Introduction
A translocation event that occurs between
chromosomes 9 and 22, t(9;22)(q34;q11), results in the Philadelphia
(Ph) chromosome, and consequently the BCR-ABL fusion gene, which is
transcribed and translated into a hybrid protein (1). This fusion gene comprises almost the
entire coding region of the Abelson (ABL) gene on chromosome 9, and
the changeable coding region of the breakpoint cluster region (BCR)
gene on chromosome 22 (2). In chronic
myelogenous leukemia (CML), the breakpoint region lies between
exons 12 and 16, and is termed the major breakpoint cluster region
(M-bcr). The translocation events involving the M-bcr transcripts
e13a2 (b2a2) and e14a2 (b3a2) encode a 210 kDa protein termed P210.
Translocations that occur within the minor(m)-bcr region retain
only the first exon of the BCR gene transcripts, e1a2, which yields
a smaller P190 gene product. A rare P230 fusion protein, with a
micro (µ)-bcr breakpoint between exons 19 and 20, has also been
described in a number of variant cases (3). In addition, some rare breakpoints have
also been sporadically reported, which may be grouped into 4
categories: The BCR breakpoints originating within introns that lie
outside the M-bcr, m-bcr, or µ-bcr fused to ABL a2; the BCR
breakpoints occurring within exons fused to ABL a2; the typical BCR
breakpoints M-bcr, m-bcr, or µ-bcr fused to ABL breakpoints located
downstream of a2; and the transcripts containing intervening
sequences between BCR and ABL a2 (4).
The National Comprehensive Cancer Network (NCCN)
practice guidelines recommend that cytogenetics, fluorescence in
situ hybridization (FISH) and quantitative polymerase chain
reaction (qPCR) are used for the diagnosis of the chronic phase of
adult CML (5). Cytogenetics and FISH
detect the translocation event, yet cannot determinate the
breakpoints on the chromosome, whereas qPCR is able to detect these
breakpoints. However, special primers or probes are required when
qPCR is used to detect these breakpoints on the chromosome.
Numerous qPCR probes and primers have been designed to detect the
common BCR-ABL transcript, but false negative results may present
in patients with atypical transcripts.
The present study reported the data from a CML
patient whose cytogenetics and FISH analyses of bone marrow
revealed a karyotype of 46, XY, t(9,22)(q34;q11); however, qPCR
failed to detect the BCR-ABL fusion gene. The PCR products by
multiplex RT-PCR assay identified a band of ~300 bp, which were
subsequently confirmed as the BCR-ABL e14a3 transcript through
sequence analysis. In addition, the biological role of the rare
fusion event was reviewed, and the ability of qPCR assays to detect
this rare transcript was discussed in the present study.
Case report
A 40 year-old male, whose white blood cell (WBC)
count had been elevated for several days, was referred to The
Second Hospital of Anhui Medical University, Hefei, China, in
November 2015 for evaluation of his markedly high leukocyte level
of 46.42×109/l. No fever, hidrosis and musculoskeletal
pain were found in the course of his physical examination. The
peripheral blood test revealed hemoglobin (HB) levels of 132 g/l, a
platelet count (PLT) of 275×109/l (67.6% neutrophils,
21.3% lymphocytes and 9.2% basophil cells). The bone marrow
examination revealed marked hyper-cellularity, with an increased
number of megakaryocytes and evidence of myeloid hyperplasia and
basophilia. The chromosomal analysis presented a karyotype of 46,
XY, t(9;22)(q34;q11). The FISH analysis produced a positive result
for the BCR-ABL translocation. Based on these findings the patient
was diagnosed with CML, and was treated with 400 mg/day imatinib.
The patient appeared to respond well to this treatment, as
demonstrated in Fig. 1.
