Introduction
Autophagy, as a major catabolic pathway in which the
cell degrades macromolecules and damaged organelles, was involved
in the process of cell proliferation, differentiation, survival and
homeostasis (1–2). The characteristics of autophagy were the
autophagosome with double layer membrane wrapping the long lived
proteins and organelles, was the process that during starvation,
many organisms retrieve nutrients via degradation of their own
intracellular components (3).
Autophagy is common in numerous eukaryotic cells and relative to
cell death (4). Previous studies have
shown that autophagy appeared in numerous tumors, including
colorectal cancer (CRC), esophageal carcinoma, breast cancer, lung
cancer and pancreatic cancer. However, the initiation mechanism of
autophagy in CRC was remained unclear. Yang et al (5) found that autophagy not only improves the
adaptability and promotes the proliferation of tumor cells, but
also induces cell death. At present, there are few studies
investigating the association between autophagy and the
proliferation and apoptosis of CRC. Numerous factors were active in
the generating process of autophagy, with the complicated
regulation mechanism (3,5). Beclin-1, as the first identified
mammalian autophagic gene, was an important regulator and may drive
autophagosome formation by binding to VPS34 (6). In the present study, Beclin-1 was
silenced by RNA interference (RNAi) and the mRNA and protein
expression was detected to investigate the effect of Beclin-1 on
proliferation and apoptosis in CRC cells, providing additional
evidence to support the treatment of CRC with gene therapy.
Materials and methods
Cell lines and cell culture
The human colorectal cancer HCT116 and SW620 cell
lines were obtained from the Institute of Biochemistry and Cell
Biology (Shanghai, China). HCT116 and SW620 cells were cultured
with high-glucose Dulbecco's modified Eagle's medium (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal
bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and
penicillin-streptomycin (10,000 U/ml; 1:100; Gibco; Thermo Fisher
Scientific, Inc.) in a Heracell™ VIOS 160i CO2 incubator
(Thermo Fisher Scientific, Inc.), and were maintained in a 5%
CO2 atmosphere at 37°C. The cells were replaced with new
medium every 2 or 3 days and passaged when the confluence reached
90% using 0.25% Trypsin-ENTA (Gibco; Thermo Fisher Scientific,
Inc.).
Small interfering (si)RNA
transfection
The siRNA targeting Beclin-1 was constructed by
Shanghai GenePharma Co., Ltd. (Shanghai, China). The siRNA
sequences were as follows: Beclin-1 sense,
5′-CACCGGACAACAAGTTTGACCATGCTTCAAGAGAGCATGGTCAAACTTGTTGTCCTTTTTTG-3′
and antisense,
5′-GATCCAAAAAAGGACAACAAGTTTGACCATGCTCTCTTGAAGCATGGTCAAACTTGTTGTCC-3′.
The sequences were then cloned into the plasmid of pSilencer 3.1
H1-neo (Addgene, USA) to generate the plasmid pSilencer-Beclin-1.
HCT116 cells were seeded onto 6-cm petri dishes (5×105
cells/well) and stably transfected at 80% confluence with siRNA.
All the dishes were divided into 3 groups, consisting of the
targeting siRNA (TS) and nonspecific siRNA (NS) groups and the
control group (CG), with 3 dishes for each group. The cells in the
TS group were transfected with siRNA targeting Beclin-1, while the
cells in the NS group were transfected with non-targeting siRNA,
using Lipofectamine® 3000 Transfection Reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). After 72 h, the cells
were replaced with new medium containing 400 µg/ml G418 (Gibco;
Thermo Fisher Scientific, Inc.). The surviving cells were grown and
selected with medium containing 400 µg/ml G418.
RNA extraction and reverse
transcription-polymerase chain reaction (RT-PCR)
Two weeks subsequent to transfection and selection
with G418, the total cellular RNA was extracted with
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). The extracted RNA was analyzed with ethidium bromide-stained
1% agarose gel by electrophoresis. Beclin-1 mRNA was tested by
RT-PCR using SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd.,
Dalian, China). The primer sequences were constructed by Thermo
Fisher Scientific, Inc., as follows: Beclin-1 sense,
5′-GGATGGATGTGGAGAAAGGCAAG-3′ and antisense,
5′-TGAGGACACCCAAGCAAGACC-3′; β-actin sense,
5′-ATCGTGCGTGACATTAAGGAGAAG-3′ and antisense,
5′-AGGAAGGAAGGCTGGAAGAGTG-3′. The PCR conditions are shown in
Table I, and the amplification
conditions are shown in Table II.
The fold-change in gene expression was calculated using the
2−ΔΔCq method (7).
 | Table I.Reverse transcription-PCR system
(20-µl reaction system). |
Table I.
Reverse transcription-PCR system
(20-µl reaction system).
| Composition | Volume, µl |
|---|
| Ultrapure water | 7 |
| 2X PCR Mix | 10 |
| Primer |
|
|
Forward | 1 |
|
Reverse | 1 |
| Template cDNA | 1 |
| Total volume | 20 |
 | Table II.Polymerase chain reaction
amplification conditions. |
Table II.
Polymerase chain reaction
amplification conditions.
| Step | Temperature, °C | Time | Cycle number |
|---|
| Predegeneration | 95 | 5 min |
|
| Degeneration | 95 | 30 sec | 40 |
| Annealing | 58–61 | 30 sec |
|
| Extension | 72 | 1 min |
|
| Extension
terminal | 72 | 30 min | 1 cycle |
| Storage | 4 | Indefinitely |
|
Western blot analysis
All cells were washed twice with PBS, centrifuged at
1,000 × g for 5 min, and resuspended in radioimmunoprecipitation
assay lysis and extraction buffer (Thermo Fisher Scientific, Inc.),
containing 1 mM phenylmethylsulfonyl fluoride protease inhibitor
(Thermo Fisher Scientific, Inc.) on ice for 30 min to completely
lyse. Loading buffer (5X) was added and boiled at 100°C for 10 min.
