Introduction
Breast cancer is the most common cancer and the
first most common cause of cancer-associated mortality in females
worldwide (1). The current treatments
for breast cancer include surgery, chemotherapy, radiotherapy and
hormone therapy, and a small number of patients currently undergo
targeted therapy (2).
The klotho gene family includes klotho α (KLA) and
klotho β (KLB). KLA is an aging-suppressor gene that encodes a type
I membrane protein that is 1,014 amino acids in length (3). The human KLA locus has been assigned to
13q12 (3–5), and has been demonstrated to be a tumor
suppressor in human breast cancer (6). KLB is a single-pass transmembrane
protein of 1,043 amino acids in length that is located on 4p14; it
shares 41.2% homology with KLA. KLB is predominantly expressed in
the liver, adipose tissue and pancreas (7), and serves an important role in the
synthesis and excretion of bile acids. A mutant mouse that lacks
KLB demonstrates increased synthesis and excretion of bile acids
via the elevation of the levels of cytochrome P450 family 7
subfamily A member 1 (CYP7A1) and cytochrome P450 family 8
subfamily B member 1 (CYP8B1) mRNA, which encode the rate-limiting
enzymes for the synthesis of bile acids (8,9).
Fibroblast growth factor (FGF)19 acts as a metabolic
regulator (10). Endocrine FGF19
functions through the FGF receptor (FGFR) and a co-receptor (either
KLA or KLB) (11). KLB may reduce the
level of FGF21 through interacting with FGFR4 and FGFR1c (9,12,13). This suggests that KLB is involved in
the signal transduction of FGFR4, and serves an important role in
the metabolic activity of FGFR4. A previous study demonstrated that
the co-expression and activation of KLB in a complex with FGFR4
induced liver cell apoptosis and inhibited hepatoma cell
proliferation through activating the signal transduction of
extracellular signal-related kinase 1/2, and reducing the signal
transduction of protein kinase B (Akt) (14).
KLB was identified to suppress tumor growth in
hepatocellular carcinoma via the regulation of the Akt/glycogen
synthase kinase 3β (GSK-3β)/cyclin D1 signaling pathway (15). In another previous study, it was
identified that FGFR4 may suppress the development of breast cancer
(16). In addition, KLB shares 41.2%
homology with KLA, and KLA was identified to be a tumor suppressor
in human breast cancer (6).
Therefore, the present study hypothesized that KLB may be involved
in carcinogenesis and may act as a tumor suppressor in breast
cancer. The expression and activities of KLB in the mammary
glandular and in breast cancer have not yet been elucidated. In the
present study, the expression and loss of heterozygosity of KLB in
invasive ductal carcinoma was investigated.
Materials and methods
Tissue microarray (TMA)
An invasive ductal carcinoma TMA (cat. no.,
OD-CT-RpBre01-006; Outdo Biotech Co., Ltd., Shanghai, China) was
performed. The tissue microarray contained tissues from 82 cases,
as 328 specimen cores (each with a diameter of 1 mm and a height of
4 µm), and the integrity of the microarray was >95%. For all 82
cases, each case included two cores of invasive ductal carcinoma
and two cores of paired adjacent non-tumorous breast tissues. All
clinical and pathological data of the specimens were provided by
Outdo Biotech Co., Ltd., including age, tumor size, whether any
axillary lymph node metastasis was present, pathological grade
according to the World Health Organization 2012 classification of
breast tumors (17) and the staining
results of estrogen receptor (ER), progesterone receptor (PR),
human epidermal growth factor receptor-2 (HER2) and Ki-67. A total
of 48 cases were classified as grade 2 and 34 cases were grade 3.
Overall, 37 cases demonstrated lymph node metastasis and 45 cases
demonstrated no lymph node metastasis. The age range of patients
was 33–88 years (median, 54.5 years) and all patients were
female.
Immunohistochemistry
The microarray was washed in 100% xylene (Beijing
Chemical Works, Beijing, China) to remove the paraffin and then
rehydrated through serial dilutions of alcohol (100, 90, 80 and
70%; Beijing Chemical Works) followed by rinsing in water. This was
followed by the quenching of endogenous peroxidase activity using a
0.3% solution of hydrogen peroxide (Beijing Chemical Works) in
methanol for 30 min. For antigen retrieval, the section was boiled
in 0.01 mol/l sodium citrate buffer (pH 6.0; Beijing Chemical
Works) in a microwave oven at 560 W for 15 min. The section was
blocked with 1% normal goat serum (cat no. ZLI-9021; ZSGB-BIO,
Beijing, China) in PBS for 1 h at room temperature then incubated
with anti-β klotho antibody (rabbit polyclonal anti-human; cat no.
