Introduction
Renal cell carcinoma (RCC) occurs in 2–3% of adult
malignancies, with ~337,860 new cases and 143,406 mortalities
worldwide in 2012 (1). The global
incidence of RCC increased gradually between 1997 and 2007
(2). In China, there are ~66,466
cases of RCC annually, making it the third most prevalent
genitourinary cancer (3). Several
risk factors have been associated with RCC, including smoking,
alcohol use, hypertension and obesity (4). However, numerous individuals exposed to
these risk factors in their lifetime do not develop RCC.
Accumulating evidence has suggested that genetic and environmental
factors perform important roles in the development of RCC (5). MicroRNAs (miRNAs/miRs) have recently
been implicated in RCC tumorigenesis (6,7).
miRNAs are a class of small, single-stranded,
noncoding RNAs that modulate gene expression by binding to the 3′
untranslated region (3′UTR) of their target genes, causing
translational suppression and/or mRNA degradation (8,9).
Accumulating evidence has suggested that miRNAs perform important
roles in a broad range of biological processes, including cellular
proliferation, apoptosis, differentiation and cancer development
(10). To regulate mRNA and protein
expression levels, miRNAs bind to the 3′UTR of their target mRNAs.
Thus, single nucleotide polymorphisms (SNPs) in the 3′UTR may
impede existing binding sites or create novel binding sites,
resulting in the misregulation of target genes, which may affect
the tumor risk of an individual (11,12). SET
domain containing (lysine methyltransferase) 8 (SET8; also termed
PR-Set7, SETD8 or KMT5A) is a SET domain-containing
methyltransferase family member, and is modulated by miR-502
binding to its 3′UTR (13,14). SET8 encodes a histone H4 lysine 20
monomethyl transferase that has been implicated in modulating cell
cycle progression and development (14–16). It
was previously reported that SET8 is recruited to DNA replication
foci through its interaction with proliferating cell nuclear
antigen and is required for proper DNA replication (17,18). The
precise modulation of SET8 levels is important for proper cell
cycle progression, and the inability to modulate SET8 expression
results in severe cell cycle defects (17–20).
Previous studies have revealed that SET8 is highly expressed in
several types of tumors, including breast cancer (21), small cell lung cancer (22), hepatocellular carcinoma (11) and ovarian cancer (23). Furthermore, SET8 expression levels
were identified to be associated with the rs16917496 SNP in the
SET8 3′UTR (21).
In the present study, the rs16917496 SNP in patients
with ccRCC in a case-control study was genotyped to assess its
association with the risk of cancer. In addition, the association
between this SNP and SET8 expression, and the roles of SET8 in
renal carcinoma 786-O cell proliferation, migration and invasion
were examined using RNA interference.
Materials and methods
Tissue specimens and DNA
extraction
Blood samples were obtained from 140 patients with
ccRCC, which were all treated at The Fourth Affiliated Hospital of
Hebei University (Hebei, China) between December 2006 and December
2010. The ccRCC patients included 59 males and 81 females, mean age
56.8 years (range, 36–79 years). Blood samples were also acquired
from age-matched healthy controls. Total DNA was extracted using a
Wizard Genomic DNA Extraction kit (Promega Corporation, Madison,
WI, USA) according to the manufacturer's protocol and stored at
−20°C. The present study was approved by the Ethics Committee of
The Fourth Affiliated Hospital of Hebei Medical University and
written informed consent was acquired from all recruited
patients.
PCR amplification and sequence
analysis
The primers for amplification were
5′-CCTGGTCAGTGGTCAGCAAAT-3′ (sense) and 5′-CTGGGAAACACGCTCAAAATC-3′
(antisense) for rs16917496 in the 3′UTR of SET8 (National
Center for Biotechnology Information database: http://www.ncbi.nlm.niih.gov/snp). PCR was
performed on DNA isolated from blood samples using a PCR Master Mix
kit according to the manufacturer's instructions (Promega
Corporation), The PCR condition consisted of incubation for 2 min
at 95°C followed by 35 cycles of 30 sec at 95°C, 30 sec at 55°C and
45 sec at 72°C, with a final extension step at 72°C for 5 min. The
PCR product was used for sequencing. Cycle sequencing was performed
using the Dye Terminator Cycle Sequencing Ready Reaction kit
(Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA,
USA), according to the manufacturer's protocol, and the products
were analyzed on the ABIPRISM Genetic Analyzer 3100 (Applied
Biosystems; Thermo Fisher Scientific, Inc.). Polymorphisms were
identified by repeated analyses of the two strands.
Measurement of SET8 levels in ccRCC
tissue
SET8 protein expression was determined by
immunostaining, which was performed on serial histopathological
sections from RCC tissue. RCC tissues were fixed in 10% formalin
overnight at room temperature and paraffin-embedded. The thickness
of the RCC tissue sections was 5 µm. The primary antibody against
SET8 (Abcam, Cambridge, UK; cat. no. ab3798) was applied to
sections at a dilution of 1:100 overnight at 4°C, and sections were
subsequently incubated with a biotinylated secondary antibody at a
dilution of 1:10,000 (ProteinTech Group, Inc., Chicago, IL, USA;
cat. no. SA00003-1) for 1 h at room temperature. The sections were
then incubated with horseradish peroxidase (HRP)-conjugated
streptavidin (Origene Technologies, Inc., Beijing, China; cat. no.
K156617 J) at 37°C for 30 min and developed using
3,3-diaminobenzidine.
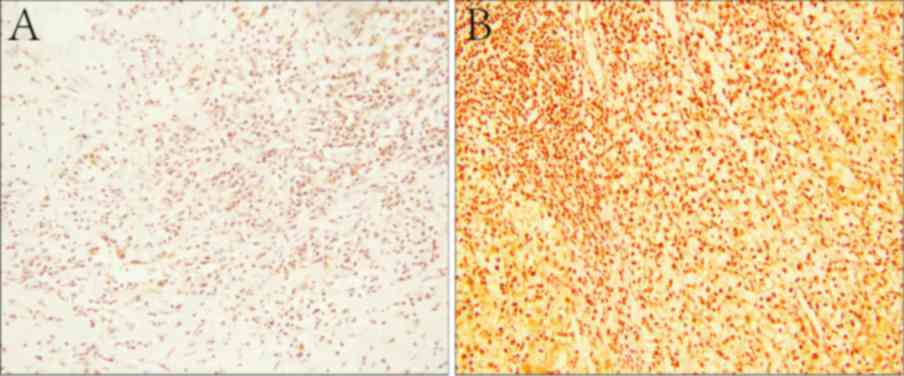
Stained slides were scored by two independent
experienced pathologists without knowledge of the patients'
clinical data. Immunostaining results were semi-quantified using
HSCORE as reported previously (24,25) under
a light microscope at magnification of ×200. Briefly, the score was
calculated based on estimates of the percentage of positively
stained renal cells in each intensity category (0, 1+, 2+, 3+, 4+).