The This was then centrifuged at 12,000 × g for 10 min and the
supernatant was collected. The total protein concentration was
detected using the bicinchoninic acid method (Beyotime Institute of
Biotechnology, Haimen, China).
A total of, 10 µg protein from each sample was
separated by SDS-PAGE, with a 5% stacking gel and 10% separating
gel, and transferred to polyvinylidene fluoride membranes (Merck
KGaA, Darmstadt, Germany). The membranes were then blocked with
Tris-buffered saline containing 0.05% Tween-20 (TBST) and 5%
non-fat milk for 1 h at room temperature. Rabbit polyclonal
anti-Beclin-1 (dilution, 1:200; catalog no., sc-11427; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) and mouse monoclonal
anti-β-actin (dilution, 1:200; catalog no., sc-47778; Santa Cruz
Biotechnology, Inc.) antibodies were added to incubate at 4°C
overnight. Following washing with TBST, the membranes were
incubated with goat anti-rabbit (dilution, 1:3,000; catalog no.,
111-035-003; Jackson ImmunoResearch, Inc., West Grove, PA, USA) and
peroxidase-conjugated goat anti-mouse (dilution, 1:3,000; catalog
no., 115-035-003; Jackson ImmunoResearch, Inc.) secondary
antibodies for 1 h at room temperature. Subsequent to washing with
TBST, the membranes were developed by electrochemiluminescence
using the Enhanced Chemiluminescence system (Merck KGaA, Darmstadt,
Germany). Image J software (version 2.1; National Institutes of
Health, Bethesda, MD, USA) was used analyze the results and
calculate the expression of Beclin-1 as a ratio of Beclin-1 to
β-actin. Following this, the reduction in protein expression was
presented as the ratio of TS or NS to CG (100%).
Apoptosis analysis by Cell Counting
Kit (CCK)-8 detection
One day prior to the transfection, HCT116 and SW620
cells were seeded onto a 96-well plate, with 8×103 cells
and 100 µl medium without penicillin-streptomycin each well, in at
least 3 wells per group. CCK-8 detection was performed at 0, 24, 48
and 72 h following transfection. Prior to detection, 10 µl CCK-8
liquid was added to each well and incubated at 37°C in a 5%
CO2 atmosphere for 1 h. The optical density (OD) of each
well was detected at 450 nm using a Synergy HTX multi-mode
microplate reader, (BioTek China, Beijing, China) and the average
OD was calculated. The relative survival rate was calculated as
follows: Relative survival rate (%)=(OD of TS group)/(OD of
CG)x100.
Cell apoptosis and cell cycle analysis
by flow cytometry (FCM)
The apoptosis incidence in HCT116 and SW620 cells
was detected using fluorescein isothiocyanate (FITC) Annexin V
Apoptosis Detection kit (Pharmingen™; BD Biosciences, Franklin
Lakes, NJ, USA), according to the specification. One day prior to
transfection, the cells were seeded onto the 6-well plate, with
7.5×105 cells and 2 ml medium without
penicillin-streptomycin in each well, and at least 3 wells per
group. The cells were trypsinized using Trypsin-ENTA and
centrifuged at 3,000 × g for 5 min at 25°C, 48 h after
transfection. The cells were collected and 500 µl binding buffer
was added to resuspend the cells, and then 5 µl each of Annexin
V-FITC and propidium iodide (PI) were added. The cells were
incubated at room temperature in the dark for 15 min. The
percentage of cells with different DNA content was calculated. The
apoptotic cells were quantified and represented as a percentage of
the total, including those stained positive by Annexin V-FITC and
negative by PI.
For cell cycle analysis, the cells were trypsinized
and centrifuged at 3,000 × g for 5 min at 25°C, then washed with
PBS and fixed with 70% precooled ethanol at 4°C overnight. The
cells were washed with PBS and stained with PI (5 mg/ml) containing
100 µl RNAse A (180 µg/ml; Thermo Fisher Scientific, Inc.). The
cells were then incubated at room temperature in the dark for 30
min. PI was excited at 488 nm and fluorescence was analyzed at 630
nm using a spectrophotometer. Analytical DNA FCM (CytoFLEX; Beckman
Coulter, Inc., Brea, CA, USA) was used for cell-cycle
phase-distribution analysis.
Proliferation analysis by MTT
assay
HCT116 and SW620 cells in each group were seeded
onto a 96-well plate, with 1×104 cells per well. The
cells were treated with 10 µl MTT reagent at 24, 48 and 72 h using
the Vybrant® MTT Cell Proliferation Assay kit (Thermo
Fisher Scientific, Inc.), according to the manufacturer's
instructions. The absorbent value was measured at 450 nm with
enzyme immunoassay analyzer (Bio-Rad Laboratories, Inc., Hercules,
CA, USA) and the proliferation activity was calculated.
Statistical analysis
The data were analyzed by SPSS 13.0 (SPSS, Inc.,
Chicago, IL, USA) and presented as the mean ± standard deviation.
Three repeats were performed for each assay. One-way analysis of
variance was used to analyze the differences between groups. A
t-test was conducted for comparison between groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
Silencing on Beclin-1 expression
Using RNAi, Beclin-1 expression was partially
silenced in HCT116 and SW620 cells. The mRNA level was evaluated by
RT-PCR and the protein level was evaluated by western blot
analysis.
