Introduction
The chromosomal abnormality t(6;9)(p23;q34)
presentsin~0.5–4% of patients with acute myeloid leukemia (AML)
(1), primarily in those with M2 and
M4 AML (2–4). AML is a disease that has unique
characteristics and patients typically have a poor prognosis.
Currently, ~400 cases of AML with t(6;9)(p23;q34) have been
reported since it was first identified in 1976, and most of these
are reports of small numbers of cases (1,4–12). This chromosomal abnormality has also
been demonstrated in patients with myelodysplastic syndromes (MDS)
and myeloproliferative neoplasms. The DEK
proto-oncogene/nucleoporin (DEK/NUP214) fusion gene results from
t(6;9)(p23;q34) in AML as reported (1,3,11,13).
Case report
Initial patient details
The patient was a 46-year-old female who presented
with a fever on June 26, 2013 and admitted to Institute of
Hematology and Blood Diseases Hospital (Tianjin, China). The
patient had a white blood cell (WBC) count of
22.8×109/l, a hemoglobin level of 71 g/l and a platelet
count of 101×109/l. Bone marrow smears with Wright's
staining, as described previously (14), revealed that bone marrow hyperplasia,
myeloblasts and immature monocytes comprised 59.5% of the bone
marrow, with visible Auer rods. The immunophenotype was analyzed
using multi-color flow cytometry, as described previously (15), and the results showed blasts highly
expressed CD13, CD33 and CD117, and dim expressed CD34, CD38, CD64,
myeloperoxidase (MPO) and human leukocyte antigen D related
(HLA-DR).
Chromosomal analysis
A chromosomal analysis was performed using the
R-banding technique as described previously (16), and the samples were karyotyped
according to the International System for Human Cytogenetic
Nomenclature (17). The results of
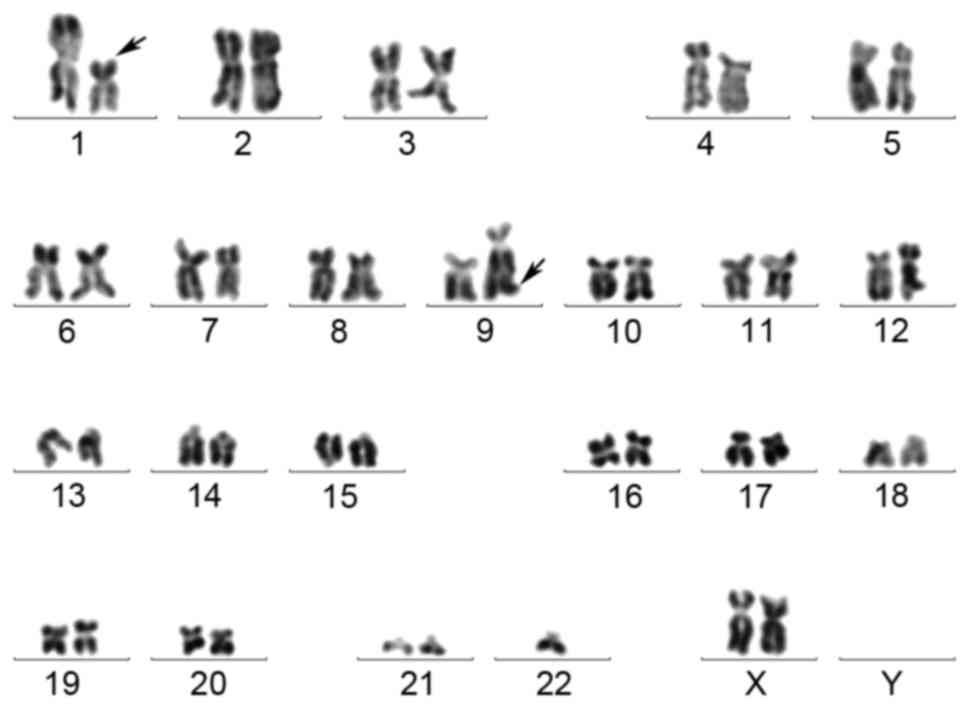
the chromosomal analysis were as follows (Fig. 1): 46, XX, t(1;9)(p22;q34)[9]/46; XX,
t(1;9)(p22;q34),-2,+22[1]/45, XX, t(1;9)(p22;q34),-21[1]/46, XX[9].