The intensity of staining of the antibody was analyzed by HSCORE.
The HSCORE was calculated using the following equation:
HSCORE=(i+1) π, where i=1, 2, 3 or 4, and π varies between 0 and
100%. A score of >100% was defined as high expression and ≤100%
was defined as low expression (Fig.
1).
Cell culture and transfection
The renal carcinoma 786-O cell line was purchased
from the Institute of Biochemistry and Cell Biology, Chinese
Academy of Sciences (Shanghai, China) and cultured in RPMI-1640
medium supplemented with 10% fetal bovine serum (FBS), 50 IU/ml
penicillin and 50 mg/ml streptomycin (all Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) at 37°C in a standard
humidified incubator containing 5% CO2.
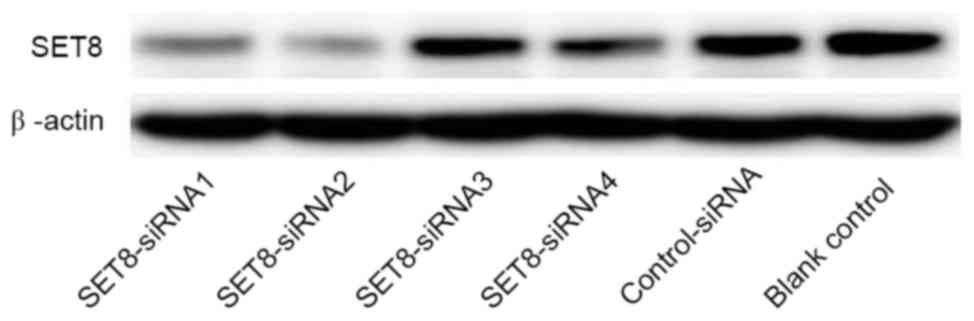
The renal carcinoma 786-O cells were transfected
with psi-H1-SET8 small interfering (si)RNA or psi-H1
plasmids (GeneCopoeia, Inc., Rockville, MD, USA) using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. The target sequences of
four siRNAs against SET8 were as follows: SET8-siRNA1,
5′-CAGAAUCGCAAACUUACGGA-3′; SET8-siRNA2,
5′-GAAUGAAGAUUGACCUCAUCG-3′; SET8-siRNA3,
5′-GCCUAGGAAGACUGAUCAAU-3′; and SET8-siRNA4,
5′-GGCGCUCACUGAAGUGUAUG-3′. Successful knockdown of SET8 was
confirmed by western blot analysis using an anti-SET8 antibody
(Abcam; cat. no. ab3798).
Western blot analysis
Radioimmunoprecipitation assay lysis buffer (150 mM
NaCl, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, Ph
7.9, 10 mM NaF, PMSF, and 1X protease inhibitors (Roche
Diagnostics, Basel, Switzerland) was used to isolate total protein
from all the experimental renal carcinoma 786-O cells. The Bradford
assay was used to determine protein concentration. Western blot
analysis was performed as described previously (26). Briefly, 40 µg of total protein for
each lane was separated on a 10% denaturing polyacrylamide gel and
transferred to a polyvinylidene difluoride membrane (Roche
Diagnostics). Immunoblots were probed with a mouse monoclonal
anti-SET8 antibody at 1:500 (Abcam; cat. no. ab3798) or β-actin at
1:20,000 (Santa Cruz Biotechnology, Inc., Dallas, TX, USA; cat. no.
SC-47778A). Membranes were blocked in TBS-Tween-20 containing 5%
nonfat dry milk for 1 h at room temperature and incubated overnight
with primary antibody at 4°C. A HRP-conjugated anti-mouse IgG
antibody was used as the secondary antibody (ProteinTech Group,
Inc, Chicago, IL, USA; cat. no. SA00003-1) for 2 h at room
temperature. Signals were detected using FluorChem® HD2
(Alpha-InnoTec, San Leandro, CA, USA).
RNA extraction and reverse
transcription-polymerase chain reaction (RT-PCR)
Total RNA was extracted from cultured cells using
TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) one-step method
according to the manufacturer's protocol. RNA (2 µg) was used for
reverse transcription to synthesize template cDNA a using RevertAid
First-Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions.
GAPDH gene was used as an endogenous control. The
primer sequences are listed in Table
I. PCR was performed using the Quantstudio™ Dx PCR instrument
(Thermo Fisher Scientific, Inc.) and iQ™ SYBR-Green Super mix
(Bio-Rad Laboratories, Inc., Hercules, CA, USA), which contained 5
ng cDNA and 10 pM of each primer. PCR reaction started with 1 cycle
of 95°C for 10 min, followed by 40 cycles of three steps as 94°C
for 30 sec, 58°C for 30 sec, and 72°C for 30 sec. The PCR products
were electrophoresed on a 1.5% agarose gel and visualized with
ethidium bromide staining, and the data were normalized to the
endogenous control gene GAPDH using the 2−ΔΔCq method
(27).
 | Table I.Reverse transcription-polymerase
chain reaction primers. |
Table I.
Reverse transcription-polymerase
chain reaction primers.