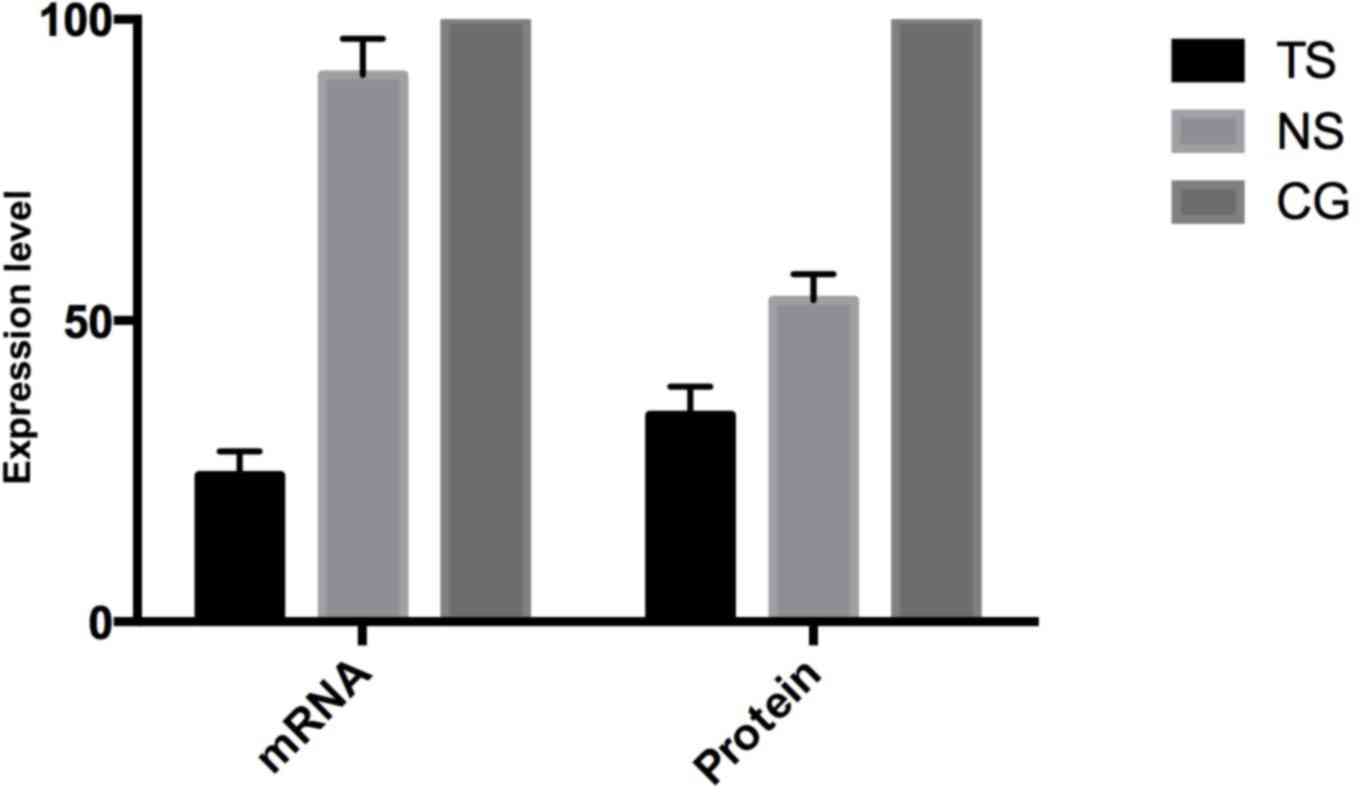
Subsequent to transfecting HCT116 cells, the
Beclin-1 mRNA in TS group was evidently decreased by 85.7%
(P<0.05) compared with the CG, while Beclin-1 expression in the
NS group was decreased by 9.2% (Fig.
1). At the protein level, the Beclin-1 expression in TS was
decreased by 65.7% compared with the CG, while the expression in
the NS group was decreased by 46.6%, consistent with the mRNA
level.
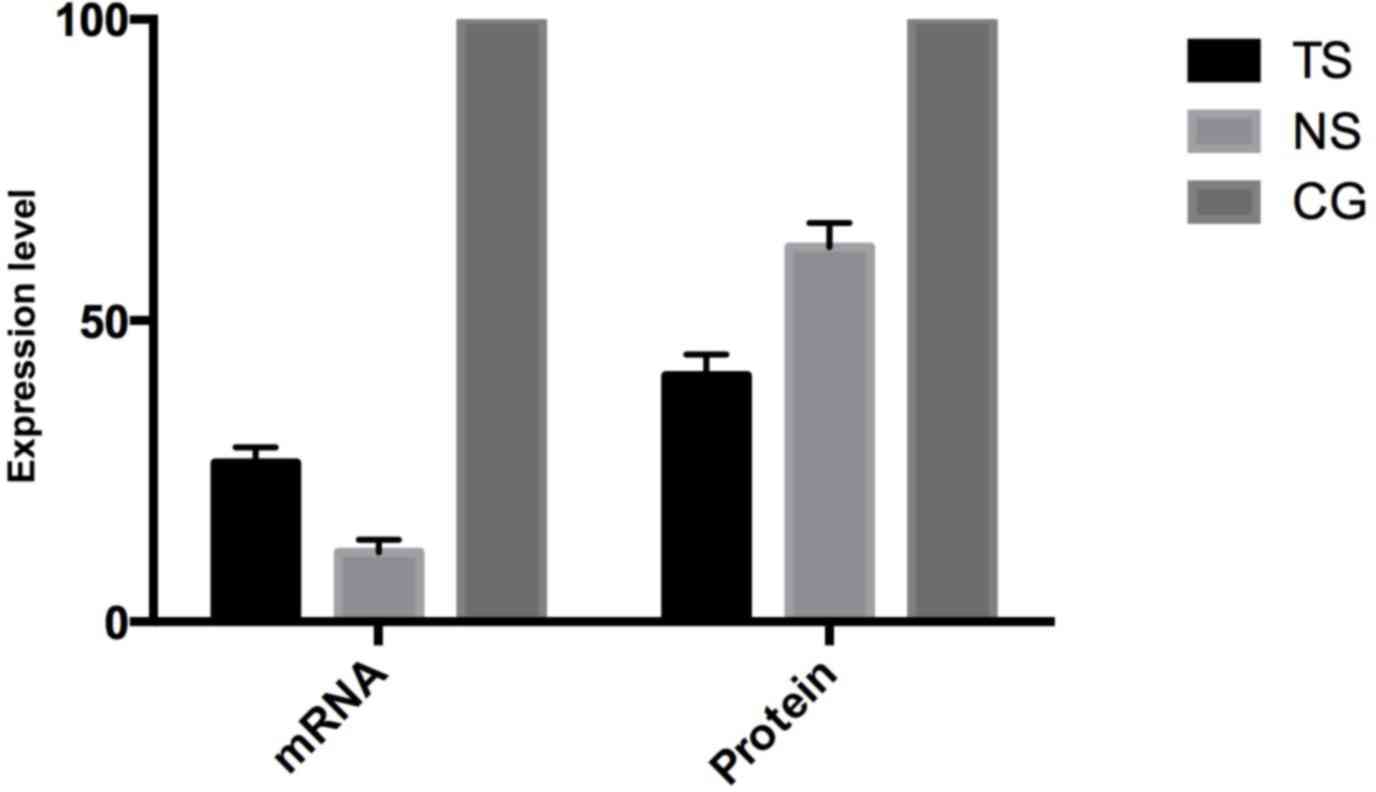
Subsequent to transfecting SW620 cells, Beclin-1
mRNA expression in the TS group was evidently decreased by 73.6%
(P<0.05) compared with the CG, while in the NS group Beclin-1
expression was decreased by 8.5% (Fig.