Materials and methods
Cytogenetic analysis was performed on bone marrow
aspirates according to the standard laboratory protocol. A total of
twenty G-banded metaphases and two karyotypes were evaluated. LSI
BCR/ABL dual color, dual fusion translocation probe set (Abbott
Molecular Inc., Des Plaines, IL, USA) was used to performed FISH
analysis, following the protocol of the manufacturer. Total RNA was
extracted from EDTA-anticoagulated bone marrow samples using an
UNlQ-10 Column RNA Isolation kit (Sangon Biotech Co., Ltd.,
Shanghai, China), following the protocol of the manufacturer. A
multiplex RT-PCR was carried out as described previously (6). A qPCR assay was performed to detect the
BCR-ABL fusion gene, using a quantitative fluorescent RNA detection
kit (Shanghai Shenyou Bio Technology Co., Ltd., Shanghai, China).
The human leukemia sup-b15 (e1a2), nalm-6 (BCR-ABL negative) cell
lines, were purchased from JENNIO Biological Technology (Guangzhou,
China), and BV173 (e13a2) cell line, was provided by Dr Mengqing
Gao (Shanghai Jiao Tong University School of Medicine, Shanghai,
China), were used as controls. Multiplex RT-PCR products were
sequenced by Sangon Biotech Co., Ltd. The sequence of Multiplex