| Gene | Primer
sequence | Amplicon, bp |
|---|
| c-Myc | F:
5′-CCTACCCTCTCAACGACAGC-3′ | 179 |
|
| R:
5′-TTCCTCCTCAGAGTCGCTGC-3′ |
|
| MMP-7 | F:
5′-TGGGAACAGGCTCAGGACTAT-3′ | 413 |
|
| R:
5′-AATGGGTAGGAGTCCCCATGA-3′ |
|
| GAPDH | F:
5′-CAAGGTCATCCATGACAACTTTG-3′ | 496 |
|
| R:
5′-GTCCACCACCCTGTTGCTGTAG-3′ |
|
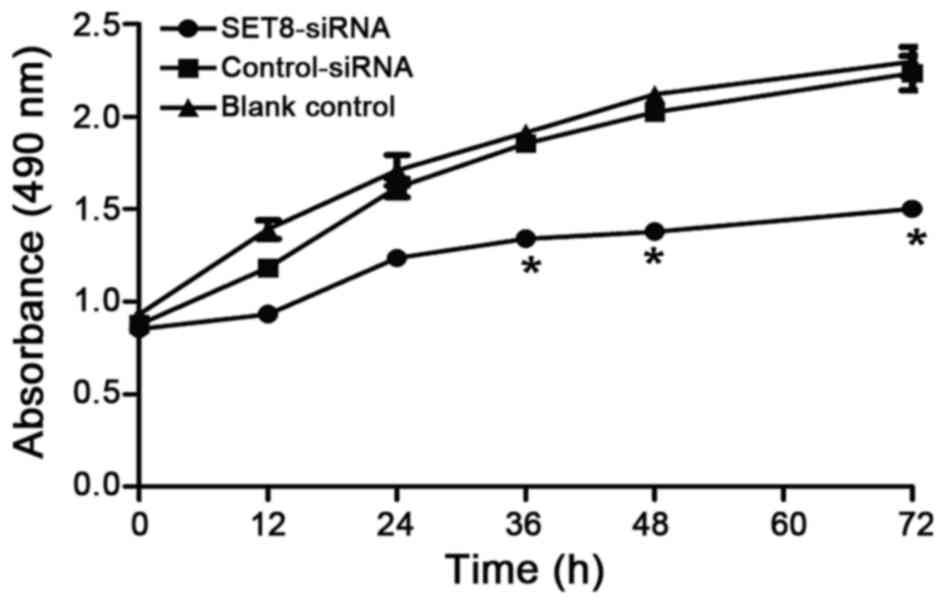
Cell proliferation assay
Cell proliferation was analyzed by MTT
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) assay as described
previously (26). Cells were seeded
in sextuplicate at 1×103 cells/well on 96-well
microplates and transfected with SET8 siRNA-2 or negative control
plasmids using Lipofectamine 2000. The cells were incubated with
100 µl MTT (0.5 mg/ml; Sigma-Aldrich; Merck KGaA) for 4 h at 37°C
after various time periods of SET8-knockdown (0, 12, 24, 36, 48 and
72 h). Following centrifugation (room temperature, 10 min at 1,000
× g), 100 µl of 0.04 mol/l HCl-isopropanol was added to the cells.
The absorbance was measured at 490 nm using an ELISA microplate
reader. The experiment was repeated three times.
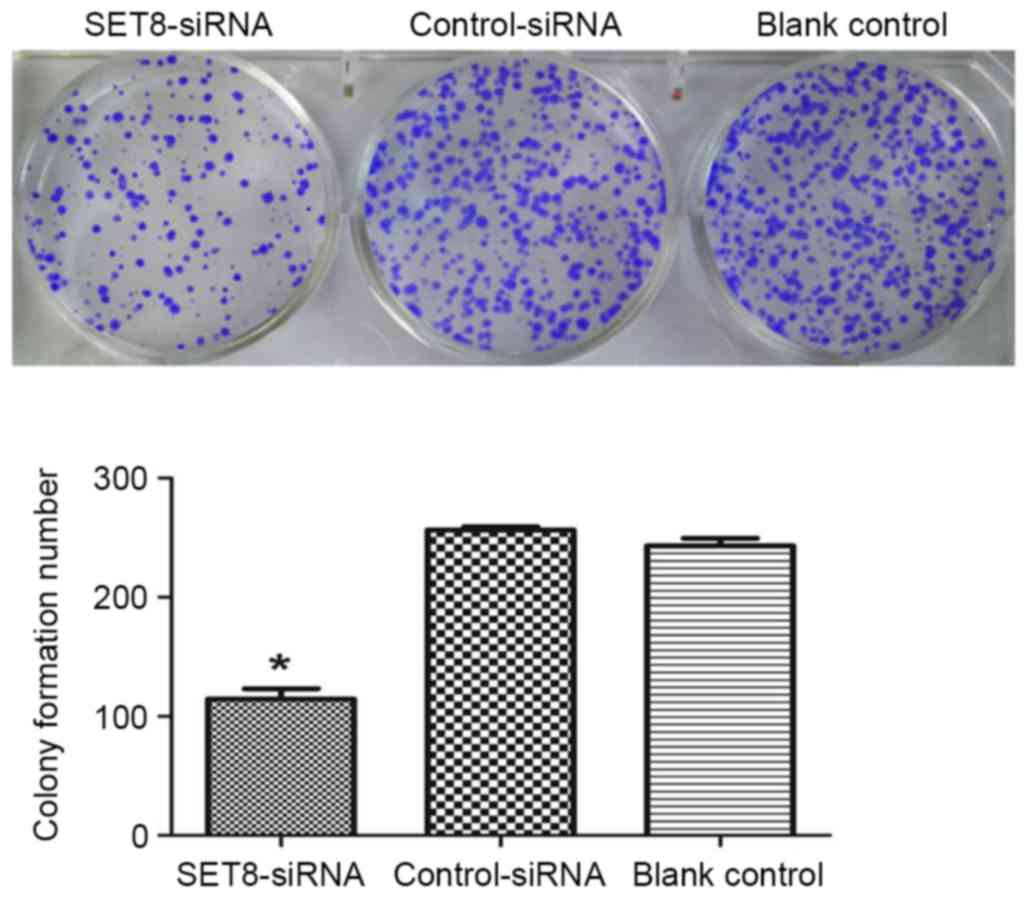
Colony formation assay
The renal carcinoma 786-O cells were seeded on
6-well plates at a density of 500 cells/well at 48 h
post-transfection. After 10 days at 37°C in 5% CO2, the
cells were fixed with 95% methanol and stained with 0.1% crystal
violet for 15 min at room temperature. Colonies of >50 cells
were scored under a light microscope at a magnification of ×100.
The colony formation ratio (%) was calculated as follows: (Number
of cell colonies/500) ×100.
Wound healing assay
At 48 h post-transfection, the cell monolayer was
scraped in a straight line using a 200-µl pipette tip to create a
scratch once they reached 100% confluence. The medium was removed
and the cells were washed twice in PBS. Wound healing results were
observed under a light microscope at a magnification of ×100.
Images were captured at 0 and 12 h after scratching. At least five
fields were analyzed for each scratch, and the migration index was
calculated as the width of a scratch divided by the initial width
of the same scratch, as previously described (28).
