhibitor of nuclear DNA replication in
eukaryotes. Trends Biochem Sci. 7:29–32. 1982. View Article : Google Scholar
|
|
23
|
Harper JV: Synchronization of cell
population in G1/S and G2/M phases of the cell cycle. Methods Mol
Biol. 296:157–166. 2005.PubMed/NCBI
|
|
24
|
Cude K, Wang Y, Choi HJ, Hsuan SL, Zhang
H, Wang CY and Xia Z: Regulation of the G2-M cell cycle progression
by the ERK5-NFkappaB signaling pathway. J Cell Biol. 177:253–264.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Watanabe N, Broome M and Hunter T:
Regulation of the human WEE1Hu CDK tyrosine 15-kinase during the
cell cycle. EMBO J. 14:1878–1891. 1995.PubMed/NCBI
|
|
26
|
Malumbers M and Barbacid M: To cycle or
not to cycle: A critical decision in cancer. Nat Rev Cancer.
1:222–231. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Malumbers M and Barbacid M: Mammalian
cyclin-dependent kinases. Trends Biochem Sci. 30:630–641. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ecans T, Rosethal ET, Youngblom J, Distel
D and Hunt T: Cyclin: A protein specificed by maternal mRNA in sea
urchin eggs that is destroyed at each cleavage division. Cell.
33:389–396. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Salaun P, Rannou Y and Prigent C: Cdk1,
Plks, Auroras and Neks: The mitotic bodyguards. Adv Exp Med Biol.
617:41–56. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yacoub A, Augen L, Kelley MR, Doetsch PW
and Deutsch WA: A Drosophila ribosomal protein contains
8-oxoguanine and abasic site DNA repair activities. EMBO J.
15:2306–2312. 1996.PubMed/NCBI
|
|
31
|
Deutsch WA, Yacoub A, Jaruga P, Zastawny
TH and Disdaroglu M: Characterization and mechanism of action of
Drosophila ribosomal protein S3 DNA glycosylase activity for
the removal of oxidatively damage DNA bases. J Biol Chem.
272:32857–32860. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ko SI, Park JH, Park MJ, Kim J, Kang LW
and Han YS: Human ribosomal protein S3 (hRpS3) interacts with
uracil-DNA glycosylase (hUNG) and stimulates its glycosylase
activity. Mutat Res. 648:54–64. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hegde V, Wang M and Deutsch WA: Human
ribosomal protein S3 interacts with DNA base excision repair
protein hAPE/Ref-1 and hOGG1. Biochemistry. 43:14211–14217. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bernstein KA and Baserga SJ: The small
subunit processome is required for cell cycle progression at G1.
Mol Biol Cell. 15:5038–5046. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Guo X, Shi Y, Gou Y, Li J, Han S, Zhang Y,
Huo J, Ning X, Sun L, Chen Y, et al: Human ribosomal protein S13
promotes gastric cancer growth through down-regulating p27 (Kip1).
J Cell Mol Med. 15:296–306. 2011. View Article : Google Scholar : PubMed/NCBI
|