Somatic mutations were detected by targeted next-generation
sequencing (18,19): The patient was negative for FLT3/ITD,
FLT3/TKD, C-KIT Exon 8, C-KIT D816, CEBPα/TAD, CEBPα/BZIP,
DNMT3A/ZNF and DNMT3A/Mtase mutations. In order to identify the
specific fusion gene formed by the chromosomal abnormalities in the
patient, transcriptome sequencing (RNA-seq) was performed on the
patient's bone marrow mononuclear cells and peripheral blood cells
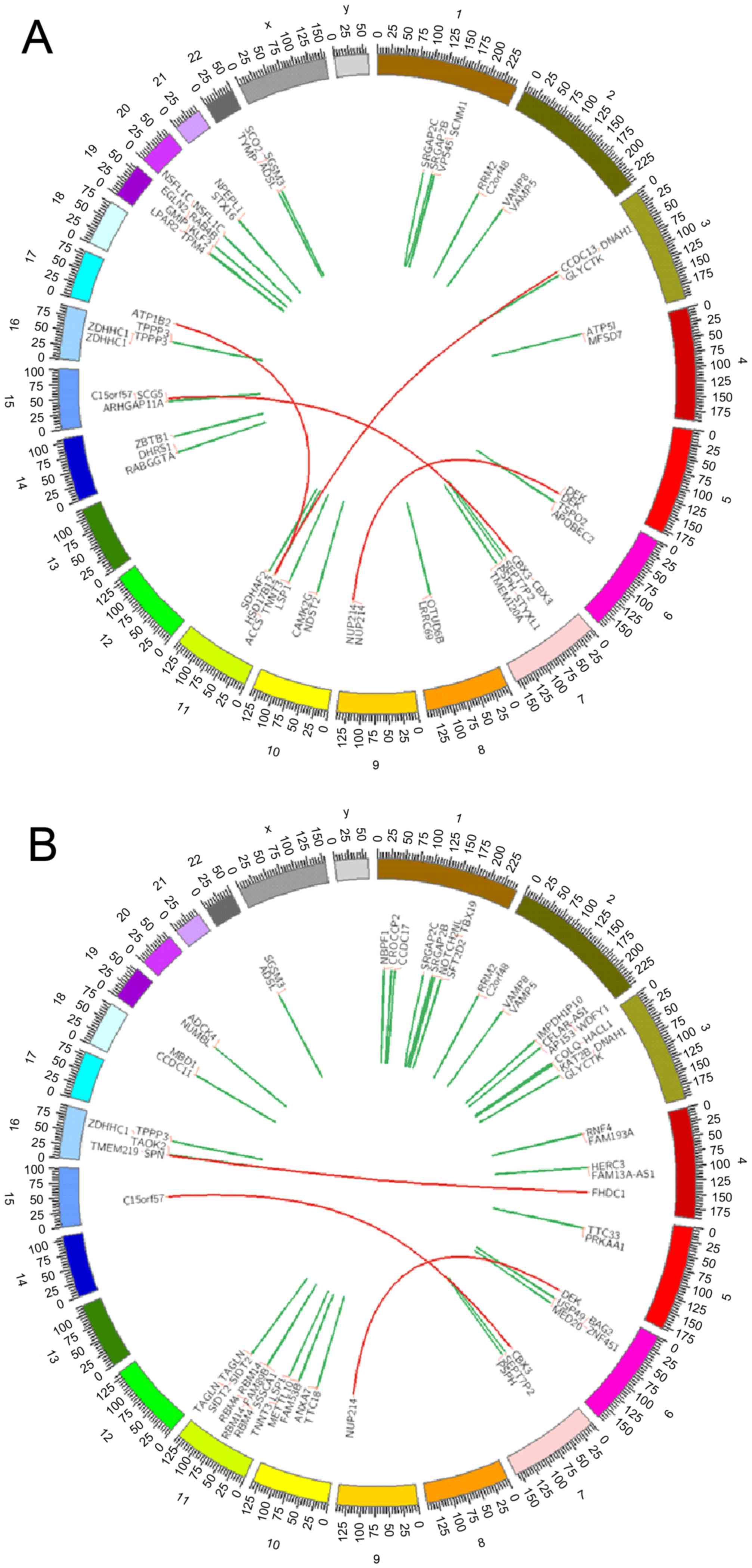
in the complete remission (CR) state. Expression of DEK/NUP214
fusion gene was demonstrated in the bone marrow and peripheral
blood mononuclear cells, which suggested that the patient had a
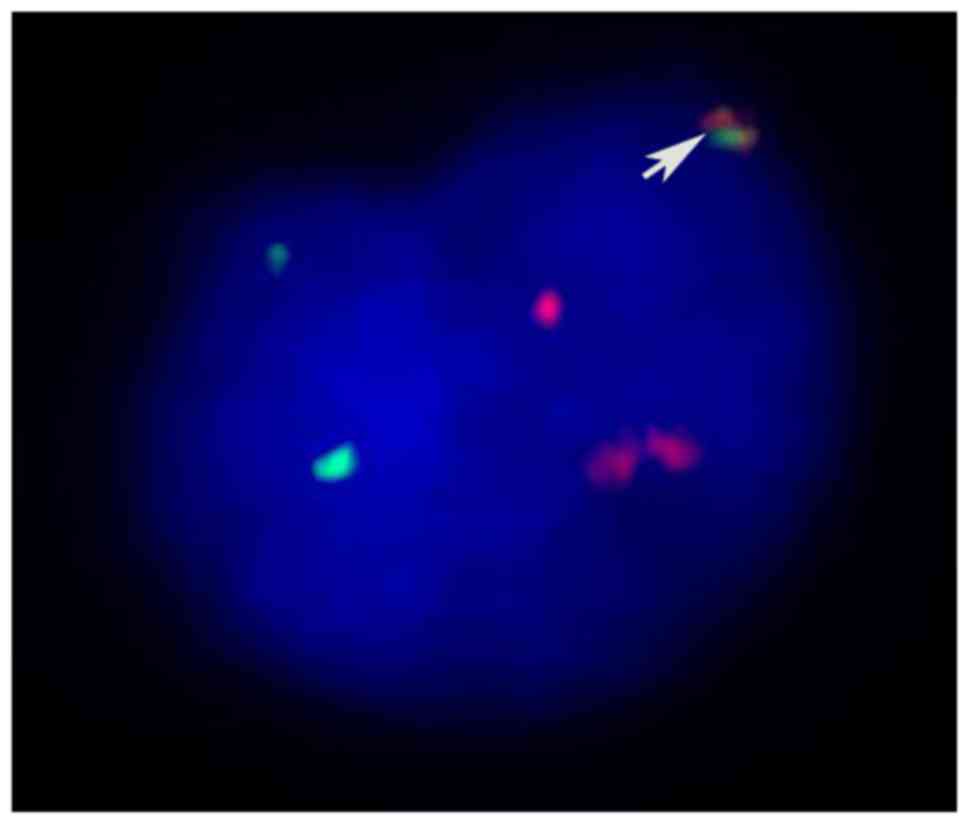
variant translocation of t(6;9)(p23;q34) (Fig. 2). Fluorescent in situ
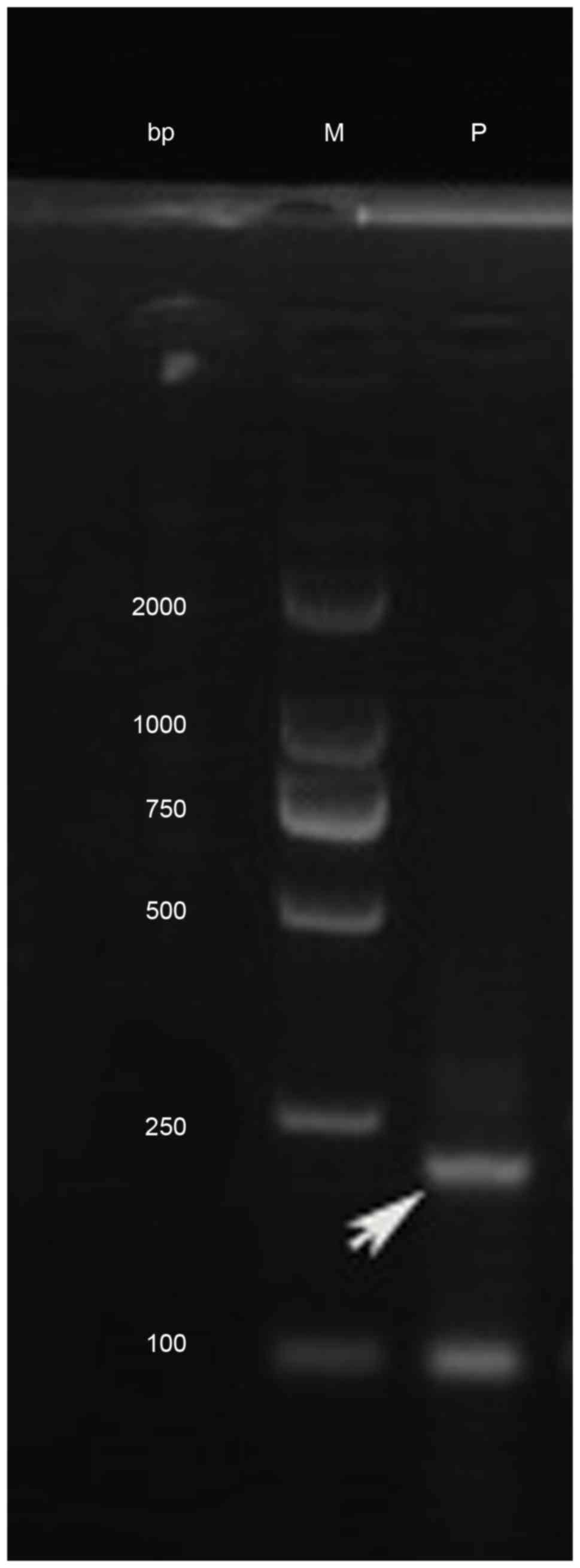
hybridization (FISH) using the DEK/NUP214 probes, nested polymerase
chain reaction (PCR) and agarose gel electrophoresis were performed
to confirm the existence of the DEK/NUP214 fusion gene (Figs. 3 and 4).