Cell invasion assay
Cell invasion assays were performed using a
Transwell assay (pore size, 8 µm; Corning Incorporated, Corning,
NY, USA). The insert was coated with 30 µl Matrigel (BD
Biosciences, Franklin Lakes, NJ, USA) mixed with RPMI-1640
serum-free medium in a 1:5 dilution for 30 min at 37°C. At 24 h
post-transfection, 100,000 cells were resuspended in 100 µl
serum-free medium and plated in the upper chamber. The bottom
chamber was filled with 500 µl RPMI-1640 containing 10% FBS.
Following incubation for 24 h at 37°C, the upper chamber was
removed, and non-penetrating cells were gently wiped away. The
remaining cells were stained with 0.1% crystal violet for 15 min at
room temperature and counted under a light microscope (Axio
Observer D1, Gemney) in five representative areas at 400x,
irrespective of staining intensity or cell number. The experiment
was repeated three times. The cell invasion inhibition rate (%) =
[1-(the number of invasive cells in the experimental group / the
number of invasive cells in the control group)] ×100.
Statistical analysis
Data are presented as the mean ± standard deviation.
A χ2 test was used to analyze dichotomous values. The
odds ratio (OR) and 95% confidence interval (CI) were calculated
using an unconditional logistic regression model. Student's t-test
was performed to analyze results of MTT, colony formation,
migration and invasion assays. All statistical analyses were
performed using SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA).
P<0.05 was considered statistically significant for all
statistical tests.
Results
SET8 genotype is associated with ccRCC
risk
A total of 140 patients with ccRCC and 130 controls
were genotyped for the rs16917496 SNP. The SET8 CC, CT and TT
genotype frequencies in patients with ccRCC, and control samples
were 14, 47 and 79 and 30, 32 and 68, respectively. The
distribution of the rs16917496 genotype followed a Hardy-Weinberg
equilibrium. The overall frequencies and genotype distributions of
the rs16917496 polymorphism in patients with ccRCC and controls are
presented in Table II. The C allele
frequencies of rs16917496 in patients with ccRCC (26.79%) were
significantly lower compared with that in controls (35.38%)
(P=0.031), and the presence of the C allele significantly decreased
the risk of developing ccRCC (OR=0.668; 95% CI, 0.463–0.964). The
CC genotype was associated with a decreased risk of ccRCC compared
with the CT (P=0.003; OR=0.318; 95% CI, 0.146–0.691), TT (P=0.011;
OR=0.402; 95% CI, 0.197–0.819) and CT+TT (P=0.004; OR=0.370; 95%
CI, 0.186–0.736) genotypes.
 | Table II.Association between the rs16917496
single nucleotide polymorphism and clear cell renal cell cancer
risk. |
Table II.
Association between the rs16917496
single nucleotide polymorphism and clear cell renal cell cancer
risk.
| Type | Case, n (%) | Controls, n
(%) | χ2 | P-value | OR | 95% CI |
|---|
| Genotype |
|
|
|
|
|
|
| CC | 14 (10.0) | 30 (23.1) |
|
| 1.000 |
|
| CT | 47 (33.6) | 32 (24.6) | 8.659 | 0.003a | 0.318 | 0.146–0.691 |
| TT | 79 (56.4) | 68 (52.3) | 6.515 | 0.011b | 0.402 | 0.197–0.819 |
|
CT+TT | 126 (90.0) | 100 (76.9) | 8.451 | 0.004c | 0.370 | 0.186–0.736 |
| Allelotype |
|
|
|
|
|
|
| C | 75 | 92 |
|
| 1.00 |
|
| T | 205 | 168 | 4.666 | 0.031d | 0.668 | 0.463–0.964 |
Effect of rs16917496 SNP on SET8
expression
To identify the association between the rs16917496
SNP and SET8 expression, SET8 expression was measured by
immunostaining in 140 ccRCC tissues. All samples were analyzed for
SET8 staining and an HSCORE was calculated. Patients with the SET8
CC genotype had lower SET8 expression compared with patients with
the CT (χ2=4.238; P=0.038) or TT (χ2=5.741;
P=0.017) genotypes (Table III).
 | Table III.Distribution frequency of SET8
expression levels for each genotype by χ2 test. |
Table III.
Distribution frequency of SET8
expression levels for each genotype by χ2 test.
|
| Expression level,
n |
|
|
|---|
|
|
|
|
|
|---|
| Genotype | Low | High | χ2 | P-value |
|---|
| CC | 8 | 6 |
|
|
| CT | 15 | 39 | 4.283 | 0.038a |
| TT | 18 | 54 | 5.741 | 0.017b |
Associations between SET8 expression
and clinicopathological variables
SET8 was diffusely distributed throughout the
nucleus of ccRCC tumor cells, as determined by immunostaining
(Fig. 1B). Among all of the samples
analyzed, 99 cases (70.7%) demonstrated high SET8 protein
expression, while 41 samples (29.3%) exhibited low expression. In
addition, the χ2 test was used to assess the association
between SET8 protein expression and various clinicopathological
variables (Table IV). Notably, low
SET8 protein expression was negatively associated with ccRCC
Tumor-Node-Metastasis (TNM) (29)
staging (P=0.002), tumor size (P=0.039) and lymph node metastasis
(P=0.014). However, no significant association was observed between
SET8 expression and age or gender (P>0.05). These results
demonstrated that SET8 expression is highly induced in human ccRCC,
indicating a potential role for SET8 in ccRCC development and
progression.
 | Table IV.Association between SET8 expression
and clear cell renal cell carcinoma clinicopathological
features. |
Table IV.
Association between SET8 expression
and clear cell renal cell carcinoma clinicopathological
features.
|
|
| SET8 expression,
n |
|
|
|---|
|
|
|
|
|
|
|---|
|
Characteristics | No. of cases | Low | High | χ2 | P-value |
|---|
| Age, years |
|
|
| 0.761 | 0.383 |
|
<55 | 57 | 19 | 38 |
|
|
|
≥55 | 83 | 22 | 61 |
|
|
| Gender |
|
|
| 1.048 | 0.306 |
|
Male | 59 | 20 | 39 |
|
|
|
Female | 81 | 21 | 60 |
|
|
| TNM
classification |
|
|
| 9.952 | 0.002 |
| I | 96 | 36 | 60 |
|
|
|
II+III+IV | 44 | 5 | 39 |
|
|
| Size of tumor
(diameter, cm) |
|
|
| 4.241 | 0.039 |
|
<5 | 95 | 33 | 62 |
|
|
| ≥5 | 45 | 8 | 37 |
|
|
| LN metastasis |
|
|
| 6.010 | 0.014 |
|
Negative | 99 | 35 | 64 |
|
|
|
Positive | 41 | 6 | 35 |
|
|
SET8-knockdown inhibits proliferation,
colony formation, migration and invasion of renal carcinoma 786-O
cells
For SET8-knockdown, four psi-H1-SET8
siRNAs were transfected into renal carcinoma 786-O cells. As
presented in Fig. 2, SET8
siRNA2 in the psi-H1 plasmid markedly reduced SET8 protein
levels compared with the other SET8 siRNAs. Therefore, the
SET8 siRNA2 construct was selected for subsequent
analyses.
