Introduction
Lung cancer is one of the most prominent causes of
cancer-associated mortality in the world (1). Although the molecular network of lung
carcinogenesis was partly understood, the high mortality rate has
not markedly changed. Of the lung cancer cases diagnosed, ~85% are
non-small cell lung cancer (NSCLC), which has a poor prognosis and
is difficult to treat (2). Despite
years of research (3,4), the survival rate of patients with NSCLC
remains poor. Therefore, understanding the pathogenesis of lung
cancer is important to those patients with NSCLC.
CXC receptor 4 (CXCR4), considered as the only
receptor of stromal-derived-factor-1 (also termed CXCL12), is an
α-chemokine receptor (5). A number of
studies have hypothesized that CXCR4 and CXCL12 increase the
adhesive ability of tumors, as well as the degradation of the
extracellular matrix (ECM) and basement membrane, which is
beneficial to the invasion and metastasis of cancer cells (6–9). Kim et
al (10) reported that LECs
promote the migration of CXCR4-positive tumor cells by secretion of
CXCL12. In turn, previous studies demonstrated that vascular
endothelial growth factor increases the CXCR4 expression of
endothelial cells, increases CXCL12 endothelial cell activity,
promotes the generation of blood and lymphatic vessels and induces
tumor cells to specific organs (11,12).
Epidermal growth factor receptor (EGFR), a type of transmembrane
glycoprotein, exists on the cell surface and is encoded by the
proto-oncogene c-erb-B1 (13).
Ligand-mediated EGFR signaling, such as the
RAS/extracellular-signal-regulated kinase (ERK) and
phosphoinositide 3-kinase (PI3K)/protein kinase B pathways,
regulates various cellular processes, including cell survival,
death, growth, proliferation and motility (14). T798M mutations lead to EGFR
overexpression or over-activity in breast cancer (15). Patients with NSCLC who exhibit
co-expression of CXCR4 and EGFR achieve a more aggressive clinical
progression of cancer (16).
Furthermore, studies have revealed that two receptors, CXCR4 and
EGFR, can enhance the capacity of cell chemotaxis and increase the
invasion and metastatic ability of tumors (16,17).
However, the molecular mechanism by which CXCR4 and EGFR promote
invasion and migration remains unclear.
Invasion and metastasis are the main characteristics
of tumors and are responsible for the poor prognosis of advanced
NSCLC, which involves multiple processes, including the
downregulation of adhesion between cells and the ECM and
degradation of the ECM, leading to invasion of new tissues and
finally resulting in colonization (18). Matrix metalloproteinases (MMPs) are a
large family of zinc-dependent peptidases, which perform important
roles in tumor metastasis by degradation of the ECM proteins
(19). MMP-9 is one of the important
MMPs highly expressed in lung tumor cells, and its expression is
associated with invasiveness, tumor growth and angiogenesis
(19,20). MMP-9 expression increases in the
tissues of various malignant cancers, including lung, breast and
head and neck cancers (21–23). Positive immunostaining of MMP-9 in
NSCLC has an independent prognostic value for the diagnosis of
distant metastasis or local recurrence (24). The present study aimed to detect the
association between CXCR4 and MMP-9. The objective of the present
study was to verify whether overexpression of CXCR4 promotes
invasion and migration of NSCLC via EGFR and matrix
metallopeptidase 9 (MMP-9) and to detect any association between
CXCR4, EGFR and MMP-9.
Materials and methods
Antibodies
Monoclonal antibodies against the following were
used: CXCR4 (catalog no. ab1760; Abcam, Cambridge, UK); EGFR
(catalog no. sc-3049; Santa Cruz Biotechnology, Inc., Dallas, TX,
USA); β-tubulin (catalog no. 560340; BD Pharmingen; BD Biosciences,
San Jose, CA, USA); and MMP-9 antibody (5G3; catalog no. ab119906;
Abcam). All the antibodies were diluted at 1:1,000. Anti-mouse
(catalog no. sc-2371; Santa Cruz Biotechnology, Inc., Dallas, TX,
USA) and anti-rabbit secondary antibodies (catalog no. sc-516087;
Santa Cruz Biotechnology, Inc.), conjugated to horseradish
peroxidase for immunoblotting or conjugated to fluorescein
isothiocyanate, were obtained from Jackson ImmunoResearch
Laboratories, Inc., West Grove, PA, USA.
Cell culture and transfection
The human lung adenocarcinoma A549 cell line
(catalog no. 86012804; Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) was cultured at 37°C in a humidified atmosphere of 5%
CO2. Cells were maintained in RPMI-1640 (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) with 10% fetal bovine
serum (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), 1%
penicillin and streptomycin (Gibco; Thermo Fisher Scientific,
Inc.). Briefly, for constitutive expression of CXCR4,
5×105 A549 cells were transfected using
Lipofectamine® 2000 reagent, with CXCR4 in a pcDNA3
vector (catalog no. 13031; Addgene, Inc., Cambridge, MA, USA), and
the stably transfected cells were selected with G418 to yield the
CXCR4 cell line. Similarly, the mock-transfected cell line was
generated from A549 cells using an empty vector.
For RNA interference analysis, the sequences of the
small interfering RNAs (siRNAs) were as follows: CXCR4 siRNA sense,
5′-CCGACCUCCUCUUUGUCAUTT-3′; negative control siRNA sense,
5′-UUCUCCGAACGUGUCACGUTT-3′. siRNA targeting human CXCR4 was
delivered into CXCR4 cells using Lipofectamine® 2000
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. In addition, the non-targeting
(scramble) siRNA pool was used at the same concentration (100 nM)
as a control for the RNA interference assays. At 48 h
post-transfection (37°C), cells were subjected to migration and
invasion assays as described below or cell lysates were collected
and analyzed by western blot analysis.
Migration and invasion assays
In the Transwell migration assay, as previously
described (21,25), the underside of the Transwell insert
(pore size, 8 µm; Costar; Corning Incorporated, Corning, NY, USA)
was pre-coated with collagen type I (Col-I, 1 µg/ml) or fibronectin
(10 µg/ml). Subsequently, 2×105 A549 and CXCR4 cells
were loaded onto the upper chamber of the Transwell and the lower
chamber was filled with serum-free medium. Cells were incubated for
4 h at 37°C, fixed with 4% paraformaldehyde for 10–15 min at room
temperature and stained with 0.1% crystal violet for 30 min at
37°C. Non-migrating cells retained on the upper side were removed
by wiping with a cotton swab. Cells that had migrated through the
filter were counted and averaged from three randomly selected
optical microscopic fields (20X objective).