RT-PCR product was clarified by BLASTN (National Center for
Biotechnology Information; blast.ncbi.nlm.nih.gov/Blast.cgi). All the data of
patient were collected according to the approval of the
institutional review board and informed consent was obtained.
Results
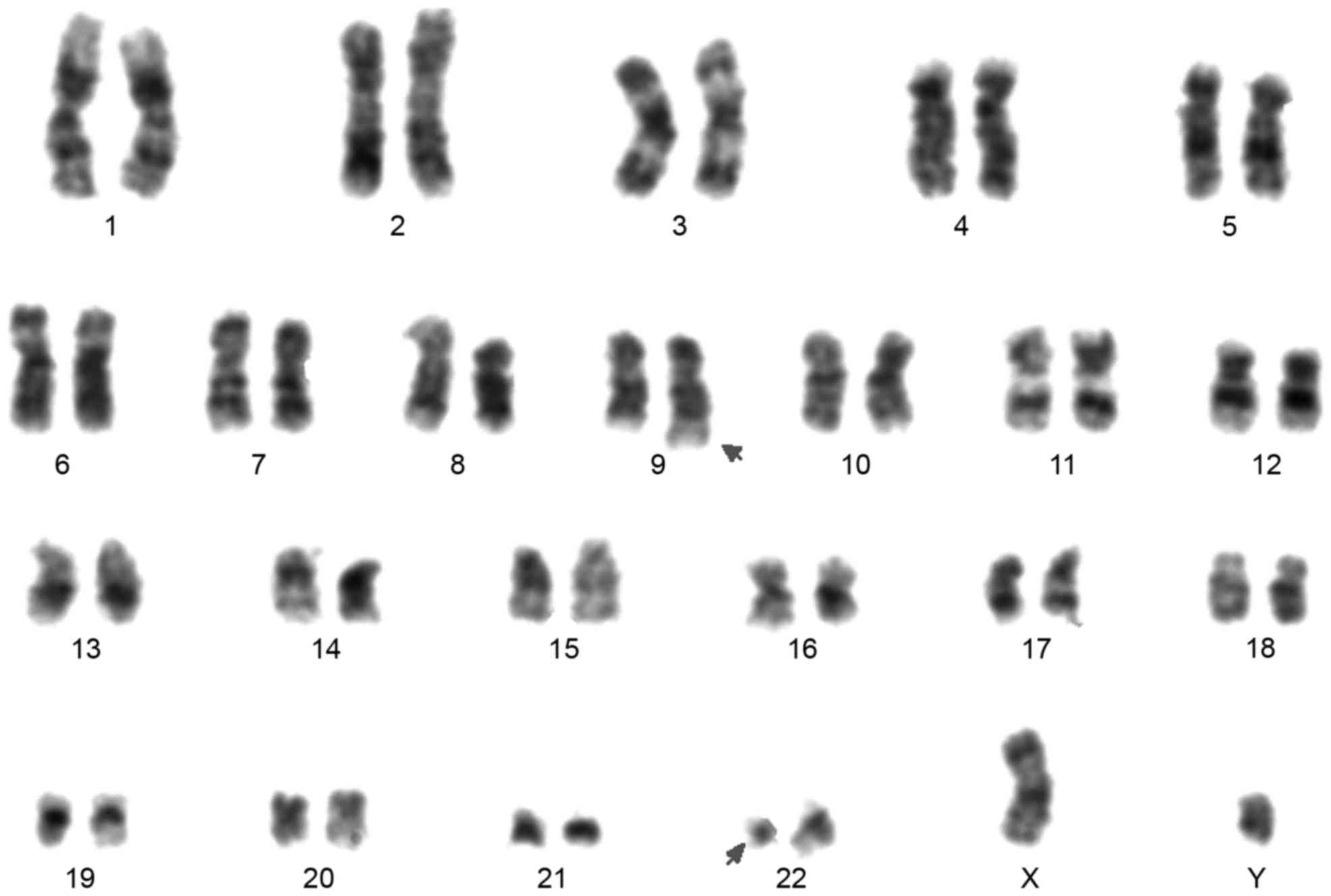
The chromosomal analysis of the patient presented a
karyotype of 46, XY, t(9;22)(q34;q11) in all 20 metaphases
examined, as demonstrated in Fig. 2.
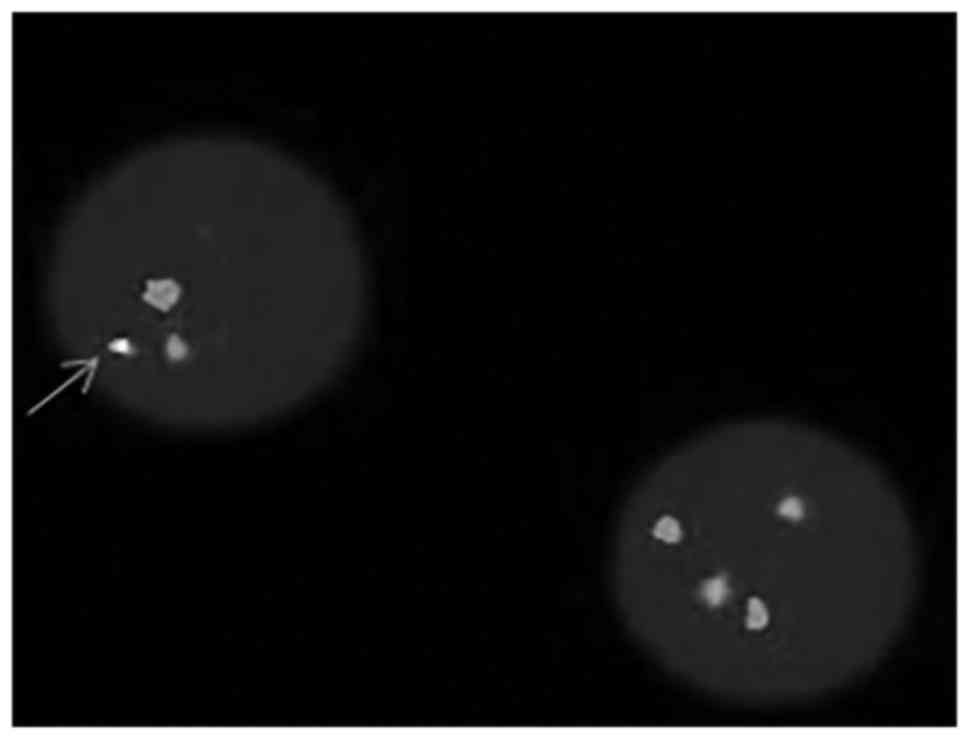
FISH analysis detected the BCR-ABL translocation, as displayed in
Fig. 3. Commercial kits of qPCR
failed to detect the BCR-ABL fusion gene. The multiplex RT-PCR
performed on the RNA did not detect the classic BCR-ABL P210
transcript in the cells obtained from the bone marrow of the
patient, or the sup-b15 or nalm-6 cell lines, but was detected in
the BV173 cell line with the e13a2 transcript. However, the
multiplex RT-PCR product of the patient demonstrated a band
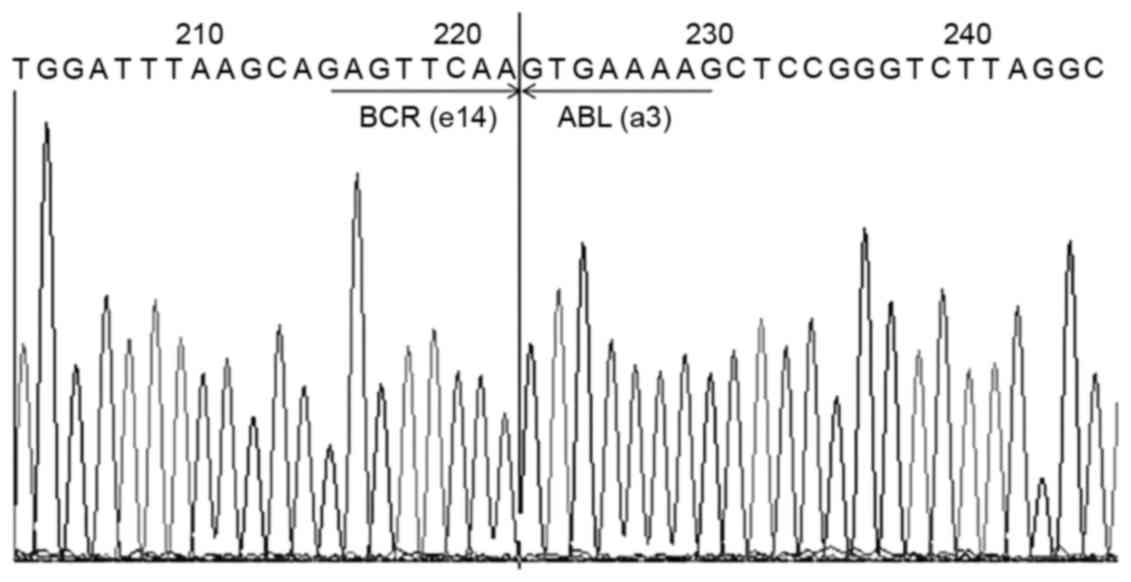
measuring ~300 bp in 2% agarose gel, as demonstrated in Fig. 4. The multiplex RT-PCR product was
sequenced by Sangon Biotech Co., Ltd., and confirmed the presence
of the e14a3 BCR-ABL transcript (b3a3) by BLASTN analysis, which is
showed in Fig. 5.
Discussion
In patients with CML with the Ph chromosomal
abnormality, almost all fusion transcripts are e13a2 and e14a2
(M-bcr), which produce a 210 kDa protein when transcribed (7). In rare variant patients, e1a2 (m-bcr)
and e19a2 (µ-bcr) transcripts are found. The breakpoint of these
transcripts usually occurs in ABL exon2, but occasionally arises in
ABL exon3. The protein transcribed by exon2 contains tyrosine
kinase (SH1), SH2 and SH3 domains. In comparison with the proteins
transcripted by exon2, the structure of the protein transcribed by
exon3 lacked two-thirds of the N-terminal in the SH3 domain. A
previous study determined that in vivo, SH3 domains
negatively regulate the kinase domain (SH1) and activate the Signal
transducer and activator of transcription 5-signaling pathway,
which is an essential leukemogenic factor (8). Furthermore, mouse models have
demonstrated lower tissue invasiveness and longer leukemic cell
cycles when BCR-ABL mutants lack the SH3 domain (9). Consequently, it is expected that
patients who exhibit BCR-ABL without exon2 may present with a
different clinical course.