SET8 was reported to be associated with the
development of several tumors (23,30,31). So,
we next examined whether SET8-knockdown affects renal
carcinoma 786-O cell proliferation. Renal carcinoma 786-O cells
were transfected with psi-H1-SET8 siRNA, psi-H1 (empty
vector) or blank control and MTT assays were performed to determine
the proliferation capacity of the cells. Compared with the
psi-H1-transfected cells and blank control cells, the proliferation
rate of renal carcinoma 786-O cells was significantly decreased
between 36 and 72 h following SET8 siRNA-2 transfection (P<0.05;
Fig. 3). The effects of
SET8-knockdown on cell colony formation were studied in
vitro. As presented in Fig. 4,
SET8-knockdown significantly inhibited colony formation as compared
with empty psi-H1 or the blank control. To investigate the
underlying mechanisms by which SET8 regulates proliferation and
colony formation, RT-PCR was performed to examine the expression of
proliferation-associated genes. Compared with cells transfected
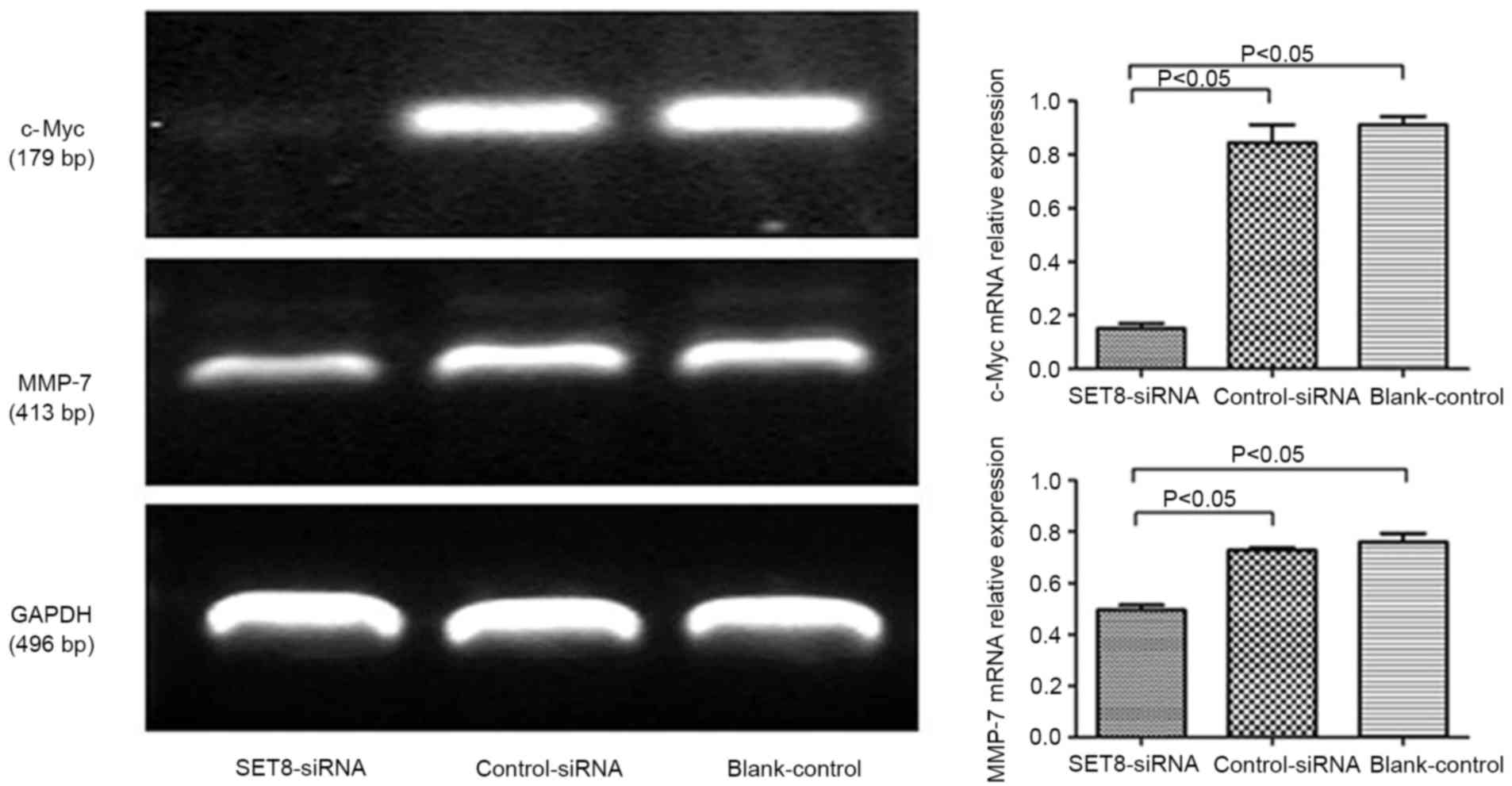
with empty psi-H1, c-Myc mRNA levels significantly decreased upon
SET8 knockdown (Fig. 5). These
results indicated that SET8-knockdown inhibits renal carcinoma
786-O cell proliferation and colony formation. It was speculated
that SET8-knockdown suppresses cell growth by decreasing c-Myc mRNA
expression.