109454; 1:500 dilution; LifeSpan BioSciences, Inc., Seattle, WA,
USA) overnight at 4°C. The section was laid at room temperature for
30 min prior to additional analysis the following day. Next, the
section was incubated with peroxidase-conjugated goat anti-rabbit
IgG secondary antibody (cat no. ZB-2301; 1:250 dilution; ZSGB-BIO)
for 30 min. The reaction was visualized using 3,3′-diaminobenzidine
(Beijing Chemical Works) under light microscopy (magnification,
×40) to control optimal dyeing. The section was then counterstained
with hematoxylin (Beijing Chemical Works), dehydrated in graded
ethanol and xylene, and embedded using permount TM mounting medium
(Beijing Chemical Works). The stained TMA was scanned into digital
format with the Leica sSCN400 program (Leica Microsystems GmbH,
Wetzlar, Germany).
The staining was scored according to the staining
intensity and the percentage of the positive tumor cells. The
staining intensity was divided into 0, 1, 2 and 3 points based on
color (no color, faint yellow, brownish yellow and brown,
respectively). The percentage of positive tumor cells was divided
into four levels (<5, 5–25, 26–50, 51–75 and 76–100%),
corresponding to the assignment of 0, 1, 2, 3 and 4 points,
respectively. The product of the scores for staining intensity and
the percentage of the positive tumor cells was then used to divide
the results into four groups as follows: Negative (−), score 0;
mild (+), score 1–4; moderate (++), score 5–8; and marked (+++),
score 9–12.
Microdissection of breast
specimens
The use of all specimens was approved by the Ethics
Committee of Capital Medical University (Beijing, China). Invasive
ductal carcinoma tissues, paired adjacent non-tumorous breast
tissues and lymph nodes were obtained from 42 patients who were
diagnosed with primary breast invasive ductal carcinoma. The
specimens were collected from between January 2007 to December 2012
at Da Xing Hospital of Capital Medical University, and all patients
provided written informed consent. No chemotherapy, radiotherapy or
hormone therapy was administered to the patients prior to
therapeutic resection. All the tissues were fixed in 10% formalin
(Beijing Chemical Works) at room temperature for 24 h, embedded in
paraffin (Beijing Chemical Works) and cut into slices; the
paraffin-embedded tissues were cut into 6 sections, each with a
diameter of 4 µm. Next, all sections were stained using hematoxylin
and eosin. All cases were reviewed by two pathologists who
confirmed the diagnosis of invasive ductal carcinoma and the
grading of the tumors according to the 2012 World Health
Organization criteria (17). A total
of 2 cases were classified as grade 1, 16 cases were classified as
grade 2 and 24 cases were classified as grade 3. Overall, 28 cases
demonstrated lymph node metastasis and 14 cases demonstrated no
lymph node metastasis. Isolating normal epithelial cells and tumor
cells with a needle under an inverted microscope, normal epithelial
cells and tumor cells were collected and placed into Eppendorf
tubes. The same tissue from the same 6 sections were put into the
same tube. The genomic DNA was then extracted using
QIAamp®DNA mini kit (cat no. 51304; Qiagen GmbH, Hilden,
Germany) for loss of heterozygosity (LOH) examination.
LOH
A total of 2 microsatellite markers, D4S251 and
D4S3040, were selected due to their close proximity to the KLB
locus. The 5′ polymerase chain reaction (PCR) primers were labeled
with 6-carboxyfluorescein. The following PCR primers were used:
D4S251 forward, 5′-TATGTATATATGTGTGCGTGCG-3′ and reverse,
5′-TATGTATATATGTGTGCGTGCG-3′; and D4S3040 forward,
5′-AGCCTAAGCCTATCACAATCCAG-3′; and reverse,
5′-CTGATTGGAACCAAGATGTATATATG-3′ (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). PCR was performed as follows:
10 min at 94°C, followed by 30 sec at 95°C, then 40 cycles of 30
sec each at 55°C and 30 sec at 72°C, followed by an extension of 10
min at 72°C, in a 20-µl reaction mixture containing 0.2 µM of each
primer, 2.5 ng/µl DNA and 10 µl 2X EasyTaq® PCR SuperMix
for PAGE (Beijing TransGen Biotech Co., Ltd., Beijing, China). The
PCR products were electrophoresed in an ABI Prism 3730 system
(Applied Biosystems; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol and the data were analyzed using the
GeneMapper 3.2 software (Applied Biosystems; Thermo Fisher
Scientific, Inc.).