The diagnosis of the patient was AML M4 with the DEK-NUP14 fusion
gene.
Chemotherapy and outcome
Daunorubicin (DNR; 60 mg/day, days 1–3) and cytosine
arabinoside (Ara-C, 200 mg/day, days 1–7) were administered to the
patient for induction chemotherapy from June 29, 2013. The patient
did not achieve CR according to response criteria of International
Working Group (20). Mitoxantrone
(MTZ), Ara-C, and cyclophosphamide (CTX) were administered for
re-induction chemotherapy from July 24, 2013, which contained MTZ
15 mg/day (day 1) and 10 mg/day (days 2–3), Ara-C 200 mg/day (days
1–7) and CTX 800 mg/day (days 1 and 4). From September 25, 2013, a
third course of chemotherapy was administered, which included 3
mg/day homoharringtonine (days 1–7), 200 mg/day Ara-C (days 1–7)
and 20 mg/day aclarubicin (days 1–7). The patient did not achieve
CR. The patient was offered the FLAG regimen [Fludarabine (Flu;45
mg/day, days 1–5), Ara-C (1.5 g/day, days 1–5) and granulocyte
colony-stimulating factor (G-CSF 300 µg/day, days 0–5)], from
November 16, 2013. Bone marrow examination revealed that the
patient achieved CR on December 16, 2013. From December 17, 2013,
the patient received a second FLAG regimen at the same dose as a
consolidation treatment. The patient suffered from a severe
pulmonary infection in the second FLAG chemotherapy regimen and
refused further treatment. Instead, the patient chose outpatient
follow-up only. The patient was re-admitted on December 24, 2014
because of fever; a bone marrow smear revealed that the percentage
of myeloblasts was 6.5% and immature monocytes accounted for 13.5%.
From December 26, 2014, the re-induction chemotherapy CAG regimen
(100 mg/day Ara-C, days 1–7;20 mg/day Acla, days 1–6; and 300
µg/day G-CSF, days 1–7) was used. The patient achieved CR2. Two
cycles of Ara-C (150 mg/day, days 1–5) and Acla (20 mg/day, day
1–5) chemotherapy were administered, with the last treatment
administered in March 2015. Bone marrow examination on November 12,
2015 demonstrated that the patient relapsed again and succumbed in
June 2016. The overall survival was ~36 months from diagnosis.