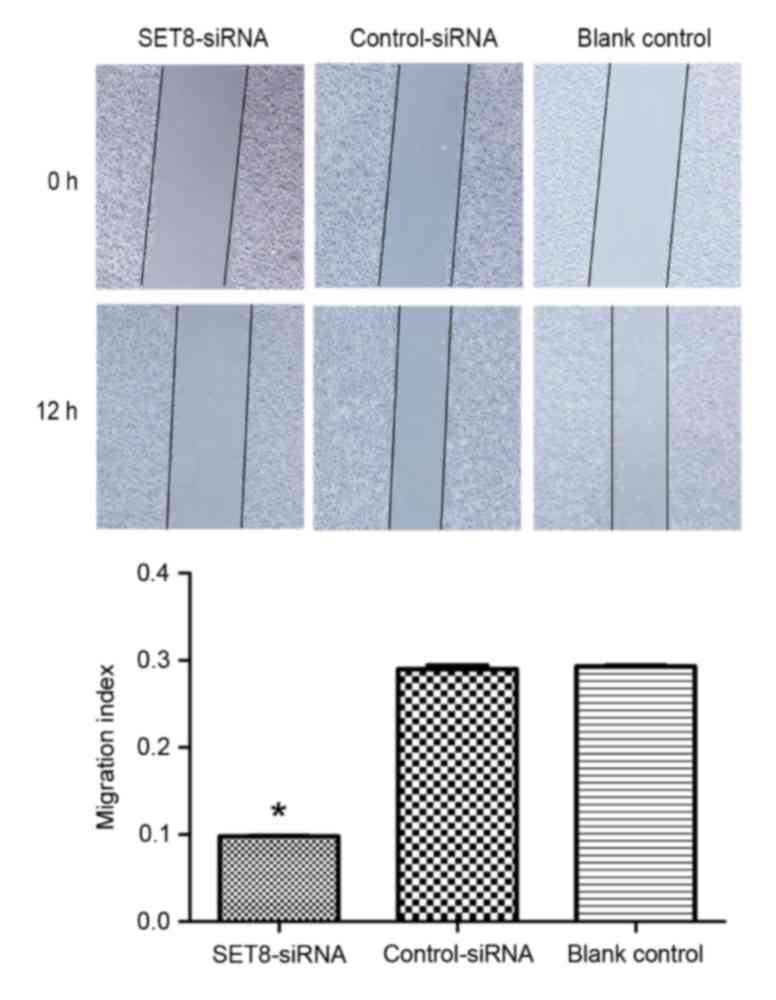
To determine whether SET8 affects cell migration or
invasion, wound healing and Transwell assays were performed,
respectively. As presented in Fig. 6,
SET8-knockdown significantly decreased cell migration capacity
compared with cells transfected with empty vector or blank control
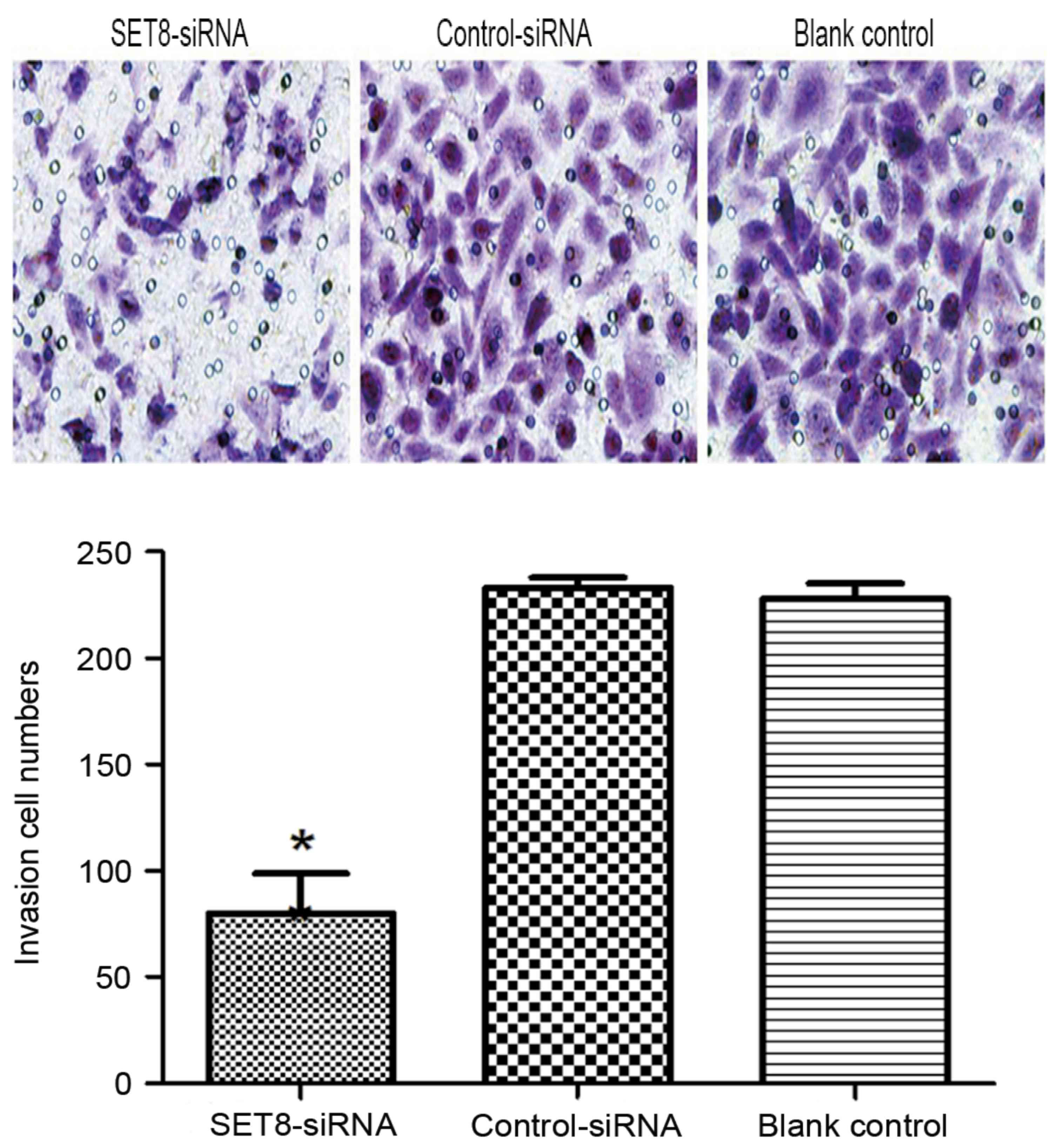
cells (P<0.05). Cell invasion was examined using Transwell
assays and it was revealed that SET8-knockdown markedly decreased
cell invasiveness compared with cells transfected with empty vector
or blank control cells (P<0.05; Fig.
7). To determine the mechanisms by which SET8 regulates
invasion and migration, RT-PCR was performed. Compared with cells
transfected with control-siRNA, SET8-knockdown cells exhibited
significantly decreased matrix metalloproteinase-7 (MMP-7) levels
(Fig. 5). These results indicated
that SET8-knockdown inhibited cell migration and invasion. It was
suggested that SET8-knockdown suppresses cell migration and
invasion by decreasing MMP-7 mRNA expression.