Statistics
The statistical analyses were performed with SPSS
19.0 (IBM Corp., Armonk, NY, USA). The correlation between KLB
expression and the clinicopathological parameters was evaluated
using the χ2 test and Spearman's correlation test.
Two-tailed P<0.05 was considered to indicate a statistically
significant difference.
Results
KLB expression is significantly
decreased in invasive ductal carcinoma
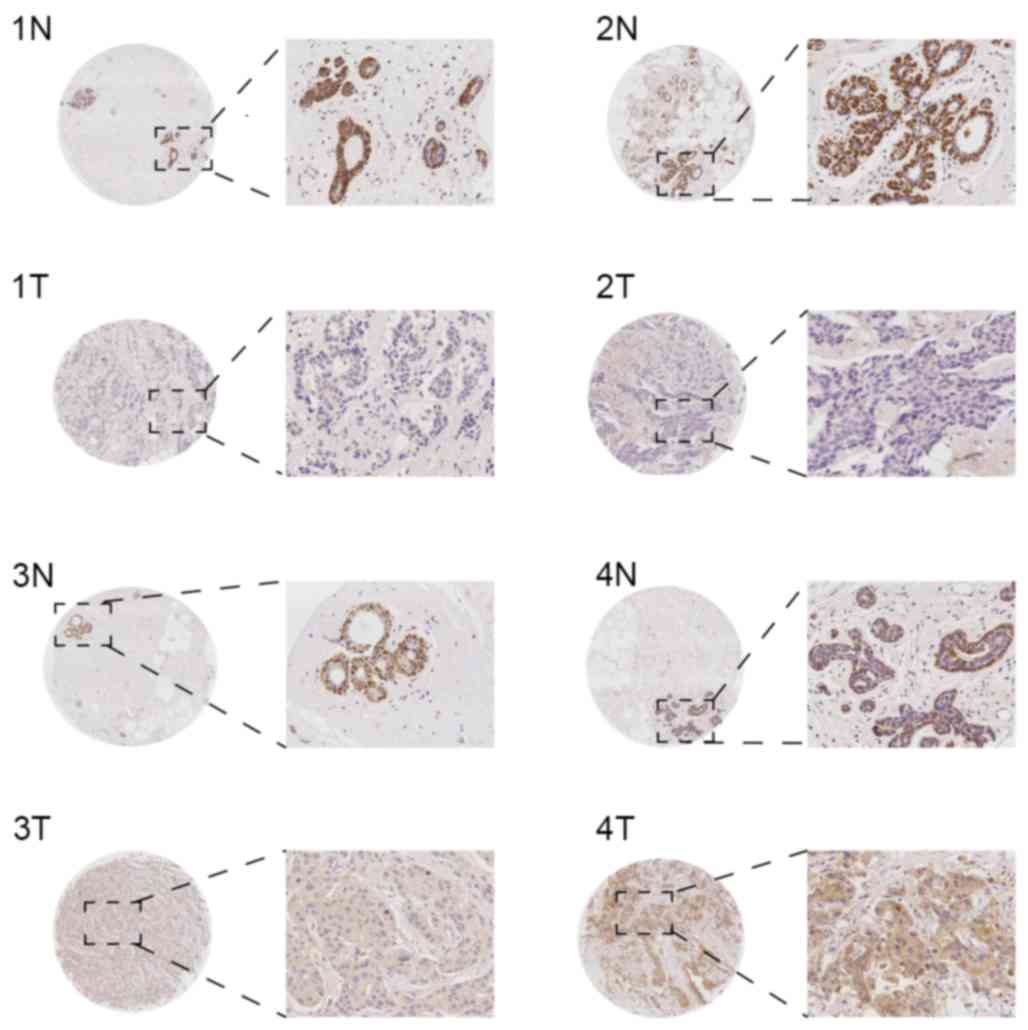
In the invasive ductal carcinoma tissues from the
TMA, KLB was primarily expressed in the cytoplasm, and in a small
number of specimens, KLB was expressed simultaneously in the cell
membrane. In general, the expression was identified to be weak, and
the rates of negative, mild, moderate and marked expression were
43.90 (36/82), 42.69 (35/82), 10.98 (9/82) and 2.44 (2/82),
respectively. In the paired adjacent non-tumorous breast tissues,
KLB was primarily expressed in the cell cytoplasm of myoepithelial
cells, the expression was marked (+++), and the positive expression
rate was 100% (82/82) in the samples. However, mammary glandular
epithelial cytoplasm exhibited weak expression (Fig. 1). Overall, KLB expression was
significantly decreased in invasive ductal carcinoma tissue
compared with the paracancerous tissue (P<0.01).