To our knowledge, there have been 14 reports,
including the patient of the present study, of patients with the
BCR-ABL e14a3 transcripts (5,10–15)
(Table I). The median age of these
patients is 44.5 years old: Patients with BCR-ABL e14a3 transcripts
appear to be younger than patients with the common transcript at
the time of diagnosis. Notably, 12 patients were male, suggesting
that the BCR-ABL e14a3 transcript is more common in males. In
addition, these 14 patients exhibited a less aggressive form of the
disease and lower WBC counts, ranging from 9–300×109/l.
It was also noted that the deletion of exon2 did not affect the
curative effects of imatinib, and 7 patients appeared to respond
well to treatment with this drug (5,10). The
patient in the present study also appeared to have a good response
to imatinib. Combining these data with previous studies (5,10,16), patients with the BCR-ABL e14a3
transcript may expect a good clinical course.
 | Table I.Summary of CML patients and the
respective BCR-ABL e14a3 transcripts. |
Table I.
Summary of CML patients and the
respective BCR-ABL e14a3 transcripts.
| Case no. | Age/gender | Phase of CML | WBC count
(×109/l) | Chromosome
karyotype | BCR-ABL
transcript | Clinical outcome | Duration of follow-up
(months) | Splenomegaly | References |
|---|
| 1 | 39/F | CP | 9 | T (9,22)13Q | E14A3 | Alive | Unknown | NA | (10) |
| 2 | 19/M | CP | 42 | T (9,22) | E14A3 | Dead | 34 | NA | (11) |
| 3 | 23/M | CP | 95.8 | T (9,22) | E14A3 | Alive | 60 | NO | (12) |
| 4 | 51/M | CP | NA | T (9,22) | E14A3 | Alive | 36 | NO | (16) |
| 5 | 69/M | CP | 18 | T (4,9,22) | E14A3 | Alive | 126 | NO | (16) |
| 6 | 69/M | CP | 29 | T (9,22) | E14A3 | Alive | Unknown | NO | (13) |
| 7 | 81/M | CP | 28 | T (9,22) | E14A3 | Alive | Unknown | NA | (5) |
| 8 | 30/M | CP | NA | T (9,22) | E14A3 | Alive | Unknown | NO | (14) |
| 9 | 52/M | CP | 229 | T (9,22) | E14A3 | Alive | 112 | YES | (9) |
| 10 | 41/M | AP | 26 | T (9,22) | E14A3 | Alive | 66 | YES | (9) |
| 11 | 41/M | CP | 115 | T (9,22) | E14A3 | Alive | 101 | YES | (9) |
| 12 | 48/F | CP | 300 | T (9,22) | E14A3 | Dead | 53 | YES | (9) |
| 13 | 48/M | CP | 98.2 | T (9,22) | E14A3 | Alive | 30 | YES | (9) |
| 14 | 40/M | CP | 46 | T (9,22) | E14A3 | Alive | 2 | NO | Present study |
The present study observed that the multiplex RT-PCR
produced negative results for e13a2 and e14a2, but exhibited a band
measuring ~300 bp in the agarose gel. qPCR analysis to detect the
BCR-ABL fusion gene was then performed, but this failed to detect
the BCR-ABL fusion gene. However, the Ph chromosome was detected by
cytogenetics and FISH analyses. To additionally investigate the
discordant result, the multiplex RT-PCR products were sequenced and
analyzed by BLASTN, which confirmed the atypical transcript e14a3
in the patient of the present study. Although qPCR failed to detect
this transcript, the multiplex RT-PCR assay previously described
succeeded in detecting the BCR-ABL e14a3 transcript. Based on these
findings, the present study suggested that multiplex RT-PCR assay
reported by Pallisgaard et al (6) may provide evidence that PCR products
amplified by R6B group presents a band with ~300 bp, and associated
with BCR-ABL e14a3 transcript.