In vitro invasion assays were performed using
Transwell inserts (Corning Incorporated), which consisted of cell
culture inserts containing 8 µm pore size filters. Briefly, the
Transwell inserts were coated with Matrigel (1 mg/ml; BD
Biosciences) or Col-I (1.2 mg/ml, Sigma-Aldrich; Merck KGaA,
catalog no. SAB4200678), and allowed to gel at 37°C for 1 h.
Subsequently, A549 cells (3×105) in 200 µl serum-free
Dulbecco's modified Eagle's medium (Biowest, Nuaillé, France) were
added to the top of the Transwell. Serum-free medium was then added
to the lower chamber and incubated for 24 h at 37°C. Cells were
fixed and stained with 0.1% crystal violet as previously. Matrigel
and associated cells were removed with a cotton swab. Cells that
had penetrated the Matrigel and had reached the underside of the
filter membrane were then counted and averaged from 3 randomly
selected microscopic fields with an optical microscope
(magnification, ×200).
Western blotting
The conditioned medium without FBS was concentrated
in a 50 ml centrifuge tube. Cells (106) were extracted
using lysis buffer [50 mmol/l Tris (pH 7.5), 500 mmol/l NaCl, 1%
Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 10 mmol/l MgCl2
and complete protease inhibitor mixture (Roche Molecular
Diagnostics, Pleasanton, CA, USA)]. Protein concentrations were
determined with the bicinchoninic acid protein assay kit (Pierce;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. Protein (10 µg per lane) was separated using 10%
SDS-PAGE. Following transferal onto nitrocellulose membranes (EMD
Millipore, Billerica, MA, USA). Non-specific binding was blocked by
the 5% skimmed milk solution at 37°C for 45 min. Then proteins were
probed with primary antibodies against CXCR4, EGFR or β-tubulin
overnight at 4°C and secondary horseradish peroxidase-coupled
antibodies at room temperature for 45 min. Following 3 washes with
phosphate-buffered saline, blots were developed using an enhanced
chemiluminescence system (GE Healthcare Life Sciences, Chalfont,
UK).
Gelatinolytic zymography
Gelatinolytic zymography of MMP-9 activity was
performed using SDS-PAGE (7.5% gel) containing 2.56 mg/ml gelatin.
A mix of one part sample with one part Tris-glycine SDS 2X Sample
Buffer was left to stand for 10 min at room temperature. Gels were
run until completion, indicated by the bromophenol blue tracking
dye reaching the bottom of the gel, incubated in the zymogram
renaturing buffer [2.5% Triton X-100, 50 mmol/l Tris-HCl, 5 mmol/l
CaCl2 (pH 7.6)]. Subsequently, gentle agitation for 30
min at room temperature was performed and the renaturing buffer was
replaced with 1X zymogram developing buffer [0.5 M Tris-HCl (pH
7.45), 2.0 M NaCl, 50 mM CaCl2, 0.2% Brij-35]. Gels were
gently agitated for 30 min at room temperature and fresh 1X
zymogram developing buffer was added prior to incubation at 37°C
for 4 h. Staining was performed using 0.5% (w/v) Coomassie Blue
R-250 (Beijing Solarbio Science & Technology Co., Ltd.,
Beijing, China) for 30 min. Gels were destained with an appropriate
Coomassie Blue R-250 destaining solution (methanol, acetic acid and
water, 50:10:40). Areas of protease activity appeared as clear
bands against a dark blue background where the protease had
digested the substrate.
Patients and specimens
Tissue from surgery was collected from 61 patients
from the People's Hospital of Zhangjiajie City (Zhangjiajie,
China), who were confirmed by pathological diagnosis of lung cancer
without radiotherapy and chemotherapy treatment. The study was
approved by the ethics committee of the People's Hospital of
Zhangjiajie City and patients provided written informed consent.
The study included 36 males and 25 females, whereby 41 individuals
were ≥60 years old and 20 individuals were <60 years old. Of
these patients, according to the classification standard of the
World Health Organization of lung cancer tumor histological grade
(26), 16 cases of poorly
differentiated carcinoma were identified, as well as 45 cases of
well/moderately differentiated carcinoma. Following surgery, the
tissues were formalin-fixed and paraffin-embedded. The
characteristics of the patients are summarized in Table I.
 | Table I.Summary of baseline patient
characteristics (n=61). |
Table I.
Summary of baseline patient
characteristics (n=61).
| Parameter | Value |
|---|
| Age |
|
| Range,
years | 41–82 |
| Median,
years | 61.1 |
| <60,
n (%) | 20 (33) |
| ≥60, n
(%) | 41 (67) |
| Sex, n (%) |
|
|
Male | 36 (59) |
|
Female | 25 (41) |
| Tumor
differentiation, n (%) |
|
|
Well/moderate | 45 (74) |
|
Poor | 16 (26) |
| Lymph node
metastasis, n (%) |
|
| No | 31 (51) |
|
Yes | 30 (49) |
H&E staining and
immunohistochemical technique
The lesions were visualized with hematoxylin and
eosin (H&E) staining. Immunohistochemical analysis for CXCR4,
EGFR and MMP-9 was performed with 5-µm formalin-fixed,
paraffin-embedded tissue sections using a standard
immunohistochemical technique as previously described (27).
Survival analysis
The duration of overall survival was calculated from
the date of first diagnosis of the disease to the date of mortality
or the last follow-up. The Kaplan-Meier method was used for
survival analysis, and Cox proportional hazards models were used to
evaluate the association between the expression of CXCR4, EGFR and
MMP-9 and the prognosis of patients with NSCLC. Significant
prognostic variables, including patient age, tumor stage and type
of treatment were included in these models. Cox regression plots
were constructed for CXCR4+ vs. CXCR4−,
EGFR+ vs. EGFR− and MMP-9+ vs.
MMP-9−.
Statistical analysis
SPSS 19.0 statistical software was used for the
analysis of the image data (IBM SPSS, Armonk, NY, USA). The data
were presented as the mean ± standard deviation. The χ2
test was applied to analyze the association between CXCR4, EGFR and
MMP-9 and the clinical features of NSCLC. Spearman's rank
correlation analysis was applied to detect the correlation among
CXCR4, EGFR and MMP-9. P<0.05 was considered to indicate a
statistically significant difference.