2). On the protein level, Beclin-1 expression in the TS group
was decreased by 59.1% compared with the CG, while in the NS group
Beclin-1 expression was decreased by 37.8%, which is consistent
with the mRNA level.
Apoptosis analysis by CCK-8
detection
HCT116 and SW620 cells were transfected and the
apoptotic percentage was detected by CCK-8 after 0, 24, 48 and 72
h.
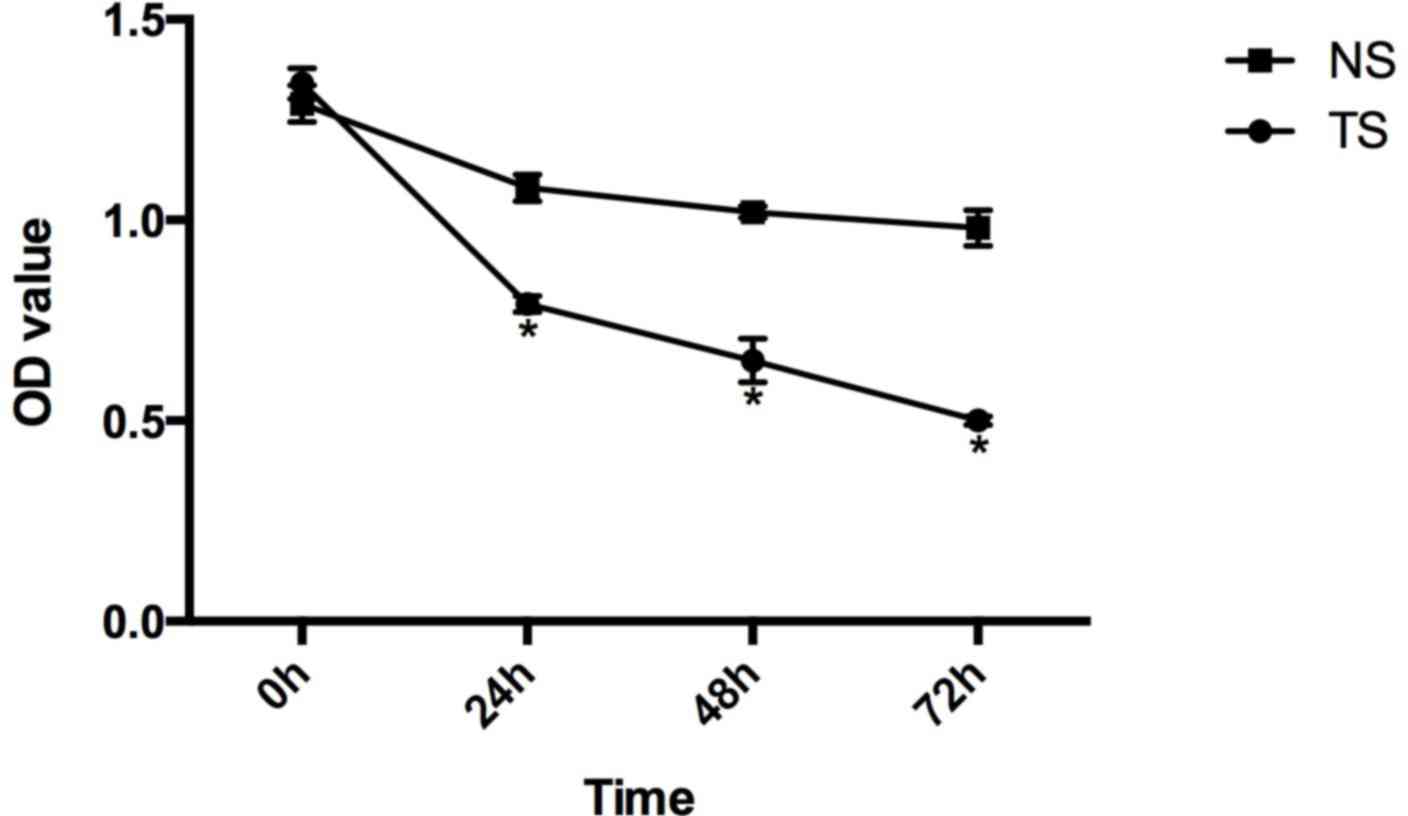
Compared with the CG of HCT116 cells, the relative
survival rates of the TS group at 0, 24, 48 and 72 h subsequent to
transfection were 1.34±0.038, 0.79±0.020, 0.65±0.054 and
0.50±0.011, respectively, and in the NS group these were
1.29±0.046, 1.08±0.033, 1.02±0.014 and 0.98±0.044 (Fig. 3).