Downregulation of KLB is associated
with increased lymph node involvement and higher pathological
grade
The correlation between KLB expression and a variety
of clinical features was analyzed. As summarized in Table I, decreased KLB expression was
frequently associated with lymph node involvement (P=0.008) and
pathological grade (P=0.022). Using correlation analysis, decreased
KLB expression was correlated with increased lymph node involvement
(r=−0.234) and higher pathological grade (r=−0.254). However, the
expression of KLB was not associated with the age of the patient
(P=0.551) or with the size of the tumor (P=0.352).
 | Table I.Association between KLB expression and
clinicopathological features of patients with invasive ductal
carcinoma. |
Table I.
Association between KLB expression and
clinicopathological features of patients with invasive ductal
carcinoma.
|
| Score/klotho β
expression, n |
|
|---|
|
|
|
|
|---|
| Features | 0/− | 1–4/+ | 5–8/++ | 9–12/+++ | Overall P-value |
|---|
| Age, years |
|
|
|
|
|
|
>55 | 12 | 19 | 5 | 0 | 0.551 |
| ≤55 | 23 | 19 | 3 | 1 |
|
| Sizes (cm) |
|
|
|
|
|
|
>5 | 11 | 3 | 1 | 1 | 0.352 |
|
>2–5 | 22 | 31 | 8 | 1 |
|
| ≤2 | 2 | 2 | 0 | 0 |
|
| Grade |
|
|
|
|
|
| 2 | 16 | 24 | 6 | 2 | 0.022 |
| 3 | 19 | 12 | 3 | 0 |
|
| Lymph node
involvement |
|
|
|
|
|
|
Negative | 11 | 25 | 8 | 1 | 0.008 |
|
Positive | 22 | 13 | 1 | 1 |
|
| Estrogen
receptor |
|
|
|
|
|
|
Negative | 14 | 8 | 6 | 1 | 0.894 |
|
Positive | 13 | 17 | 4 | 0 |
|
|
Unknown | 10 | 8 | 0 | 1 |
|
| Progesterone
receptor |
|
|
|
|
|
|
Negative | 15 | 17 | 5 | 1 | 0.450 |
|
Positive | 10 | 11 | 3 | 1 |
|
|
Unknown | 10 | 9 | 0 | 0 |
|
| Human epidermal
growth factor receptor-2 |
|
|
|
|
|
|
Negative | 6 | 2 | 2 | 1 | 0.558 |
|
Positive | 21 | 25 | 7 | 0 |
|
|
Unknown | 9 | 8 | 0 | 1 |
|
| Ki-67 |
|
|
|
|
|
| 1 | 4 | 13 | 4 | 0 | 0.162 |
| 2 | 17 | 11 | 2 | 1 |
|
| 3 | 5 | 1 | 1 | 0 |
|
|
Unknown | 10 | 10 | 1 | 2 |
|
However, KLB expression was not associated with the
expression of ER (P=0.894), PR (P=0.450), HER2 (P=0.558) or Ki-67
(P=0.162). The data suggest that KLB expression was also not
associated with the secretion of estrogen or progestin, tyrosine
kinase activities, or the proliferative index.
KLB locus is subject to LOH in
invasive ductal carcinoma
To examine the mechanisms that regulate the
differential expression of KLB in tumors compared with normal
tissue, the occurrence of LOH at this gene locus was examined.