Results
CXCR4 overexpression enhances cell
motility and invasion
To investigate the role of CXCR4 in invasiveness and
metastatic potential, CXCR4 was overexpressed in the lung cancer
A549 cell line, which normally expresses background levels of CXCR4
protein (28). The CXCR4 cell line
represents a stable population of cells derived by transfection
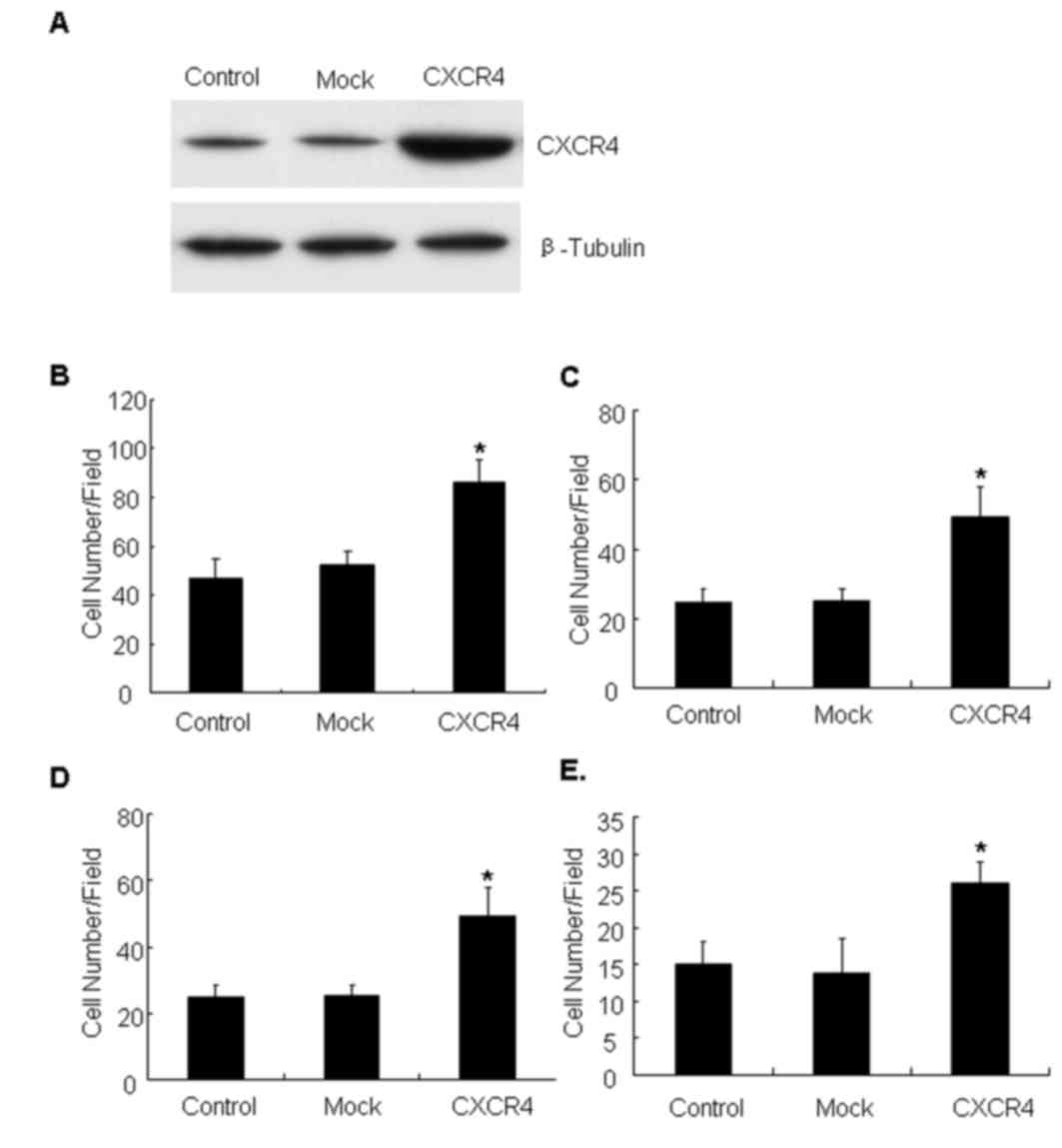
with an experimental plasmid (Fig.
1A). The migration and invasion ability of CXCR4-overexpressing
cells was evaluated, and it was identified that CXCR4 cells
migrated faster than the parental or mock controls on Col-I
(Fig. 1B) and fibronectin substrates
(Fig. 1C) in Transwell assays. The
invasive capacity of the cell line through the ECM barrier was then
examined. The results demonstrated that CXCR4 cells obtained a
marked increase in invasion compared with control cells towards
Matrigel (Fig. 1D) and Col-I
(Fig. 1E). Together, these data
indicated that CXCR4 expression enhanced motility and invasive
ability of lung carcinoma cells.
CXCR4 promotes expression of EGFR and
elevates MMP-9 production
To define the mechanism of CXCR4-enhanced cell
motility, critical proteins known to regulate cell migration and
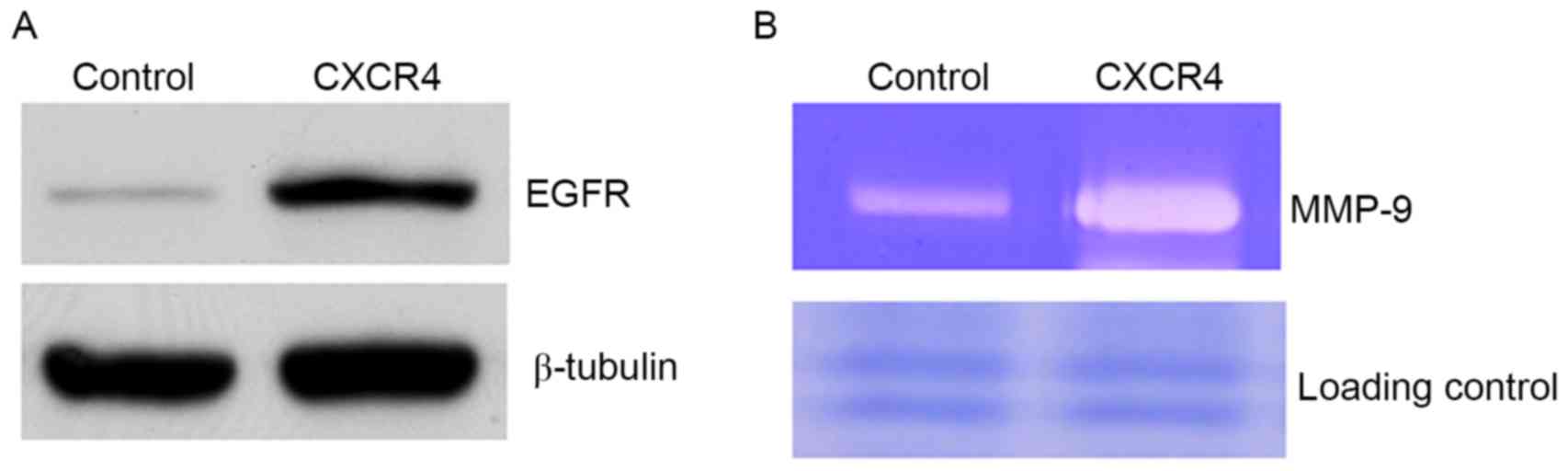
invasion were examined. EGFR was identified to be increased in
CXCR4 cells compared with controls (Fig.