Materials and methods
RNA-seq
Total RNA was extracted from frozen mononuclear
cells as described previously (21).
A total of 5 µg of RNA was used for RNA-seq, which was performed
using an IlluminaHiSeq 2000 Sequencer (Illumina, Inc., San Diego,
CA, USA). The RNA-seq was performed by BGITech Solutions Co., Ltd.
(Shenzhen, China).
Detection of the DEK/NUP214 fusion
transcript
To analyze the expression of DEK/NUP214, the total
RNA of frozen mononuclear cells was isolated using the RNeasy mini
kit according to the manufacturer's protocol (Takara Biotechnology
Co., Ltd., Dalian, China). Reverse transcription was carried out
using TaqMan reverse transcriptase reagents (Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) according to the
manufacturer's instructions (21).
Primer pairs were purchased from Invitrogen (Thermo Fisher
Scientific, Inc.) and the obtained products were subjected to
direct sequencing (Invitrogen; Thermo Fisher Scientific, Inc.). The
primer sequences for the first step were as follows: DEK/NUP214
forward, 5′-TGCCAATGTTAAGAAAGCAGATAG-3′ and reverse,
5′-GGCAAGGATTTGGTGTGAGAT-3′. The primer sequences for the second
step were as follows: DEK/NUP214 forward,
5′-AGCAGCACCACCAAGAAGAAT-3′ and reverse,
5′-GTCTCTCGCTCTGGCACAAG-3′. DEK/NUP214 fusion transcripts were
amplified using the ABI Prism 7500 Real Time PCR system (Thermo
Fisher Scientific, Inc.). cDNA was subjected to 40 cycles of
denaturing (94°C, 15 sec) and annealing (60°C, 60 sec) using the
Leukemia Related Fusion Gene Detection kit, (Shanghai Yuanqi
Bio-Pharmaceutical Co., Ltd., Shanghai, China). PCR products were
run on a 1% agarose gel.
FISH
FISH was performed according to the manufacturer's
protocol. Kreatech FISH probes targeting DEK/NUP214 (cat. no.
KBI-10306) were obtained from Leica Biosystems (Amsterdam,
Netherlands). Automated in situ hybridization staining was
performed using ThermoBrite, an automatic instrument (Leica
Biosystems). Data analysis was performed using ISIS software
(version 5.4.6; MetaSystems GmbH, Altlussheim, Germany).
Discussion
AML with t(6;9)(p23;q34) is a rare type of AML where
the bone marrow morphology is frequently accompanied by increased
levels of basophils. Auer rods and fine particles are also visible
in certain blast cells. The immunophenotypic analysis from a
previous study demonstrated that CD9, CD13, CD33, HLA-DR, CD38,
CD45, CD34, CD15 and terminal deoxynucleotidyl transferase (TdT)
were expressed in the patient with this translocation (22). AML with t(6;9) translocation, which
accounts for ≤70% more cases compared with other types of AML, is
associated with the FLT3/ITD mutation (22,23). The
2008 revision of World Health Organization classification have
defined AML with at(6;9) translocation as a recurrent genetic
abnormality of AML (24).
Few AML patients with t(6;9)(p23;q34) have been
reported to have other chromosomal or karyotype changes.
Furthermore, no cases of DEK/NUP214-positive AML without
t(6;9)(p23;q34) have been reported until now, to the best of our
knowledge. Slovak et al (4)
reported the cases of 69 patients with AML or MDS with
t(6;9)(p23;q34) in 2006. These patients were identified among 7690
patients with evaluable karyotypes, accounting for 0.9%. The t(6;9)
translocation was the sole clonal karyotypic aberration in 61/69
patients (88%) in this study; only 4 pediatric and 4 adult cases
(12%) had secondary anomalies. A total of3 patients (5%) had
complex translocations involving 3 or 4 chromosomes. The typical
translocation of t(6;9)(p23;q34) was not found in the chromosomal
karyotype analysis of the patient in the present study; however,
the t(1;9)(p22;q34) translocation was revealed to form the
DEK/NUP214 fusion gene.