Discussion
In the present study, the association between the
rs16917496 SNP in the miR-502 binding site of the SET8 3′UTR
and SET8 expression was assessed, and its implications in ccRCC
development were investigated. Logistic regression analysis
revealed that the rs16917496 SNP was associated with ccRCC risk.
Therefore, the association between the rs16917496 SNP and SET8
expression was examined. Consistent with the previous study by Song
et al (21), the present study
revealed that the SET8 CC genotype was associated with low
SET8 protein expression. Furthermore, the association between SET8
expression and clinicopathological features was explored, and it
was revealed that SET8 expression was associated with TNM staging,
tumor size and lymph node metastasis. Finally,
SET8-knockdown inhibited proliferation and invasion of renal
carcinoma 786-O cells, potentially through Wnt/β-catenin signaling.
The present data suggested that altering SET8 expression, at least
partially by the rs16917496 SNP in the miR-502 binding site of the
SET8 3′UTR, was associated with ccRCC development and
progression. SET8-knockdown inhibited renal carcinoma 786-O
cell proliferation, migration and invasion potentially through
Wnt/β-catenin signaling.
Accumulating evidence has suggested that
polymorphisms within miRNA-binding sites may affect miRNA
regulation of target gene expression and consequently modify cancer
risk and outcome (11,32–35). The
rs16917496 SNP in the miR-502 binding site of SET8 has been
associated with the risk of several tumor types, including
hepatocellular carcinoma (10), small
cell lung cancer (20) and
non-Hodgkin's lymphoma (30).
Consistent with the study by Song et al (18), the results of the present study
revealed that the CC genotype was associated with low protein
expression and low ccRCC risk. The present results also revealed
that SET8 expression was associated with TNM staging, tumor size
and lymph node metastasis of patients with ccRCC. These data
demonstrated that the rs16917496 SNP in the miR-502 binding site of
SET8 mediated SET8 expression and consequently modified
ccRCC cancer risk.
As a methyltransferase, SET8 may regulate several
signaling pathways by modulating protein lysine methyltransferases.
Of note, the major signaling pathways affected by SET8 are
Wnt/β-catenin (36) and twist
(37), which are important for
development. The Wnt/β-catenin pathway is highly conserved across
metazoans and is essential for a number of cellular functions,
including cell proliferation, migration and invasion (38). The present study demonstrated that
SET8-knockdown inhibited proliferation and invasion by mediating
the expression of the Wnt/β-catenin target genes c-Myc and MMP-7 in
renal carcinoma 786-O cells. However, expression of other Wnt
target genes, including Axin2, naked cuticle homolog 1 and lymphoid
enhancer-binding factor 1, was not detected. The full mechanism of
SET8 regulation of renal carcinoma cell proliferation and invasion
and its associated signaling pathways should be explored
further.
The present results indicated that SNPs of a miRNA
binding site were associated with ccRCC risk, but the results
require validation in other populations and in laboratory-based
functional studies. SET8 may modify cancer development and
progression through its effects on proliferation and invasion,
potentially via Wnt/β-catenin signaling. Therefore, SET8 may be a
novel target for ccRCC therapy.
Acknowledgements
The present study was supported by the project of
the Hebei Natural Science Fund (grant no. H2012206157) and the
project of the Hebei Major Medical Science (grant no. GL2011-51),
and the Project of Hebei Science and Technology Planning
(16397733D).
References
|
1
|
Remon J, Lianes P and Martinez S: Brain
metastases from renal cell carcinoma. Should we change the current
standard? Cancer Treat Rev. 38:249–257. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Salehipoor M, Khezri A, Behzad-Behbahani
A, Geramizadeh B, Rahsaz M, Aghdaei M and Afrasiabi MA: Role of
viruses in renal cell carcinoma. Saudi J Kidney Dis Transpl.
23:53–57. 2012.PubMed/NCBI
|
|
3
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chow WH, Dong LM and Devesa SS:
Epidemiology and risk factors for kidney cancer. Nat Rev Urol.
7:245–257. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Semenza JC, Ziogas A, Largent J, Peel D
and Anton-Culver H: Gene-environment interactions in renal cell
carcinoma. Am J Epidemiol. 153:851–859. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gu L, Li H, Chen L, Ma X, Gao Y, Li X,
Zhang Y, Fan Y and Zhang X: MicroRNAs as prognostic molecular
signatures in renal cell carcinoma: A systematic review and
meta-analysis. Oncotarget. 6:32545–32560. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sellitti DF and Doi SQ: MicroRNAs in renal
cell carcinoma. Microrna. 4:26–35. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Klimczak D, Pączek L, Jażdżewski K and
Kuch M: MicroRNAs: Powerful regulators and potential diagnostic
tools in cardiovascular disease. Kardiol Pol. 73:1–6. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chiang VS: Withdrawn: MicroRNAs as
potential regulators of docosahexaenoic acid benefits in
Alzheimer's disease. Nutr Neurosci. 14–Mar;2015.(Epub Ahead Of
Print). View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mennigen JA, Plagnes-Juan E,
Figueredo-Silva CA, Seiliez I, Panserat S and Skiba-Cassy S: Acute
endocrine and nutritional co-regulation of the hepatic
omy-miRNA-122b and the lipogenic gene fas in rainbow trout,
Oncorhynchus mykiss. Comp Biochem Physiol B Biochem Mol Biol.
169:16–24. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guo Z, Wu C, Wang X, Wang C, Zhang R and
Shan B: A polymorphism at the miR-502 binding site in the
3′-untranslated region of the histone methyltransferase SET8 is
associated with hepatocellular carcinoma outcome. Int J Cancer.
131:1318–1322. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu Y, Cai H, Liu J, Fan H, Wang Z, Wang
Q, Shao M, Sun X, Diao J, Liu Y, et al: A miR-151 binding site
polymorphism in the 3′-untranslated region of the cyclin El gene
associated with nasopharyngeal carcinoma. Biochem Biophys Res
Commun. 432:660–665. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Couture JF, Collazo E, Brunzelle JS and
Trievel RC: Structural and functional analysis of SET8, a histone
H4 Lys-20 methyltransferase. Genes Dev. 19:1455–1465. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fang J, Feng Q, Ketel CS, Wang H, Cao R,
Xia L, Erdjument-Bromage H, Tempst P, Simon JA and Zhang Y:
Purification and functional characterization of SET8, a nucleosomal
histone H4-lysine 20-specific methyltransferase. Curr Biol.