2A). The constitutive expression of EGFR in metastatic tumor
cells has previously been demonstrated to be associated with
enhanced MMP-9 expression and induction of invasive capacity
(25). In the present study, it was
observed that CXCR4-overexpressing cells elevated MMP-9 production
compared with controls (Fig. 2B). To
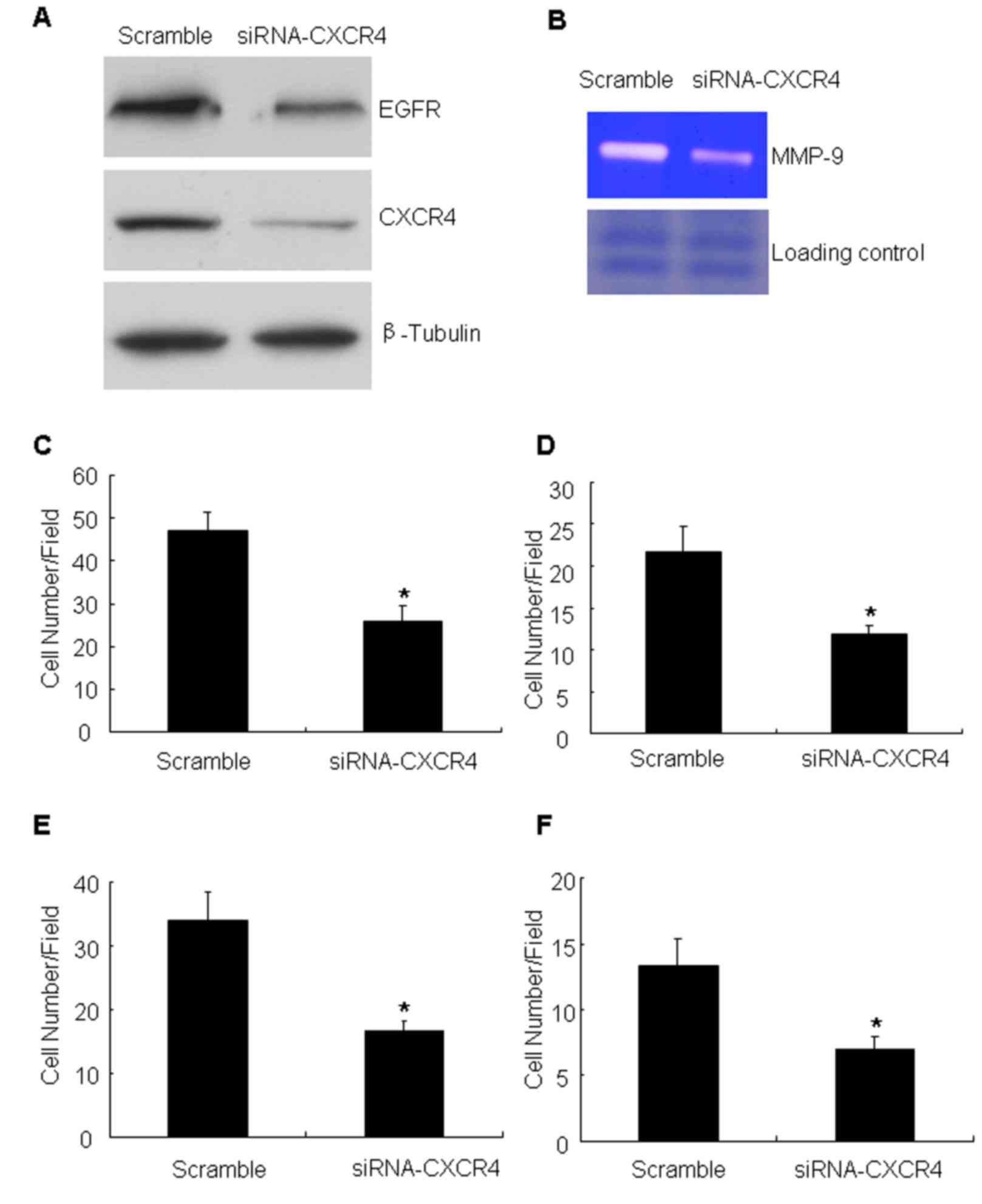
verify the aforementioned results, the effect of CXCR4 knockdown
was analyzed by targeting with specific siRNA. The siRNA was
efficient in reducing CXCR4 compared with the scramble
oligonucleotide (the non-targeting siRNA) (Fig. 3A). Having confirmed the CXCR4
knockdown, the expression of EGFR and MMP-9 production and the
migration and invasion ability of these cells was measured. The
data demonstrated that CXCR4 knockdown inhibited the expression of
EFGR (Fig. 3A). At the same time,
knockdown of CXCR4 was identified to markedly attenuate MMP-9
production compared with controls (Fig.
3B). It was also detected that the number of migrating cells
following CXCR4 suppression was significantly decreased compared
with that of the untreated cells (Fig. 3C
and D). Cell invasion assays with Matrigel barriers and Col-I
also demonstrated that suppression of CXCR4 expression
significantly reduced the number of invading cells (Fig. 3E and F).
CXCR4, EGFR and MMP-9 are highly expressed in NSCLC.