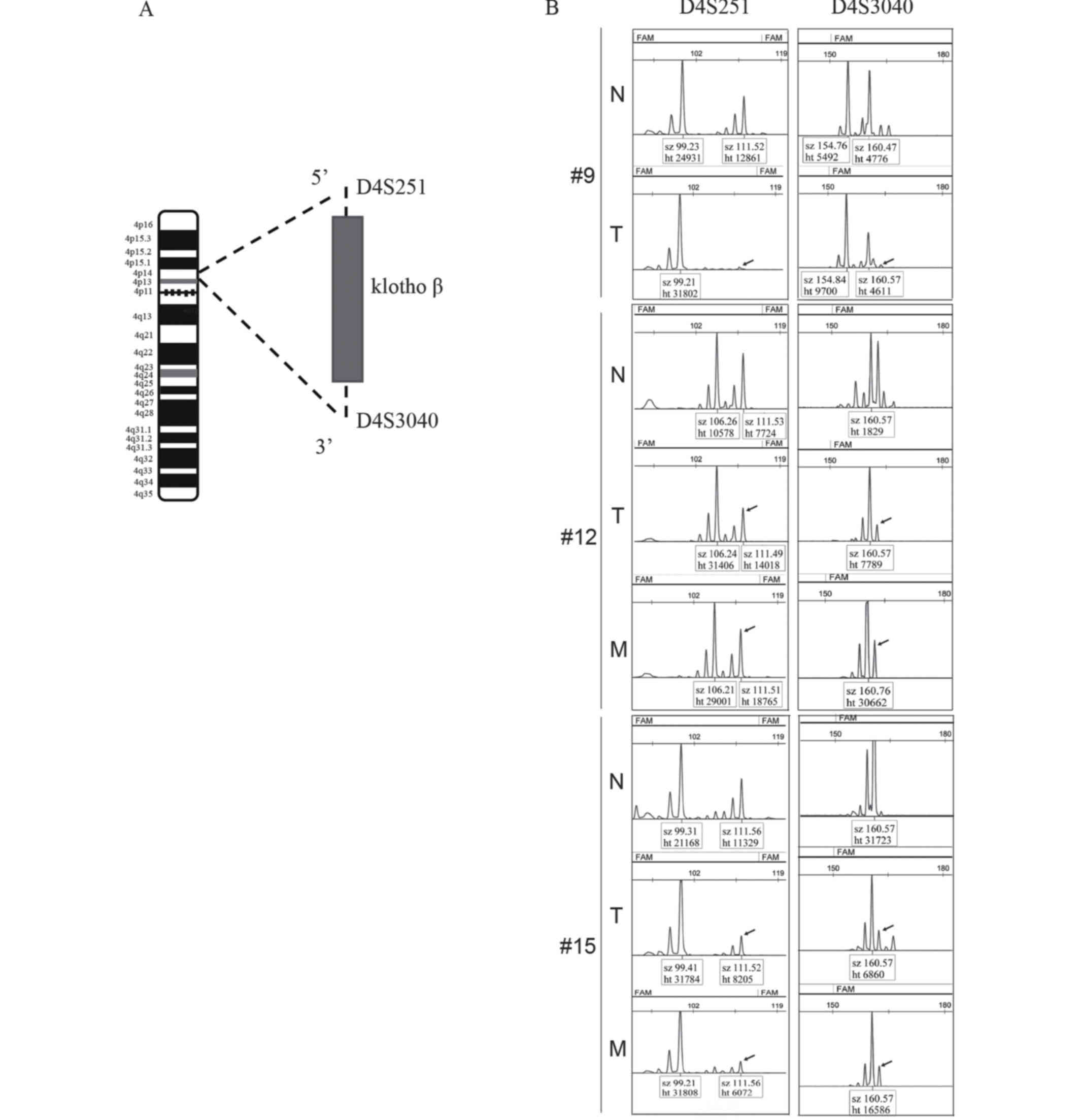
As demonstrated in Fig.
2A, the markers D4S251 and D4S3040 were used to detect LOH in
the patient samples from Da Xing Hospital of Capital Medical
University. As summarized in Table
II, compared with normal tissue, evidence of LOH was identified
in 24/42 tumors (57.14%); additionally, these two markers were lost
in 4/42 tumors (9.52%; #9, #12 and #15 in Fig. 2B). A total of 28/42 demonstrated lymph
node metastasis; 12/28 of these cases (42.86%) exhibited
accompanying LOH (Table II), and the
two aforementioned markers were lost in 2/28 tumors (7.14%; #12 and
#15 in Fig. 2B). No association was
observed between LOH and lymph node metastasis or pathological
grade (P>0.05).
 | Table II.List of LOH in tumor and metastasis
tissues at D4S251 and D4S3040 markers. |
Table II.