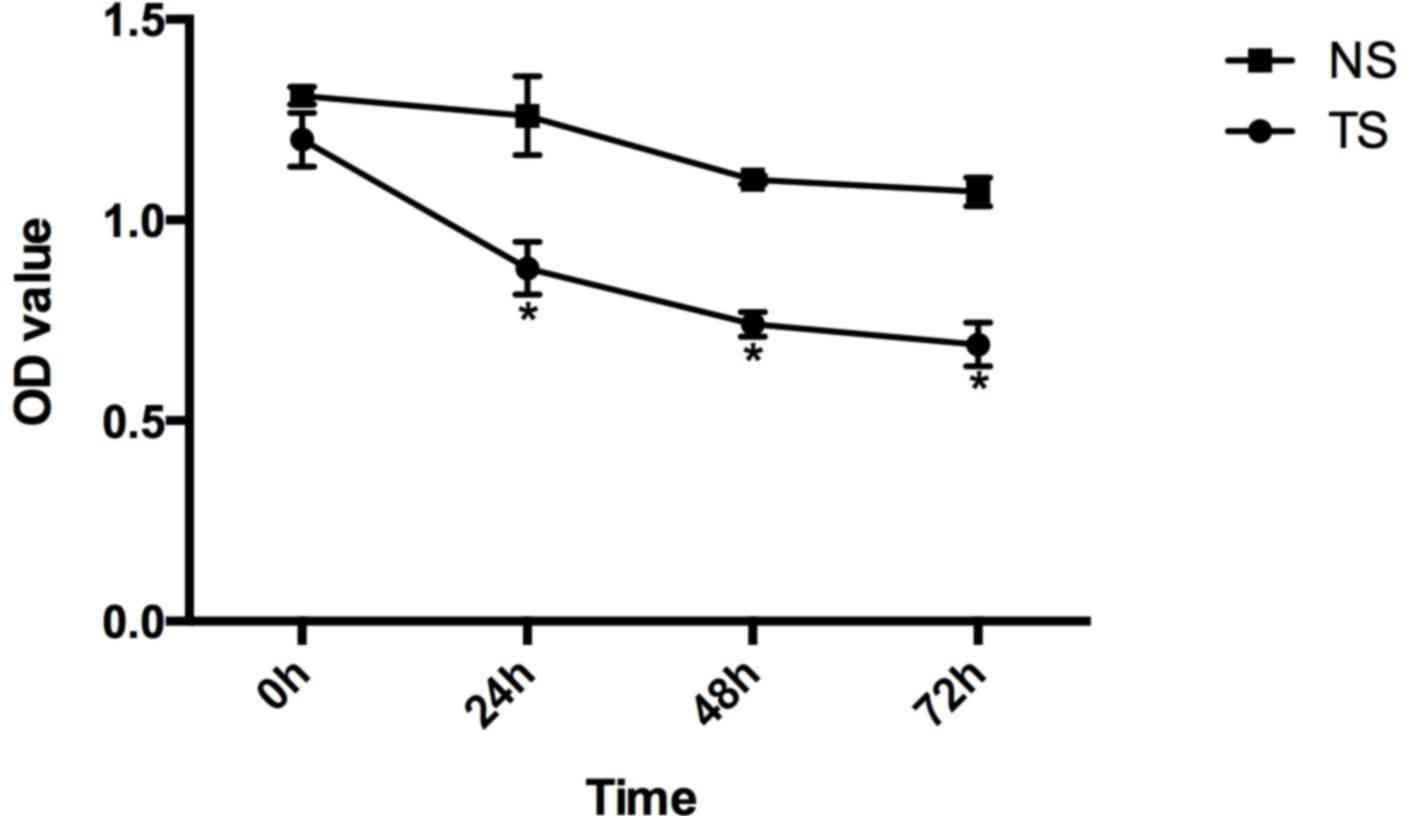
Compared with the CG of SW620 cells, the relative
survival rates of the TS group at 0, 24, 48 and 72 h subsequent to
transfection were 1.20±0.067, 0.88±0.066, 0.74±0.031 and
0.69±0.054, respectively, and in the NS group these were
1.31±0.022, 1.26±0.098, 1.10±0.011 and 1.07±0.036 (Fig. 4).
Cell apoptosis and cell cycle analysis
by FCM
By FCM, the percentages of cells in the G1 and S
phases in the TS group of HCT116 cells were 49.1 and 55.6%, and in
the CG these were 61.8 and 25.2%, respectively. However, the
percentages of cells in the G1 and S phases in the TS group of
SW620 cells were 44.5 and 59.1%, and in the CG these were 60.3 and
31.3%, respectively.
These results showed that the proportion of cells
transfected with siRNA targeting Beclin-1 in the G1 phase was
significantly decreased (P<0.05), while the proportion of cells
in the S phase was significantly increased (P<0.05). However,
there were no evident changes in cell cycle distribution and
proliferation in the NS group compared with the CG. The FCM results
were consistent with the findings of CCK-8 (data not shown).