The NCCN practice guidelines recommend that
cytogenetics, FISH and qPCR be performed to diagnose the chronic
phase of adult CML. In the present study, cytogenetics and FISH
were successful in detecting the BCR-ABL e14a3 transcript in the
patient. However, the qPCR failed, due to a lack of specific
primers, which makes repeating the qPCR every 3–6 months, as
recommend by the NCCN, useless. Presently, some commercial qPCR
kits are able to detect the e14a3 and e13a3 transcripts using the
reverse primer located in exon 4 of ABL, such as Roche Holding AG,
Basel, Switzerland and Applied Biosystems, Thermo Fisher Scientific
Co., Waltham, MA, USA (5), while
special primers that are able to detect the e14a3 transcript have
also been designed (17). However,
these assays fail to distinguish the type of BCR-ABL transcript
present. Different BCR-ABL transcripts result in different clinical
courses (18), and the impact of the
type of transcript on response and survival rate subsequent to
initial treatment with different tyrosine kinase inhibitors
demonstrates different outcomes (19). True categorization of the BCR-ABL
transcript by breakpoint is required in order to investigate and
comprehend the different types of CML, and thus provides accurate
guides for the treatment and prognosis of the disease. In
conclusion, a qPCR assay using commercial kits or primers designed
by previous studies, which would detect the exon a3 transcripts, in
order to estimate the disease progression of CML in a patient with
the e14a3 transcript. Furthermore, in the future, multiplex RT-PCR
assays may be applied to distinguish the type of BCR-ABL transcript
present, with different transcripts demonstrating different sizes.
Also, it is necessary to explore the possibility of a new qPCR
assay that detects and distinguishes different BCR-ABL transcripts
e13a2, e14a2, e13a3 and e14a3 respectively.
References
|
1
|
Rowley JD: Letter: A new consistent
chromosomal abnormality in chronic myelogenous leukaemia identified
by quinacrine fluorescence and Giemsa staining. Nature.
243:290–293. 1973. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
de Klein A, van Kessel AG, Grosveld G,
Bartram CR, Hagemeijer A, Bootsma D, Spurr NK, Heisterkamp N,
Groffen J and Stephenson JR: A cellular oncogene is translocated to
the Philadelphia chromosome in chronic myelocytic leukaemia.
Nature. 300:765–767. 1982. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bernt KM and Hunger SP: Current concepts
in pediatric Philadelphia chromosome-positive acute lymphoblastic
leukemia. Front Oncol. 4:542014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Barnes DJ and Melo JV: Cytogenetic and
molecular genetic aspects of chronic myeloid leukaemia. Acta
Haematol. 108:180–202. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jinawath N, Norris-Kirby A, Smith BD,
Gocke CD, Batista DA, Griffin CA and Murphy KM: A rare e14a3 (b3a3)
BCR-ABL fusion transcript in chronic myeloid leukemia: Diagnostic
challenges in clinical laboratory practice. J Mol Diagn.
11:359–363. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pallisgaard N, Hokland P, Riishøj DC,
Pedersen B and Jørgensen P: Multiplex reverse
transcription-polymerase chain reaction for simultaneous screening
of 29 translocations and chromosomal aberrations in acute leukemia.
Blood. 92:574–588. 1998.PubMed/NCBI
|
|
7
|
Bernards A, Rubin CM, Westbrook CA,
Paskind M and Baltimore D: The first intron in the human c-abl gene
is at least 200 kilobases long and is a target for translocations
in chronic myelogenous leukemia. Mol Cell Biol. 7:3231–3236. 1987.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nieborowska-Skorska M, Wasik MA, Slupianek
A, Salomoni P, Kitamura T, Calabretta B and Skorski T: Signal
transducer and activator of transcription (STAT)5 activation by
BCR/ABL is dependent on intact Src homology (SH)3 and SH2 domains
of BCR/ABL and is required for leukemogenesis. J Exp Med.