12:1086–1099. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nishioka K, Rice JC, Sarma K,
Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P,
Tempst P, Steward R, et al: PR-Set7 is a nucleosome-specific
methyltransferase that modifies lysine 20 of histone H4 and is
associated with silent chromatin. Mol Cell. 9:1201–1213. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu S, Wang W, Kong X, Congdon LM, Yokomori
K, Kirschner MW and Rice JC: Dynamic regulation of the PR-Set7
histone methyltransferase is required for normal cell cycle
progression. Genes Dev. 24:2531–2542. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huen MS, Sy SM, van Deursen JM and Chen J:
Direct interaction between SET8 and proliferating cell nuclear
antigen couples H4-K20 methylation with DNA replication. J Biol
Chem. 283:11073–11077. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jørgensen S, Elvers I, Trelle MB, Menzel
T, Eskildsen M, Jensen ON, Helleday T, Helin K and Sørensen CS: The
histone methyltransferase SET8 is required for S-phase progression.
J Cell Biol. 179:1337–1345. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tardat M, Murr R, Herceg Z, Sardet C and
Julien E: PR-Set7-dependent lysine methylation ensures genome
replication and stability through S phase. J Cell Biol.
179:1413–1426. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Abbas T, Shibata E, Park J, Jha S, Karnani
N and Dutta A: CRL4 (Cdt2) regulates cell proliferation and histone
gene expression by targeting PR-Set7/Set8 for degradation. Mol
Cell. 40:9–21. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Song F, Zheng H, Liu B, Wei S, Dai H,
Zhang L, Calin GA, Hao X, Wei Q, Zhang W and Chen K: An
miR-502-binding site single-nucleotide polymorphism in the
3′-untranslated region of the SET8 gene is associated with early
age of breast cancer onset. Clin Cancer Res. 15:6292–6300. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ding C, Li R, Peng J, Li S and Guo Z: A
polymorphism at the miR-502 binding site in the 3′ untranslated
region of the SET8 gene is associated with the outcome of
small-cell lung cancer. Exp Ther Med. 3:689–692. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang C, Guo Z, Wu C, Li Y and Kang S: A
polymorphism at the miR-502 binding site in the 3′ untranslated
region of the SET8 gene is associated with the risk of epithelial
ovarian cancer. Cancer Genet. 205:373–376. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Singh M, Zaino RJ, Filiaci VJ and Leslie
KK: Relationship of estrogen and progesterone receptors to clinical
outcome in metastatic endometrial carcinoma: A gynecologic oncology
group study. Gynecol Oncol. 106:325–333. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Merritt WM, Lin YG, Han LY, Kamat AA,
Spannuth WA, Schmandt R, Urbauer D, Pennacchio LA, Cheng JF, Nick
AM, et al: Dicer, Drosha, and outcomes in patients with ovarian
cancer. N Engl J Med. 359:2641–2650. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen L, Qiu J, Yang C, Yang X, Chen X,
Jiang J and Luo X: Identification of a novel estrogen receptor
beta1 binding partner, inhibitor of differentiation-1, and role of
ERbeta1 in human breast cancer cells. Cancer Lett. 278:210–219.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tan B, Li Y, Zhao Q, Fan L, Wang D and Liu
Y: Inhibition of gastric cancer cell growth and invasion through
siRNA-mediated knockdown of guanine nucleotide exchange factor
Vav3. Tumour Biol. 35:1481–1488. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Meng F, Wang F, Wang L, Wong SC, Cho WC
and Chan LW: MiR-30a-5p overexpression may overcome EGFR-inhibitor
resistance through regulating PI3K/AKT signaling pathway in
non-small cell lung cancer cell lines. Front Genet. 7:1972016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Süer E, Baltaci S, Burgu B, Aydoğdu Ö and
Göğüş Ç: Significance of tumor size in renal cell cancer with
perinephric fat infiltration: Is TNM staging system adequate for
predicting prognosis? Urol J. 10:774–779. 2013.PubMed/NCBI
|
|
30
|
Diao L, Su H, Wei G, Li T, Gao Y, Zhao G
and Guo Z: Prognostic value of microRNA 502 binding site SNP in the
3′-untranslated region of the SET8 gene in patients with
non-Hodgkin's lymphoma. Tumori. 100:553–558. 2014.PubMed/NCBI
|
|
31
|
Wang C, Wu J, Zhao Y and Guo Z: miR-502
medaited histone methyltransferase SET8 expression is associated
with outcome of esophageal squamous cell carcinoma. Sci Rep.
6:329212016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Landi D, Gemignani F, Naccarati A, Pardini
B, Vodicka P, Vodickova L, Novotny J, Försti A, Hemminki K, Canzian
F and Landi S: Polymorphisms within micro-RNA-binding sites and
risk of sporadic colorectal cancer. Carcinogenesis. 29:579–584.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gao Y, He Y, Ding J, Wu K, Hu B, Liu Y, Wu
Y, Guo B, Shen Y, Landi D, et al: An insertion/deletion
polymorphism at miRNA-122-binding site in the interleukin-1alpha 3′
untranslated region confers risk for hepatocellular carcinoma.
Carcinogenesis. 30:2064–2069. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Horikawa Y, Wood CG, Yang H, Zhao H, Ye Y,
Gu J, Lin J, Habuchi T and Wu X: Single nucleotide polymorphisms of
microRNA machinery genes modify the risk of renal cell carcinoma.
Clin Cancer Res. 14:7956–7962. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hu Z, Chen J, Tian T, Zhou X, Gu H, Xu L,
Zeng Y, Miao R, Jin G, Ma H, et al: Genetic variants of miRNA
sequences and non-small cell lung cancer survival. J Clin Invest.
118:2600–2608. 2008.PubMed/NCBI
|
|
36
|
Konac E, Varol N, Yilmaz A, Menevse S and
Sozen S: DNA methyltransferase inhibitor-mediated apoptosis in the
Wnt/β-catenin signal pathway in a renal cell carcinoma cell line.
Exp Biol Med (Maywood). 238:1009–1016. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yang F, Sun L, Li Q, Han X, Lei L, Zhang H
and Shang Y: SET8 promotes epithelial-mesenchymal transition and
confers TWIST dual transcriptional activities. EMBO J. 31:110–123.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Clevers H and Nusse R: Wnt/β-catenin
signaling and disease. Cell. 149:1192–1205. 2012. View Article : Google Scholar : PubMed/NCBI
|