The prognosis of patients with AML who have
t(6;9)(p23;q34) is poor. The CR rate is ~65% (71% in children, 58%
in adult patients), while the estimated 5-year survival rate is 28%
for children and 9% for adults based on the cases reported by
Slovak et al (4).
Acknowledgements
The present study was supported by the National
Science and Technology Pillar Program (grant no. 2014BAI09B12) and
the Tianjin Research Program of Application Foundation and Advanced
Technology (grant no. 15JCZDJC36400).
References
|
1
|
Schwartz S, Jiji R, Kerman S, Meekins J
and Cohen MM: Translocation (6;9)(p23;q34) in acute non lymphocytic
leukemia. Cancer Genet Cytogenet. 10:133–138. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bennett JM, Catovsky D, Daniel MT,
Flandrin G, Galton DA, Gralnick HR and Sultan C: Proposals for the
classification of the acute leukaemias. French-American-British
(FAB) co-operative group. Br J Haematol. 33:451–458. 1976.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Soekarman D, von Lindern M, van der Plas
SD, Selleri L, Bartram CR, Martiat P, Culligan D, Padua RA,
Hasper-Voogt KP, Hagemeijer A, et al: Dek-can rearrangement in
translocation (6;9)(p23;q34). Leukemia. 6:489–494. 1992.PubMed/NCBI
|
|
4
|
Slovak ML, Gundacker H, Bloomfield CD,
Dewald G, Appelbaum FR, Larson RA, Tallman MS, Bennett JM,
Stirewalt DL, Meshinchi S, et al: A retrospective study of 69
patients with t(6;9)(p23;q34) AML emphasizes the need for a
prospective, multicenter initiative for rare ‘poor prognosis’
myeloid malignancies. Leukemia. 20:1295–1297. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Garcon L, Libura M, Delabesse E, Valensi
F, Asnafi V, Berger C, Schmitt C, Leblanc T, Buzyn A and Macintyre
E: DEK-CAN molecular monitoring of myeloid malignancies could aid
therapeutic stratification. Leukemia. 19:1338–1344. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ishiyama K, Takami A, Kanda Y, Nakao S,
Hidaka M, Maeda T, Naoe T, Taniguchi S, Kawa K, Nagamura T, et al:
Allogeneic hematopoietic stem cell transplantation for acute
myeloid leukemia with t(6;9)(p23;q34) dramatically improves the
patient prognosis: A matched-pair analysis. Leukemia. 26:461–464.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lai YY, Li Y, Shi Y, Dang H, He Q, Feng L,
Wang Z, Wang XY, Li N, Liu Q, et al: Characteristics of 11 patients
with acute myeloid leukemia accompanied with karyotype aberration
t(6;9). Zhongguo Shi Yan Xue Ye Xue Za Zhi. 20:1293–1296. 2012.(In
Chinese). PubMed/NCBI
|
|
8
|
Sandahl JD, Coenen EA, Forestier E,
Harbott J, Johansson B, Kerndrup G, Adachi S, Auvrignon A, Beverloo
HB, Cayuela JM, et al: t(6;9)(p22;q34)/DEK-NUP214 rearranged
pediatric myeloid leukemia: An international study of 62 patients.
Haematologica. 99:865–872. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tarlock K, Alonzo TA, Moraleda PP, Gerbing
RB, Raimondi SC, Hirsch BA, Ravindranath Y, Lange B, Woods WG,
Gamis AS and Meshinchi S: Acute myeloid leukaemia (AML) with
t(6;9)(p23;q34) is associated with poor outcome in childhood AML
regardless of FLT3-ITD status: A report from the Children's
Oncology Group. Br J Haematol1. 166:254–259. 2014. View Article : Google Scholar
|
|
10
|
Wang Y, Xue Y, Chen S, Wu Y, Pan J, Zhang
J and Shen J: A clinical and laboratory study on acute myeloid
leukemia with t(6;9)(p23;q34). Zhonghua Yi Xue Yi Chuan Xue Za Zhi.
27:34–37. 2010.(In Chinese). PubMed/NCBI
|
|
11
|
Gupta M, Kumar Ashok J, Sitaram U, Neeraj
S, Nancy A, Balasubramanian P, Abraham A, Mathews V, Viswabandya A,
George B, et al: The t(6;9)(p22;q34) in myeloid neoplasms: A
retrospective study of 16 cases. Cancer Genet Cytogenet.