189:1229–1242. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Skorski T, Nieborowska-Skorska M,
Wlodarski P, Wasik M, Trotta R, Kanakaraj P, Salomoni P, Antonyak
M, Martinez R, Majewski M, et al: The SH3 domain contributes to
BCR/ABL-dependent leukemogenesis in vivo: Role in adhesion,
invasion, and homing. Blood. 91:406–418. 1998.PubMed/NCBI
|
|
10
|
Iwata S, Mizutani S, Nakazawa S and Yata
J: Heterogeneity of the breakpoint in the ABL gene in cases with
BCR/ABL transcript lacking ABL exon a2. Leukemia. 8:1696–1702.
1994.PubMed/NCBI
|
|
11
|
Polák J, Zemanová Z, Michalová K, Klamová
H, Cermák J and Haskovec C: A new case of chronic myeloid leukemia
(CML) in myeloid blast crisis with an atypical (b3/a3) junction of
the BCR/ABL gene. Leukemia. 12:2501998. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Amabile M, Martinelli G, Terragna C,
Montefusco V, Tabilio A and Tura S: An atypical (b3/a3) junction of
the bcr/abl gene lacking abl exon a2 in a patient with chronic
myeloid leukemia. Haematologica. 84:573–575. 1999.PubMed/NCBI
|
|
13
|
Paz-Y-Miño C, Arévalo M and Leone PE:
B3/A3 rearrangement in a patient with chronic myeloid leukemia.
Leuk Lymphoma. 44:375–376. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Vaniawala S, Acharya A, Parekh H and
Mukhopadhyaya PN: Rare e14a3 (b3a3) BCR-ABL fusion in chronic
myeloid leukemia in India: The threats and challenges in monitoring
minimal residual disease (MRD). Anal Cell Pathol (Amst). 36:85–92.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xiaomin G, Zhang Y, Pan J, Qiu H, Cen J,
Xue Y, Chen S, Shen H, Yao L, Zhang J, et al: Chronic myeloid
leukemia with e14a3 BCR-ABL transcript: Analysis of characteristics
and prognostic significance. Leuk Lymphoma. 56:3343–3347. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tiribelli M, Tonso A, Ferrro D, Parzilae
A, Cambrin GR, Scaravaglio P, Cilloni D, Gottardi E and Saglio G:
Lack of SH3 domain does not imply a more severe clinical course in
Ph+ chronic myeloid leukemia patients. Blood. 95:4019–4020.
2000.PubMed/NCBI
|
|
17
|
Kreuzer KA, Lass U, Bohn A, Landt O and
Schmidt CA: LightCycler technology for the quantitation of bcr/abl
fusion transcripts. Cancer Res. 59:3171–3144. 1999.PubMed/NCBI
|
|
18
|
Hanfstein B, Lauseker M, Hehlmann R,
Saussele S, Erben P, Dietz C, Fabarius A, Proetel U, Schnittger S,
Haferlach C, et al: Distinct characteristics of e13a2 versus e14a2
BCR-ABL1 driven chronic myeloid leukemia under first-line therapy
with imatinib. Haematologica. 99:1441–1457. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jain P, Kantarjian H, Patel KP, Gonzalez
GN, Luthra R, Shamanna Kanagal R, Sasaki K, Jabbour E, Romo CG,
Kadia TM, et al: Impact of BCR-ABL transcript type on response and
survival in patients with chronic-phase chronic myeloid leukemia
treated with tyrosine kinase inhibitors. Blood. 127:1269–1275.
2016. View Article : Google Scholar : PubMed/NCBI
|