A total of 61 patients (36 males and 25 females) were included in
the case series. The characteristics of the patients are summarized
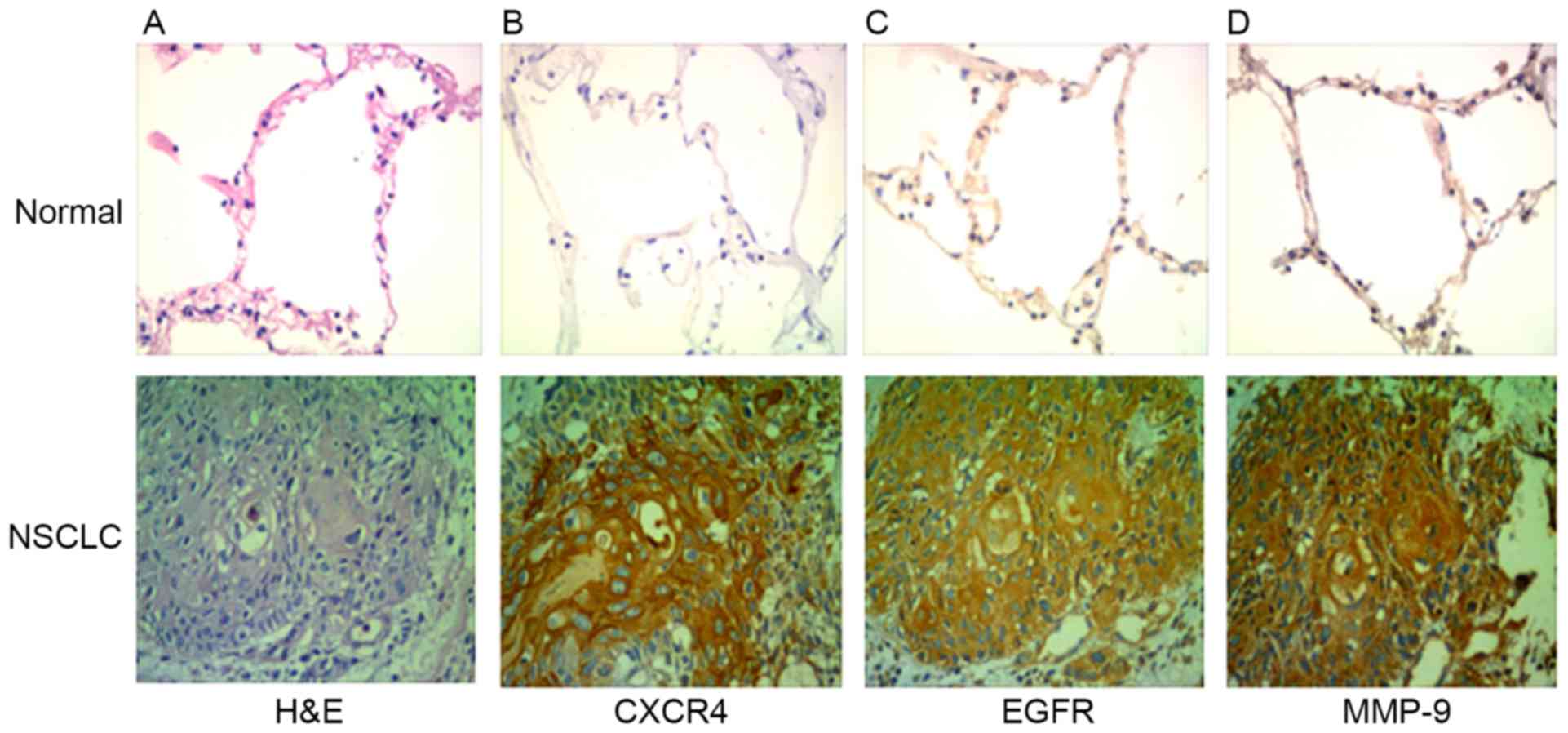
in Table I. The expression of CXCR4
and EGFR was analyzed by immunohistochemistry in clinical specimens
of lung cancer and normal lung tissue. The expression levels of
CXCR4, EGFR and MMP-9 were identified to be higher in NSCLC tissues
compared with normal lung tissue (Fig.
4). Out of the total 61 specimens, the rates of positive
expression of CXCR4, EGFR and MMP-9 in NSCLC were 69, 76 and 64%,
respectively. CXCR4, EGFR and MMP-9 expression in tissues of
patients with NSCLC was not associated age and sex (P>0.05), but
was associated with tumor differentiation and lymph node metastasis
(P<0.05; Table II).
 | Table II.Association between EGFR, CXCR4 and
MMP-9 expression and clinicopathological factors in NSCLC. |
Table II.
Association between EGFR, CXCR4 and
MMP-9 expression and clinicopathological factors in NSCLC.
|
| CXCR4, n (%) |
| EGFR, n (%) |
| MMP-9, n (%) |
|
|---|
|
|
|
|
|
|
|
|
|---|
| Parameter | Low | High | P-value | Low | High | P-value | Low | High | P-value |
|---|
| Age, years |
|
| 0.142 |
|
| 0.537 |
|
| 1 |
|
<60 | 10 (53) | 27 (64) |
| 6 (40) | 14 (30) |
| 7 (32) | 13 (33) |
|
|
≥60 | 9 (47) | 15 (36) |
| 9 (60) | 32 (70) |
| 15 (68) | 26 (67) |
|
| Sex |
|
| 0.266 |
|
| 0.13 |
|
| 0.416 |
|
Male | 8 (42) | 28 (67) |
| 6 (40) | 30 (76) |
| 11 (50) | 25 (64) |
|
|
Female | 11 (68) | 14 (33) |
| 9 (60) | 16 (24) |
| 11 (50) | 14 (36) |
|
| Tumor
differentiation |
|
| 0.025 |
|
| 0.015 |
|
| 0.033 |
|
Well | 10 (42) | 35 (64) |
| 7 (47) | 38 (61) |
| 20 (91) | 25 (64) |
|
|
Poor | 9 (58) | 7 (36) |
| 8 (63) | 8 (39) |
| 2 (9) | 14 (36) |
|
| Lymph node
metastasis |
|
| 0.002 |
|
| 0.013 |
|
| 0.008 |
|
Yes | 4 (21) | 27 (64) |
| 5 (26) | 26 (62) |
| 16 (73) | 14 (36) |
|
| No | 15 (79) | 15 (36) |
| 14 (74) | 16 (38) |
| 6 (27) | 25 (64) |
|
EGFR, CXCR4 and MMP-9 are positively
correlated in NSCLC
Using Spearman's rank correlation analysis,
significant association between EGFR and CXCR4 expression was
observed (r=0.422; P<0.05; Table
III). The expression of CXCR4 and MMP-9 in the NSCLC group
demonstrated positive correlation (r=0.438; P<0.05; Table IV), and the expression of EGFR was
also positively correlated with MMP-9 (r=0.328; P<0.05; Table V).
 | Table III.Spearman's rank correlation analysis
of clinical association between CXCR4 and EGFR. |
Table III.
Spearman's rank correlation analysis
of clinical association between CXCR4 and EGFR.
|
|
| EGFR |
|---|
|
|
|
|
|---|
|
| Expression | Low | High |
|---|
| CXCR4 | Low | 8 | 11 |
|
| High | 9 | 33 |
 | Table IV.Spearman's rank correlation analysis
of clinical association between CXCR4 and MMP-9. |
Table IV.
Spearman's rank correlation analysis
of clinical association between CXCR4 and MMP-9.
|
|
| MMP-9 |
|---|
|
|
|
|
|---|
|
| Expression | Low | High |
|---|
| CXCR4 | Low | 12 | 10 |
|
| High | 4 | 35 |
 | Table V.Spearman's rank correlation analysis
of clinical association between EGFR and MMP-9. |
Table V.