203:297–302. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Harrison CJ, Hills RK, Moorman AV,
Grimwade DJ, Hann I, Webb DK, Wheatley K, de Graaf SS, van den Berg
E, Burnett AK and Gibson BE: Cytogenetics of childhood acute
myeloid leukemia: United kingdom medical research council treatment
trials AML 10 and 12. J Clin Oncol. 28:2674–2681. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
von Lindern M, Fornerod M, van Baal S,
Jaegle M, de Wit T, Buijs A and Grosveld G: The translocation
(6;9), associated with a specific subtype of acute myeloid
leukemia, results in the fusion of two genes, dek and can, and the
expression of a chimeric, leukemia-specific dek-can mRNA. Mol Cell
Bio. 12:1687–1697. 1992. View Article : Google Scholar
|
|
14
|
Wei S, Wang S, Qiu S, Qi J, Mi Y, Lin D,
Zhou C, Liu B, Li W, Wang Y, et al: Clinical and laboratory studies
of 17 patients with acute myeloid leukemia harboring
t(7;11)(p15;p15) translocation. Leuk Res. 37:1010–1015. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ye L, Lin D, Mi YC, Zhang J, Wang Y, Wang
HJ, Wei H, Liu BC, Zhou CL, Li W and Wang JX: Comparison of
EGILl998 and WH02008 criteria for the diagnosis of mixed phenotype
acute leukemia. Zhonghua Xue Ye Xue Za Zhi. 33:286–290. 2012.(In
Chinese). PubMed/NCBI
|
|
16
|
Huang H and Chen J: Chromosome bandings.
Methods Mol Biol. 1541:59–66. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shaffer LG and Tommerup N: ISCN. An
International System of Human Cytogenetic NomenclatureBasel. S.
Karger; 2005
|
|
18
|
Weerts MJA, van Marion R, Helmijr JCA,
Beaufort CM, Krol NMG, Trapman-Jansen AMAC, Dinjens WNM, Sleijfer
S, Jansen MPHM and Martens JWM: Somatic tumor mutations detected by
targeted next generation sequencing in minute amounts of
serum-derived cell-free DNA. Sci Rep. 7:21362017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Feng J, Li Y, Jia Y, Fang Q, Gong X, Dong
X, Ru K, Li Q, Zhao X, Liu K, et al: Spectrum of somatic mutations
detected by targeted next-generation sequencing and their
prognostic significance in adult patients with acute lymphoblastic
leukemia. J Hematol Oncol. 10:612017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cheson BD, Bennett JM, Kopecky KJ, Büchner
T, Willman CL, Estey EH, Schiffer CA, Doehner H, Tallman MS, Lister
TA, et al: Revised recommendations of the International Working
Group for diagnosis, standardization of response criteria,
treatment outcomes and reporting standards for therapeutic trials
in acute myeloid leukemia. J Clin Oncol. 21:4642–4649. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li X, Li Y, Yuan T, Zhang Q, Jia Y, Li Q,
Huai L, Yu P, Tian Z, Tang K, et al: Exogenous expression of WT1
gene influences U937cell biological behaviors and activates MAPK
and JAK-STAT signaling pathways. Leuk Res. 38:931–939. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Oyarzo MP, Lin P, Glassman A, Bueso-Ramos
CE, Luthra R and Medeiros LJ: Acute myelogenous leukemia with
t(6;9)(p23;q34) is associated with dysplasia and a high frequency
of FLT3 gene mutations. Am J Clin Pathol. 122:348–358. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chi Y, Lindgren V, Quigley S and Gaitonde
S: Acute myelogenous leukemia with t(6;9)(p23;q34) and marrow
basophilia: An overview. Arch Pathol Lab Med. 132:1835–1837.
2008.PubMed/NCBI
|
|
24
|
Vardiman JW, Thiele J, Arber DA, Brunning
RD, Borowitz MJ, Porwit A, Harris NL, Le Beau MM,
Hellström-Lindberg E, Tefferi A and Bloomfield CD: The 2008
revision of the World Health Organization (WHO) classification of
myeloid neoplasms and acute leukemia: Rationale and important
changes. Blood. 114:937–951. 2009. View Article : Google Scholar : PubMed/NCBI
|