List of LOH in tumor and metastasis
tissues at D4S251 and D4S3040 markers.
| Tissue type | Total cases, n | LOH occurrence
rate, n/total n (%) | LOH occurring at
D4S251 and D4S3040 at the same time, n/total n (%) |
|---|
| Tumor | 42 | 24/42 (57.14) | 4/42 (9.52) |
| Associated
metastatic lymph node | 28 | 12/28 (42.86) | 2/28 (7.14) |
Discussion
KLB is predominantly expressed in the liver, adipose
tissue and pancreas (7); however, to
the best of our knowledge, its expression in the breast has not
been reported. It has been demonstrated that KLB, in a complex with
FGFR4, induces liver cell apoptosis and inhibits hepatoma cell
proliferation (14), and KLB was
suggested to suppress tumor growth in hepatocellular carcinoma via
the regulation of the Akt/GSK-3b/cyclin D1 signaling pathway
(15). In another previous study, it
was revealed that FGFR4 may suppress the development of breast
cancer (16). In addition, KLB shares
41.2% homology with KLA, and KLA was suggested to be a tumor
suppressor in human breast cancer (6). Therefore, the role of KLB in breast
cancer was the focus of the present study.
As demonstrated, KLB expression is significantly
decreased in invasive ductal carcinoma, and the expression was
identified to be weak; the rates of negative and mild expression
were 43.9 and 42.69%, respectively. KLB was primarily expressed in
the cytoplasm, and in a small number of specimens, it was expressed
simultaneously in the cell membrane. In the paracancerous tissue,
KLB was primarily expressed in the cell cytoplasm of myoepithelial
cells and the expression was marked, however, mammary glandular
epithelial cytoplasm exhibited weak expression. This type of KLB
expression in invasive ductal carcinoma has not been reported.
In general, tumor cell heterogeneity in the
evolutionary process is characterized by differences in morphology
and metastasis ability (18). KLB
expression in invasive ductal carcinoma tissue is negatively
correlated with lymph node metastasis and histological grade;
therefore, the weaker the expression of KLB, the greater the level
of lymph node metastasis and the higher the histological grade.
This also suggests that KLB may be associated with the
heterogeneity, progress and prognosis of invasive ductal
carcinoma.
As indicated, in the paracancerous tissue, KLB was
primarily expressed in the cell cytoplasm of myoepithelial cells
and the expression was marked. Mammary glandular epithelial
cytoplasm only exhibited weak expression, therefore, KLB may serve
as a biomarker for breast malignancies. It is known that invasive
ductal carcinoma originates in the mammary glandular epithelial
cells, as there are no myoepithelial cells in tumor tissue.
Therefore, the mechanism of this aforementioned expression
phenomenon remains to be discussed.
LOH is a common anomaly in the DNA of tumor cells,
and it may result in tumor suppressor gene inactivation and then
participate in tumor occurrence and development (19). At present, there are no studies
concerning the LOH of KLB in invasive ductal carcinoma. In the
present study, D4S251 and D4S3040 were selected as KLB markers, and
LOH was detected in 57.1% of cases. This indicates that LOH may be
the mechanism that results in KLB gene inactivation.
There have been a small number of studies
investigating KLB function in cancer, which identified that KLA may
restrain the progress of breast, lung and kidney cancer (6,7,20,21).
Additionally, KLB was suggested to suppress tumor growth in
hepatocellular carcinoma (15), and
it was revealed that FGFR4 may suppress the development of breast
cancer (16), therefore, the present
study hypothesized that KLB may inhibit the progress of breast
cancer. In our previous unpublished study, the stable breast cancer
cell line (MDA-MB-231) in which KLB is overexpressed was
successfully cultured. A series of cellular functional experiments,
including an MTT viability assay, scratch tests and a colony
formation assay, were also performed. The experimental group with
KLB overexpression was not able to inhibit cell proliferation
compared with the control group. It was hypothesized that KLB may
serve a role in the suppression of tumor growth in breast cancer,
but that this effect would be weak. In a previous study, the
differential mRNA expression of sushi, von Willebrand factor type
A, EGF and pentraxin domain containing 1, latrophilin 3, KLB,
integrin subunit α7, semaphorin 3G, tensin 1 and matrix
metalloproteinase 13 genes was examined in breast cancer using
reverse transcription-quantitative polymerase chain reaction, and
it was demonstrated that the expression of KLB decreased, but that
the amplitude was not marked (22).