Spearman's rank correlation analysis
of clinical association between EGFR and MMP-9.
|
|
| MMP-9 |
|---|
|
|
|
|
|---|
|
| Expression | Low | High |
|---|
| EGFR | Low | 10 | 12 |
|
| High | 6 | 33 |
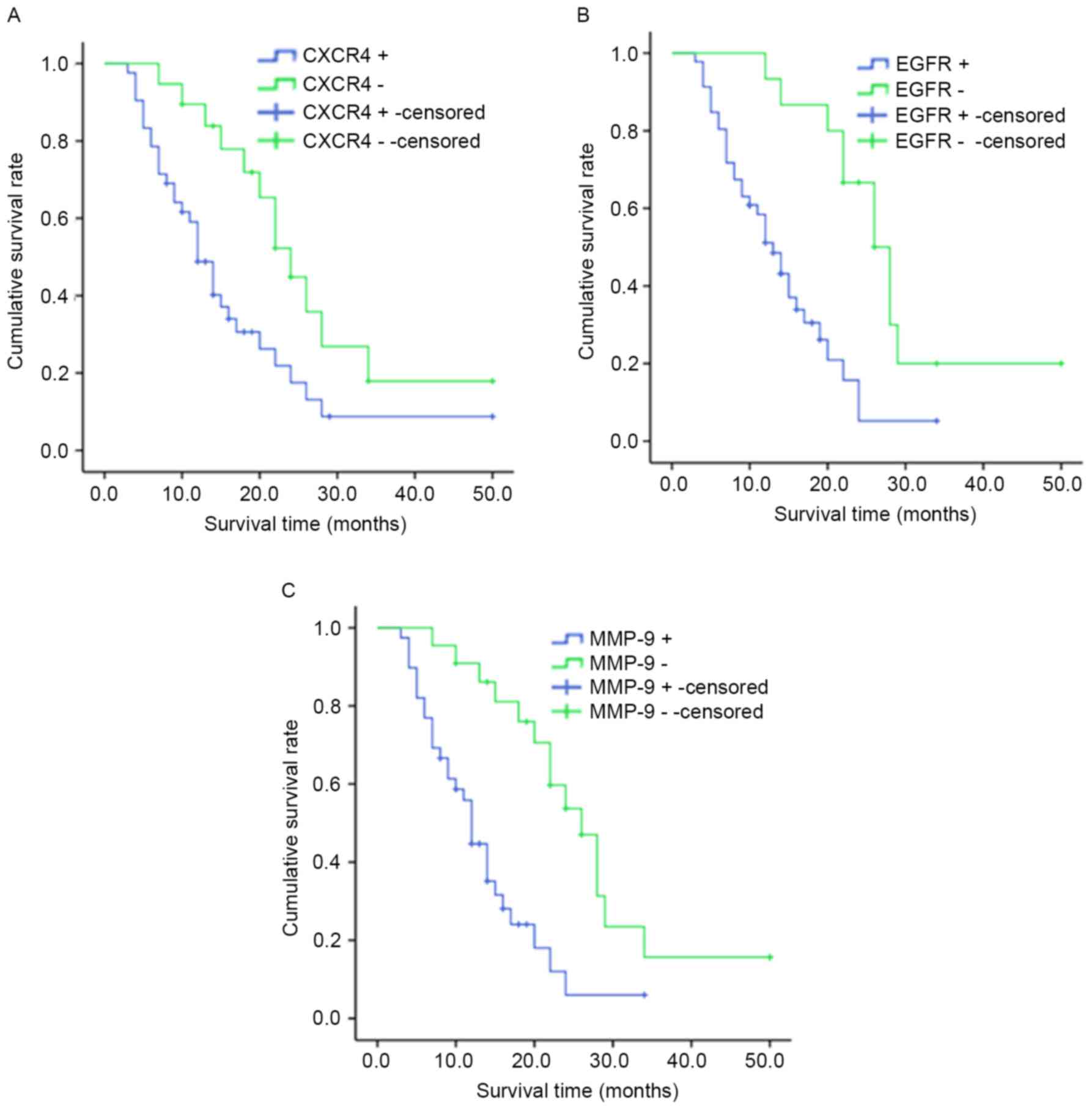
Survival analysis
The median survival time for cases with
CXCR4+ was 10.0 months [95% confidence (CI),
8.308–11.692] and 22.0 months for patients with CXCR4−
(95% CI, 14.133–29.867; P<0.05; Fig.
5A). The median survival time for patients with
EGFR+ was 10.0 months (95% CI, 7.947–12.053) and 20.0
months for patients with EGFR− (95% CI, 11.938–28.062;
P<0.05; Fig.5B). Patients with
MMP-9+ tumors (median survival time, 12.0 months; 95%
CI, 10.576–16.120) had significantly shorter overall survival times
compared with patients with MMP-9 tumors (median survival time,
26.0 months; 95% CI, 21.124–32.641; P<0.05; Fig. 5C).
Discussion
Lung cancer with high incidence and mortality is the
leading cause of cancer-associated mortality in the western world,
and the 5-year survival rate of NSCLC is estimated at ~15%
(29). In addition to surgery,
chemotherapy is a crucial element of treatment for patients with
NSCLC (30). Therefore, an emerging
understanding of the molecular pathways that characterize cell
growth, cell cycle, apoptosis, angiogenesis and invasion is of
crucial importance to the treatment of patients with lung cancer.
Previous studies reported that treating leukemia with a combination
of CXCR4 inhibitors and chemotherapeutic agents produced additive
therapeutic effects (31,32). Overexpression of EGFR is common in
head and neck squamous cell carcinoma (HNSCC) (33). Previous data have presented EGFR as a
new target for anti-HNSCC therapy (34). However, the mechanism of CXCR4 and
EGFR-stimulated tumor cell invasion and migration requires to be
elucidated.
CXCR4, a seven transmembrane G protein-coupled
receptor, is widely expressed in various cell types, including
endothelial, epithelial and hematopoietic stem cells, lymphocytes
and cancer cells (35). Previous
studies indicated that CXCR4 is involved in cellular proliferation,
migration and metastasis of solid tumors in a variety of cancers,
including breast, colorectal and gastric cancer (36–38). A
previous study has demonstrated that the CXCL12/CXCR4 axis is
involved in tumor progression, angiogenesis, metastasis and
survival (38). In the present study,
the results demonstrated that CXCR4 is highly expressed in NSCLC
and promotes cellular migration and invasion, which is consistent
with the aforementioned studies.
EGFR, is a transmembrane glycoprotein and can bind
to ligands, including transforming growth factor α and EGF, with
its extracellular domains to promote receptor dimerization and
activation of a cytoplasmic tyrosine kinase domain (39). The involvement of EGFR signaling has
been demonstrated in various cellular processes ranging from normal
growth and differentiation to oncogenesis (40–42).