Proliferation analysis by MTT
assay
The photo-absorption values in the HCT116 and SW620
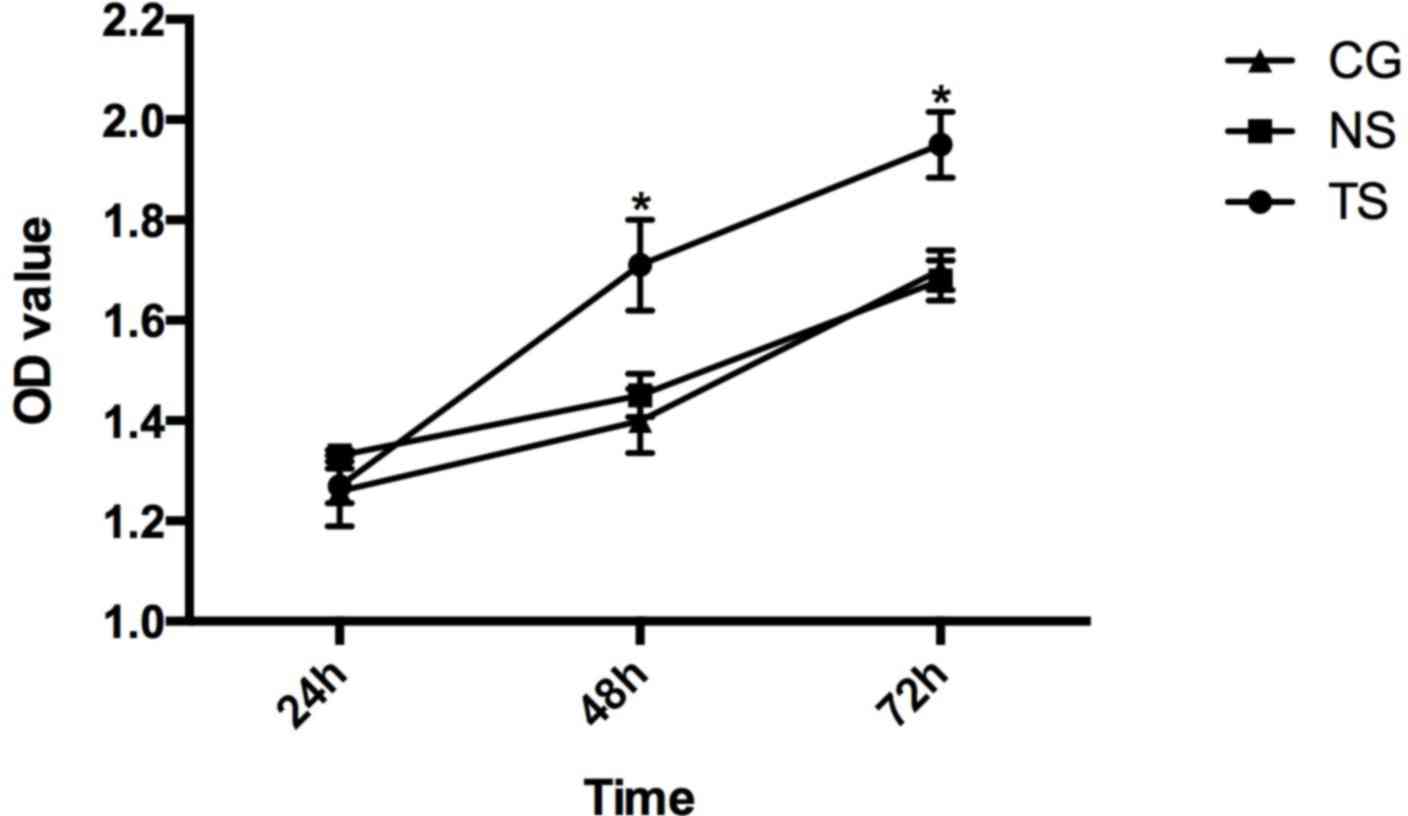
cells in each group were measured. In the TS group of HCT116 cells,
the values at 24, 48 and 72 h were 1.27±0.034, 1.71±0.090 and
1.95±0.066, respectively. In the NS group, the values were
1.33±0.012, 1.450±0.043 and 1.68±0.040. In the CG, the values were
1.26±0.071, 1.40±0.064 and 1.70±0.039 (Fig. 5).
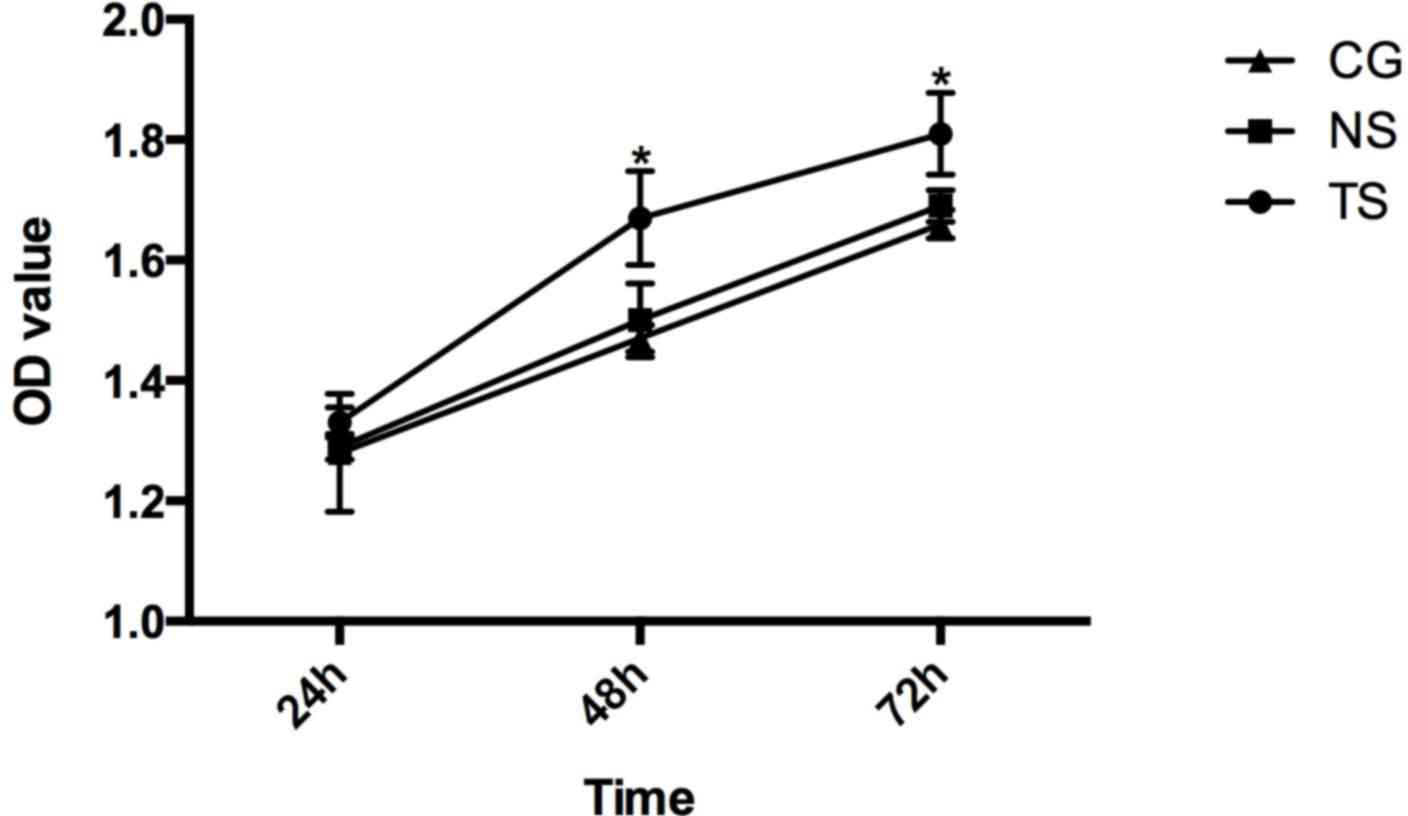
In the TS group of SW620 cells, the photo-absorption
values at 24, 48 and 72 h were 1.33±0.025, 1.67±0.078 and
1.81±0.068, respectively. In the NS group, the values were
1.29±0.021, 1.50±0.061 and 1.69±0.026. In the CG, the values were
1.28±0.098, 1.47±0.022 and 1.66±0.024 (Fig. 6).
The results of the MTT assay showed that Beclin-1
silencing promoted cell proliferation compared with the CG
(P<0.05), while there were no significant differences between
the cell proliferation in the NS group and in the CG.
Discussion
CRC is one of the most common digestive tract
cancers, with the second-highest cancer incidence in women and the
third-highest cancer incidence in men worldwide. Every year, there
are ~1.2 million patients with CRC and 600,000 of these succumb to
CRC (8). At present, the
CRC-associated incidence and mortality in China have been
increasing (9). The main therapeutic
strategies include surgical resection, radiotherapy and
chemotherapy. However, the efficacy of treatment in the early stage
of CRC is good, while it is poor in advanced CRC. Therefore, a new
therapy for treating CRC is urgently required.
Autophagy, which has been indicated as a type of
cell death different from apoptosis since the 1950s, has an
important role in the generation and development of diseases.
Autophagy is characterized by accumulation of autophagic vacuoles,
without the formation of apoptosis bodies, and chromosome
condensation, but the process is caspase-independent (10). In the generation and development of
autophagy, parts of the cytoplasm or entire organelles are
incorporated into autophagosomes, which are vesicles with a double
membrane, and these finally fuse with lysosomes, change into a
single-membrane structure and are degraded (11). Previous studies have demonstrated that
autophagic capacity in cancer cells was decreased compared with
normal cells, indicating that suppressing autophagy contributes to
cancer development (12,13).
At present, >30 associated genes, termed
autophagy-related genes, are essential for autophagic induction,