These data are consistent with the results of the present study.
Invasive ductal carcinoma originates in the mammary glandular
epithelial cells, and the results of the present study indicated
that mammary glandular epithelial cytoplasm exhibited weak KLB
expression in paracancerous tissue and that KLB expression was weak
in invasive ductal carcinoma tissue, which suggests that the
hypothesis that KLB suppresses progress in invasive ductal
carcinoma requires additional analysis. KLB has been indicated to
suppress tumor growth in hepatocellular carcinoma (15,16),
however, an additional study demonstrated that KLB expression was
frequently upregulated in HCC and that the silencing of KLB
expression decreases HCC cell growth (23), therefore, the exact role of KLB in
cancer remains unclear.
In summary, KLB expression was decreased in invasive
ductal carcinoma in the present study, and this downregulation was
correlated with a higher degree of pathology and increased lymph
node metastasis. There was a high frequency of LOH in the KLB gene
location, and LOH may be the mechanism that resulted in KLB gene
inactivation. KLB may also serve as a particular marker of
myoepithelial cells. KLB studies have only investigated its role in
HCC, but the role of KLB in the pathogenesis of other malignant
diseases should also be investigated.
Acknowledgements
The authors would like to thank Dr Haye Ding
(General Hospital of Beijing Military Region, Beijing, China) for
technical assistance. The present study was supported by the
National Natural Science Foundation of China (grant no.
81172519).
References
|
1
|
Ferlay J, Soerjomataram I, Ervik M,
Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D and
Bray F: GLOBOCAN 2012 v1.0, Estimated cancer incidence, mortality
and prevalence worldwide: IARC CancerBase No. 11
(Internet)International Agency for Research on Cancer. Lyon,
France: 2014, http://globocan.iarc.fr/pages/fact_sheets_population.aspx?country=900October
9–2014
|
|
2
|
National Cancer Institute: Breast Cancer
Treatment (PDQ®)–Patient Version. https://www.cancer.gov/types/breast/patient/breast-treatment-pdqUpdated
May 5, 2017.
|
|
3
|
Kuro-o M, Matsumura Y, Aizawa H, Kawaguchi
H, Suga T, Utsugi T, Ohyama Y, Kurabayashi M, Kaname T, Kume E, et
al: Mutation of the mouse klotho gene leads to a syndrome
resembling ageing. Nature. 390:45–51. 1997. View Article : Google Scholar
|
|
4
|
Matsumura Y, Aizawa H, Shiraki-Iida T,
Nagai R, Kuro-o M and Nabeshima Y: Identification of the human
Klotho gene and its two transcripts encoding membrane and secreted
Klotho protein. Biochem Biophys Res Commun. 242:626–630. 1998.
View Article : Google Scholar
|
|
5
|
Shiraki-Iida T, Aizawa H, Matsumura Y,
Sekine S, Iida A, Anazawa H, Nagai R, Kuro-o M and Nabeshima Y:
Structure of the mouse klotho gene and its two transcripts encoding
membrane and secreted protein. FEBS Lett. 424:6–10. 1998.
View Article : Google Scholar
|
|
6
|
Wolf I, Levanon-Cohen S, Bose S, Ligumsky
H, Sredni B, Kanety H, Kuro-o M, Karlan B, Kaufman B, Koeffler HP
and Rubinek T: Klotho: A tumor suppressor and a modulator of the
IGF-1 and FGF pathways in human breast cancer. Oncogene.
27:7094–7105. 2008. View Article : Google Scholar
|
|
7
|
Ito S, Kinoshita S, Shiraishi N, Nakagawa
S, Sekine S, Fujimori T and Nabeshima YI: Molecular cloning and
expression analyses of mouse betaklotho, which encodes a novel
Klotho family protein. Mech Dev. 98:115–119. 2000. View Article : Google Scholar
|
|
8
|
Ito S, Fujimori T, Furuya A, Satoh J and
Nabeshima Y and Nabeshima Y: Impaired negative feedback suppression
of bile acid synthesis in mice lacking betaKlotho. J Clin Invest.