Aberrant EGFR as a result of receptor mutation and/or
overexpression occurs in NSCLC (43)
laryngeal cancer (44), pancreatic
cancer (45). Our previous study
indicated that EGFR activation promotes HNSCC cell migration and
invasion by inducing an EMT-like phenotypic change and
MMP-9-mediated degradation of E-cadherin into soluble E-cadherin
(sE-cad) associated with activation of ERK-1/2 and PI3K signaling
pathways (25). In the present study,
similar results were obtained (46,47). The
results of the present study demonstrated that overexpression of
CXCR4 can lead to the expression of EGFR and enhance the production
of MMP-9, which finally results in elevating cell migration and
invasion.
In conclusion, the results of the present study
indicate that CXCR4 overexpression enhances cellular motility and
invasion via EGFR and MMP-9 in NSCLC. It was also revealed that the
expression of CXCR4, EGFR and MMP-9 is associated with pathological
grading and lymph node metastasis. Furthermore, positive
correlations were observed among CXCR4, EGFR and MMP-9 in
NSCLC.
Acknowledgements
The present study was supported by grants from the
National Nature Science Foundation of China (grant no. 81272960),
the Key Research Program from Science and Technology Department of
Hunan Province, China (grant no. 2013WK2010), the Key Research
Program from Ministry of Human Resources and Social Security of the
People's Republic of China (grant no. 2016176), the Hunan Province
Key Laboratory of Tumor Cellular and Molecular Pathology (grant no.
2016TP1015), the Construction Program of the Key Discipline in
Hunan Province, China (Basic Medicine Sciences in University of
South China, grant no. 201176), the General Research Program from
Science and Technology Department of Hunan Province, China (grant
no. 2013FJ3145), the Funds for the Development of Chemical Industry
of Forest Products in Hunan Province Key Laboratory (grant no.
JDLC201203) and the Wu Jieping Foundation Research Fund Project
(grant no. 320.6750.12245).
References
|
1
|
Stewart BW and Wild CP: World Cancer
Report 2014World Health Organization. IARC; Lyon: 2015, View Article : Google Scholar
|
|
2
|
He J and Song X: Advances in association
of estrogen and human non-small cell lung cancer. Zhongguo Fei Ai
Za Zhi. 18:315–320. 2015.PubMed/NCBI
|
|
3
|
Schvartsman G, Ferrarotto R and Massarelli
E: Checkpoint inhibitors in lung cancer: Latest developments and
clinical potential. Ther Adv Med Oncol. 8:460–473. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Saavedra D and Crombet T: CIMAvax-EGF: A
new therapeutic vaccine for advanced non-small cell lung cancer
patients. Front Immunol. 8:2692017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Singh AK, Arya RK, Trivedi AK, Sanyal S,
Baral R, Dormond O, Briscoe DM and Datta D: Chemokine receptor
trio: CXCR3, CXCR4 and CXCR7 crosstalk via CXCL11 and CXCL12.
Cytokine Growth Factor Rev. 24:41–49. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kodama J, Hasengaowa, Kusumoto T, Seki N,
Matsuo T, Ojima Y, Nakamura K, Hongo A and Hiramatsu Y: Association
of CXCR4 and CCR7 chemokine receptor expression and lymph node
metastasis in human cervical cancer. Ann Oncol. 18:70–76. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guo L, Cui ZM, Zhang J and Huang Y:
Chemokine axes CXCL12/CXCR4 and CXCL16/CXCR6 correlate with lymph
node metastasis in epithelial ovarian carcinoma. Chin J Cancer.
30:336–343. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shen B, Zheng MQ, Lu JW, Jiang Q, Wang TH
and Huang XE: CXCL12-CXCR4 promotes proliferation and invasion of
pancreatic cancer cells. Asian Pac J Cancer Prev. 14:5403–5408.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Qiu MY, Li JW and Zheng MH: The chemokine
receptors CXCR4 and CXCR7 in cancer. Chin Oncol. 20:222–226.
2010.
|
|
10
|
Kim M, Koh YJ, Kim KE, Koh BI, Nam DH,
Alitalo K, Kim I and Koh GY: CXCR4 signaling regulates metastasis
of chemoresistant melanoma cells by a lymphatic metastatic niche.
Cancer Res. 70:10411–10421. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Maderna E, Salmaggi A, Calatozzolo C,
Limido L and Pollo B: Nestin, PDGFRbeta, CXCL12 and VEGF in glioma
patients: Different profiles of (pro-angiogenic) molecule
expression are related with tumor grade and may provide prognostic
information. Cancer Biol Ther. 6:1018–1024. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Driessen WH, Fujii N, Tamamura H and
Sullivan SM: Development of peptide-targeted lipoplexes to
CXCR4-expressing rat glioma cells and rat proliferating endothelial
cells. Mol Ther. 16:516–524. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gulati S, Ytterhus B, Granli US, Gulati M,
Lydersen S and Torp SH: Overexpression of c-erbB2 is a negative
pognostic factor in anaplastic astrocytomas. Diagn Pathol.
5:182010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lemmon MA and Schlessinger J: Cell
signaling by receptor tyrosine kinases. Cell. 141:1117–1134. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rexer BN, Ghosh R, Narasanna A, Estrada
MV, Chakrabarty A, Song Y, Engelman JA and Arteaga CL: Human breast
cancer cells harboring a gatekeeper T798M mutation in HER2
overexpress EGFR ligands and are sensitive to dual inhibition of
EGFR and Her2. Clin Cancer Res. 19:5390–5401. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Al Zobair AA, Al Obeidy BF, Yang L, Yang
C, Hui Y, Yu H, Zheng F, Yang G, Xie C, Zhou F, et al: Concomitant
overexpression of EGFR and CXCR4 is associated with worse prognosis
in a new molecular subtype of non-small cell lung cancer. Oncol
Rep. 29:1524–1532. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Guo Z, Cai S, Fang R, Chen H, Du J, Tan Y,
Ma W, Hu H, Cai S and Liu Y: The synergistic effects of CXCR4 and
EGFR on promoting EGF-mediated metastasis in ovarian cancer cells.