generation, maturation and recycling (14). Beclin-1 was hypothesized to be one of
the critical molecules between apoptosis and autophagy (15). Beclin-1, 60 kDa, was functional in
suppressing tumors by promoting cellular macroautophagy (16). Certain studies indicated that Beclin-1
improved the survival rate of cells, while other studies
demonstrated that Beclin-1 acted as a tumor suppressor. Liang et
al (17) found that the Beclin-1
expression decreased the proliferation of breast cancer MCF-7
cells. In lung and liver carcinomas, heterozygous disruption of
Beclin-1 in mice increased the frequency of spontaneous lymphomas
(15,18). These results indicated that Beclin-1
had the ability to suppress tumor growth, which was associated with
regulation of the death of autophagic cells. Yu et al
(19) silenced Beclin-1 expression in
L292 cells to prevent from autophagic death triggered by treatment
with a caspase inhibitor. Shimizu et al (20) showed that the downregulation of
Beclin-1 by RNAi blocked the death of non-apoptotic cells, which
was induced by doubly knocking out B cell lymphoma-2 (Bcl-2)
-associated X protein/Bcl-2 homologous antagonist/killer in
mice.
In order to investigate the function of Beclin-1 in
autophagy, cell proliferation and apoptosis, Beclin-1 expression
was silenced in HCT116 and SW620 cells using RNAi. Using CCK-8, FCM
and an MTT assay, the association between the downregulation of
Beclin-1 expression and the proliferation and apoptosis of HCT116
and SW620 cells was investigated. The results showed that siRNA
against Beclin-1 significantly decreased Beclin-1 expression, which
led to the decrease of survival rate, as determined by CCK-8, and
the increase of apoptosis rate and cell proliferation, as
determined by FCM and MTT, respectively. The present results were
consistent with the hypothesis that Beclin-1 may be an important
molecule in adjusting autophagy and apoptosis in mammalian cells.
However, the regulation mechanism of silencing Beclin-1 expression
to promote cell apoptosis and autophagy remains unclear and
requires additional investigation.
In the present study, it was found that compared to
the NS group and CG, the decreased expression level of Beclin-1 in
TS group effectively suppressed autophagy signaling, which affected
the cell proliferation and cell cycle. As shown by FCM, the
proportion of cells transfected with siRNA targeting Beclin-1 in
the G1 phase was significantly decreased (P<0.05), while the
proportion of cells in the S phase was significantly increased
(P<0.05), indicating the proliferation of cells in the TS group
was promoted.