115:2202–2208. 2005. View
Article : Google Scholar
|
|
9
|
Ogawa Y, Kurosu H, Yamamoto M, Nandi A,
Rosenblatt KP, Goetz R, Eliseenkova AV, Mohammadi M and Kuro-o M:
BetaKlotho is required for metabolic activity of fibroblast growth
factor 21. Proc Natl Acad Sci USA. 104:pp. 7432–7437. 2007,
View Article : Google Scholar
|
|
10
|
Fukumoto S: Actions and mode of actions of
FGF19 subfamily members. Endocr J. 55:23–31. 2008. View Article : Google Scholar
|
|
11
|
Tomiyama K, Maeda R, Urakawa I, Yamazaki
Y, Tanaka T, Ito S, Nabeshima Y, Tomita T, Odori S, Hosoda K, et
al: Relevant use of Klotho in FGF19 subfamily signaling system in
vivo. Proc Natl Acad Sci USA. 107:pp. 1666–1671. 2010, View Article : Google Scholar
|
|
12
|
Kurosu H, Choi M, Ogawa Y, Dickson AS,
Goetz R, Eliseenkova AV, Mohammadi M, Rosenblatt KP, Kliewer SA and
Kuro-o M: Tissue-specific expression of betaKlotho and fibroblast
growth factor (FGF) receptor isoforms determines metabolic activity
of FGF19 and FGF21. J Biol Chem. 282:26687–26695. 2007. View Article : Google Scholar
|
|
13
|
Lin BC, Wang M, Blackmore C and Desnoyers
LR: Liver-specific activities of FGF19 require Klotho beta. J Biol
Chem. 282:27277–27284. 2007. View Article : Google Scholar
|
|
14
|
Luo Y, Yang C, Lu W, Xie R, Jin C, Huang
P, Wang F and McKeehan WL: Metabolic regulator betaKlotho interacts
with fibroblast growth factor receptor 4 (FGFR4) to induce
apoptosis and inhibit tumor cell proliferation. J Biol Chem.
285:30069–30078. 2010. View Article : Google Scholar
|
|
15
|
Ye X, Guo Y, Zhang Q, Chen W, Hua X, Liu
W, Yang Y and Chen G: βKlotho suppresses tumor growth in
hepatocellular carcinoma by regulating Akt/GSK-3β/Cyclin D1
signaling pathway. PLoS One. 8:e556152013. View Article : Google Scholar
|
|
16
|
Zhu X, Zheng L, Asa SL and Ezzat S: Loss
of Heterozygosity and DNA methylation affect germline fibroblast
growth factor receptor 4 polymorphism to direct allelic selection
in breast cancer. Am J Pathol. 177:2860–2869. 2010. View Article : Google Scholar
|
|
17
|
Lakhani Sunil R: International Agency for
Research on CancerWorld Health Organization. Lyon: International
Agency for Research on Cancer; 2012
|
|
18
|
Marusyk A, Almendro V and Polyak K:
Intra-tumour heterogeneity: A looking glass for cancer? Nat Rev
Cancer. 12:323–334. 2012. View
Article : Google Scholar
|
|
19
|
Yang CY, Lu RH and Lin CH, Jen CH, Tung
CY, Yang SH, Lin JK, Jiang JK and Lin CH: Single nucleotide
polymorphisms associated with colorectal cancer susceptibility and
loss of heterozygosity in a taiwanese population. PLoS One.
9:e1000602014. View Article : Google Scholar
|
|
20
|
Chen B, Wang X, Zhao W and Wu J: Klotho
inhibits growth and promotes apoptosis in human lung cancer cell
line A549. J Exp Clin Cancer Res. 29:992010. View Article : Google Scholar
|
|
21
|
Doi S, Zou Y, Togao O, Pastor JV, John GB,
Wang L, Shiizaki K, Gotschall R, Schiavi S, Yorioka N, et al:
Klotho inhibits transforming growth factor-beta1 (TGF-beta1)
signaling and suppresses renal fibrosis and cancer metastasis in
mice. J Biol Chem. 286:8655–8665. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kotepui M, Thawornkuno C,
Chavalitshewinkoon-Petmitr P, Punyarit P and Petmitr S:
Quantitative real-time RT-PCR of ITGA7, SVEP1, TNS1, LPHN3, SEMA3G,
KLB and MMP13 mRNA expression in breast cancer. Asian Pac J Cancer
Prev. 13:5879–5882. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Poh W, Wong W, Ong H, Aung MO, Lim SG,
Chua BT and Ho HK: Klotho-beta overexpression as a novel target for
suppressing proliferation and fibroblast growth receptor-4
signaling in hepatocellular carcinoma. Mol Cancer. 11:142012.
View Article : Google Scholar : PubMed/NCBI
|