Colloids Surf B Biointerfaces. 60:1–6. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Patel LR, Camacho DF, Shiozawa Y, Pienta
KJ and Taichman RS: Mechanisms of cancer cell metastasis to the
bone: A multistep process. Future Oncol. 7:1285–1297. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shon SK, Kim A, Kim JY, Kim KI, Yang Y and
Lim JS: Bone morphogenetic protein-4 induced by NDRG2 expression
inhibits MMP-9 activity in breast cancer cells. Biochem Biophys Res
Commun. 385:198–203. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Faraji SN, Mojtahedi Z, Ghalamfarsa G and
Takhshid MA: N-myc downstream regulated gene 2 overexpression
reduces matrix metalloproteinase-2 and −9 activities and cell
invasion of A549 lung cancer cell line in vitro. Iran J Basic Med
Sci. 18:773–779. 2015.PubMed/NCBI
|
|
21
|
Zuo J, Ishikawa T, Boutros S, Xiao Z,
Humtsoe JO and Kramer RH: Bcl-2 overexpression induces a partial
epithelial to mesenchymal transition and promotes squamous
carcinoma cell invasion and metastasis. Mol Cancer Res. 8:170–182.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gao J, Liu X, Yang F, Liu T, Yan Q and
Yang X: By inhibiting Ras/Raf/ERK and MMP-9, knockdown of EpCAM
inhibits breast cancer cell growth and metastasis. Oncotarget.
6:27187–27198. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Su L, Zhou W, Park S, Wain JC, Lynch TJ,
Liu G and Christiani DC: Matrix metalloproteinase-1 promoter
polymorphism and lung cancer risk. Cancer Epidemiol Biomarkers
Prev. 14:567–570. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cai J, Li R, Xu X, Zhang L, Wu S, Yang T,
Fang L, Wu J, Zhu X, Li M, et al: URGCP promotes non-small cell
lung cancer invasiveness by activating the NF-kappaB-MMP-9 pathway.
Oncotarget. 6:36489–36504. 2015.PubMed/NCBI
|
|
25
|
Zuo JH, Zhu W, Li MY, Li XH, Yi H, Zeng
GQ, Wan XX, He QY, Li JH, Qu JQ, et al: Activation of EGFR promotes
squamous carcinoma SCC10A cell migration and invasion via inducing
EMT-like phenotype change and MMP-9-mediated degradation of
E-cadherin. J Cell Biochem. 112:2508–2517. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Travis WD: The 2015 WHO classification of
lung tumors. Pathologe. 35 Suppl 2:S1882014. View Article : Google Scholar
|
|
27
|
Zuo J, Wen M, Lei M, Peng X, Yang X and
Liu Z: MiR-210 links hypoxia with cell proliferation regulation in
human Laryngocarcinoma cancer. J Cell Biochem. 116:1039–1049. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dai X, Mao Z, Huang J, Xie S and Zhang H:
The CXCL12/CXCR4 autocrine loop increases the metastatic potential
of non-small cell lung cancer in vitro. Oncol Lett. 5:277–282.
2013.PubMed/NCBI
|
|
29
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chang A: Chemotherapy, chemoresistance and
the changing treatment landscape for NSCLC. Lung Cancer. 71:3–10.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zeng Z, Shi YX, Samudio IJ, Wang RY, Ling
X, Frolova O, Levis M, Rubin JB, Negrin RR, Estey EH, et al:
Targeting the leukemia microenvironment by CXCR4 inhibition
overcomes resistance to kinase inhibitors and chemotherapy in AML.
Blood. 113:6215–6224. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Peled A and Tavor S: Role of CXCR4 in the
pathogenesis of acute myeloid leukemia. Theranostics. 3:34–39.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ali Sheikh MA, Gunduz M, Nagatsuka H,
Gunduz E, Cengiz B, Fukushima K, Beder LB, Demircan K, Fujii M,
Yamanaka N, et al: Expression and mutation analysis of epidermal
growth factor receptor in head and neck squamous cell carcinoma.
Cancer Sci. 99:1589–1594. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fung C and Grandis JR: Emerging drugs to
treat squamous cell carcinomas of the head and neck. Expert Opin
Emerg Drugs. 15:355–373. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Teicher BA and Fricker SP: CXCL12
(SDF-1)/CXCR4 pathway in cancer. Clin Cancer Res. 16:2927–2931.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Murakami T, Kawada K, Iwamoto M, Akagami
M, Hida K, Nakanishi Y, Kanda K, Kawada M, Seno H, Taketo MM, et
al: The role of CXCR3 and CXCR4 in colorectal cancer metastasis.
Int J Cancer. 132:276–287. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yang SX, Loo WT, Chow LW, Yang XH, Zhan Y,
Fan LJ, Zhang F, Chen L, Wang QL, Xiao HL, et al: Decreased
expression of C-erbB-2 and CXCR4 in breast cancer after primary
chemotherapy. J Transl Med. 10 Suppl 1:S32012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ying J, Xu Q, Zhang G, Liu B and Zhu L:
The expression of CXCL12 and CXCR4 in gastric cancer and their
correlation to lymph node metastasis. Med Oncol. 29:1716–1722.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Billah S, Stewart J, Staerkel G, Chen S,
Gong Y and Guo M: EGFR and KRAS mutations in lung carcinoma:
Molecular testing by using cytology specimens. Cancer Cytopathol.
119:111–117. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Okamoto I: Epidermal growth factor in
relation to tumor development: EGFR-targeted anticancer therapy.
FEBS J. 277:309–315. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lichtenberger BM, Tan PK, Niederleithner
H, Ferrara N, Petzelbauer P and Sibilia M: Autocrine VEGF signaling
synergizes with EFGR in tumor cells to promote epithelial cancer
development. Cell. 140:268–279. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Mitsudomi T and Yatabe Y: Epidermal growth
factor receptor in relation to tumor development: EGFR gene and
cancer. FEBS J. 277:301–308. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gajiwala KS, Feng J, Ferre R, Ryan K,
Brodsky O, Weinrich S, Kath JC and Stewart A: Insights into the
aberrant activity of mutant EGFR kinase domain and drug
recognition. Structure. 21:209–219. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Morrison LE, Jacobson KK, Friedman M,
Schroeder JW and Coon JS: Aberrant EGFR and chromosome 7 associate
with outcome in laryngeal cancer. Laryngoscope. 115:1212–1218.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tzeng CD, Frolov A, Howard JH, Vickers SM,
Buchsbaum DJ, Heslin MJ and Arnoletti JP: 96: Analysis of aberrant
epidermal growth factor rector (EGFR) pathway signaling in
pancreatic cancer patients: Implications for anti-EGFR therapy. J
Surg Res. 137:1912007. View Article : Google Scholar
|
|
46
|
Shen B, Zheng MQ, Lu JW, Jiang Q, Wang TH
and Huang XE: CXCL12-CXCR4 promotes proliferation and invasion of
pancreatic cancer cells. Asian Pac J Cancer Prev. 14:5403–5408.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Pei J, Lou Y, Zhong R and Han B: MMP9
activation triggered by epidermal growth factor induced FoxO1
nuclear exclusion in non-small cell lung cancer. Tumour Biol.
35:6673–6678. 2014. View Article : Google Scholar : PubMed/NCBI
|