Beclin-1 was the first identified mammalian gene
mediating autophagy (17). At
present, numerous studies find that tumor suppressors, including
Beclin-1, death-associated protein-kinase (21,22) and
phosphatase and tensin homolog (23),
have a causative role in autophagy deficiencies in cancer
formation. Therefore, autophagy draws more and more attention in
studies about the tumor generating and developing (24,25). By
inhibiting the expression of Beclin-1, the autophagy in CRC cells
was not only suppressed, but the association between autophagy and
cell proliferation and apoptosis was also indicated, which
regulates tumor growth and has an effective anti-tumor effect. The
present results provide new evidence for Beclin-1 as a target for
regulating autophagy and treating CRC. The mechanism of how
Beclin-1 is involved in autophagy and regulates cell proliferation
and apoptosis require additional investigation.
References
|
1
|
Li Z and Zhu WG: Targeting histone
deacetylases for cancer therapy: From molecular mechanisms to
clinical implications. Int J Biol Sci. 10:757–770. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ouyang L, Shi Z, Zhao S, Wang FT, Zhou TT,
Liu B and Bao JK: Programmed cell death pathways in cancer: A
review of apoptosis, autophagy and programmed necrosis. Cell
Prolif. 45:487–498. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hailey DW, Rambold AS, Satpute-Krishnan P,
Mitra K, Sougrat R, Kim PK and Lippincott-Schwartz J: Mitochondria
supply membranes for autophagosome biogenesis during starvation.
Cell. 141:656–667. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Maiuri MC, Criollo A and Kroemer G:
Crosstalk between apoptosis and autophagy within the Beclin 1
interactome. EMBO J. 29:515–516. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang Z and Klionsky DJ: Eaten alive: A
history of macroautophagy. Nat Cell Biol. 12:814–822. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kang R, Zeh HJ, Lotze MT and Tang D: The
Beclin 1 network regulates autophagy and apoptosis. Cell Death
Differ. 18:571–580. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jemal A, Center MM, DeSantis C and Ward
EM: Global patterns of cancer incidence and mortality rates and
trends. Cancer Epidemiol Biomarkers Prev. 19:1893–1907. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ren JS, Chen WQ, Shin HR, Ferlay J, Saika
K, Zhang SW and Bray F: A comparison of two methods to estimate the
cancer incidence and mortality burden in China in 2005. Asian Pac J
Cancer Prev. 11:1587–1594. 2010.PubMed/NCBI
|
|
10
|
Levine B and Kroemer G: Autophagy in the
pathogenesis of disease. Cell. 132:27–42. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yoshimori T: Autophagy: A regulated bulk
degradation process inside cells. Biochem Biophys Res Commun.
313:453–458. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gozuacik D and Kimchi A: Autophagy as a
cell death and tumor suppressor mechanism. Oncogene. 23:2891–2906.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Thompson CB: Apoptosis in the pathogenesis
and treatment of disease. Science. 267:1456–1462. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Behrends C, Sowa ME, Gygi SP and Harper
JW: Network organization of the human autophagy system. Nature.
466:68–76. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yue Z, Jin S, Yang C, Levine AJ and Heintz
N: Beclin 1, an autophagy gene essential for early embryonic
development, is a haploinsufficient tumor suppressor. Proc Natl
Acad Sci USA. 100:15077–15082. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zeng X, Overmeyer JH and Maltese WA:
Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase
complex in macroautophagy versus endocytosis and lysosomal enzyme
trafficking. J Cell Sci. 119:259–270. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liang XH, Jackson S, Seaman M, Brown K,
Kempkes B, Hibshoosh H and Levine B: Induction of autophagy and
inhibition of tumorigenesis by beclin 1. Nature. 402:672–676. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh
H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, et al:
Promotion of tumorigenesis by heterozygous disruption of the beclin
1 autophagy gene. J Clin Invest. 112:1809–1820. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yu L, Alva A, Su H, Dutt P, Freundt E,
Welsh S, Baehrecke EH and Lenardo MJ: Regulation of an ATG7-beclin
1 program of autophagic cell death by caspase-8. Science.
304:1500–1502. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shimizu S, Kanaseki T, Mizushima N, Mizuta
T, Arakawa-Kobayashi S, Thompson CB and Tsujimoto Y: Role of Bcl-2
family proteins in a non-apoptotic programmed cell death dependent
on autophagy genes. Nat Cell Biol. 6:1221–1228. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zalckvar E, Berissi H, Mizrachy L,
Idelchuk Y, Koren I, Eisenstein M, Sabanay H, Pinkas-Kramarski R
and Kimchi A: DAP-kinase-mediated phosphorylation on the BH3 domain
of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and
induction of autophagy. EMBO Rep. 10:285–292. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zalckvar E, Berissi H, Eisenstein M and
Kimchi A: Phosphorylation of Beclin 1 by DAP-kinase promotes
autophagy by weakening its interactions with Bcl-2 and Bcl-XL.
Autophagy. 5:720–722. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ying H, Qu D, Liu C, Ying T, Lv J, Jin S
and Xu H: Chemoresistance is associated with Beclin-1 and PTEN
expression in epithelial ovarian cancers. Oncol Lett. 9:1759–1763.
2015.PubMed/NCBI
|
|
24
|
Won KY, Kim GY, Lim SJ, Sung JY, Kim YW,
Park YK, Lee J and Choi HS: Autophagy is related to the hedgehog
signaling pathway in human gastric adenocarcinoma: Prognostic
significance of Beclin-1 and Gli2 expression in human gastric
adenocarcinoma. Pathol Res Pract. 211:308–315. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Galadari S, Rahman A, Pallichankandy S and
Thayyullathil F: Tumor suppressive functions of ceramide: Evidence
and mechanisms. Apoptosis. 20:689–711. 2015. View Article : Google Scholar : PubMed/NCBI
|