Introduction
Oral squamous cell carcinoma (OSCC) was the tenth
most frequently occurring solid cancer worldwide in 2015, and
tongue squamous cell carcinoma (TSCC) is one of the leading causes
of cancer-associated mortality in patients with oral cancer
globally (1,2). Despite the advances made in treatments
for tongue cancer, including radical surgery and chemoradiotherapy,
the 5-year survival rate of patients with TSCC remains poor, mainly
due to the tendency of this cancer to affect lymph nodes and
develop distant metastasis (3,4). Although
several studies regarding molecular mechanisms of TSCC progression
have been performed, little is known regarding the molecular
mechanisms underlying the regulation of metastatic dissemination
(5–7).
The cluster of differentiation 63 (CD63) gene is
located on human chromosome 12q13 and was the first tetraspan into
be characterized (8). It is a member
of the transmembrane 4 superfamily (TM4SF), which comprises
heterogeneous membrane-bound glycoproteins that are expressed in
endosomes and lysosomes and on the cell surface (9,10). CD63
was first identified to be a protein strongly expressed on the cell
surface in early-stage human melanoma cells, and was originally
known as melanoma antigen 491 (ME491); it was observed within the
complex network of internal membranes that are characteristic of
late endosomes in mammalian cells (8,11).
The trafficking of CD63 between the endosomal system
and the cell surface is mediated by the clathrin adaptor protein 2
(AP2) complexes and caveolae or clathrin-coated pit-mediated
endocytosis, a process that requires specific amino acid motifs
present in the CD63 protein (10).
Expression of transmembrane 4 L6 family member 5 (TM4SF5) is
negatively correlated with that of CD63 in mouse fibrotic and human
hepatic carcinoma tissues (12). CD63
is also known to be involved in the regulation of diverse cellular
processes, including proliferation, adhesion, motility and
differentiation (13). As malignant
melanomas progress, expression of CD63 declines and the cells
become more invasive (8,14). Indeed, when the expression of CD63 in
melanoma cells was silenced, their cell motility and
matrix-degrading ability increased (15); however, when the recombinant vector
pREP9-CD63 was transected in a CD63-negative melanoma cell line,
the cell motility and metastatic capacity were reduced and the
cells became more adhesive to the extracellular matrix (16).
CD63 was shown to serve a key role in the malignancy
of cancer types, including lung adenocarcinoma, breast cancer, and
colon cancer (17–19). Although CD63 is closely associated
with multiple types of cancer development and progression, the
functions of CD63 in TSCC have not been elucidated. Therefore, the
present study investigated the roles of CD63 in the progression and
development of TSCC using molecular and cell biology methods
including immunohistochemistry, RNA interference (RNAi), gene
transfection technology, wound healing and transwell invasion
assays and western blotting.
Materials and methods
Cell culture
The human tongue squamous cell carcinoma TCA8113
cell line and 293 cells (Shanghai Institute for Cellular Biology,
Chinese Academy of Sciences, Shanghai, China) were cultured in RPMI
1640 (Hyclone; GE Healthcare Life Science, Logan, UT, USA) and High
Glucose DMEM Pyruvate medium (Hyclone; GE Healthcare Life Science),
respectively, supplemented with 10% fetal bovine serum (FBS; Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA), 100 U/ml
penicillin, and 100 µg/ml streptomycin (Gibco; Thermo Fisher
Scientific, Inc.) in 5% CO2 at 37°C.
Patients and tissue specimens
A total of 40 human TSCC tissues and four normal
tongue tissue samples were examined. Among the 40 cases, there were
23 males and 17 females, with an age range from 36–86 years (median
age, 55 years). The patients were histopathologically and
clinically diagnosed at the Oral and Maxillofacial Surgery of the
First Affiliated Hospital of Jinzhou Medical University (Jinzhou,
China) between January of 2001 and April of 2017, and the
pathological diagnosis was verified for each case. All patients had
not received chemotherapy and radiotherapy prior to surgery. The
present study was approved by the Ethics Committees of Jinzhou
Medical University (approval nos. 20140005 and 20171105). Written
informed consent was obtained from all patients stating their
agreement to be involved in the study. All TSCC samples were staged
according to the 2002 Union for International Cancer Control
guidelines (20). Of the TSCC
specimens, 16 cases were well-differentiated, 17 were moderately
differentiated and 7 were poorly differentiated. A total of 11
(27.5%) TSCC cases exhibited lymph node metastasis. The four normal
tissue samples were collected from patients with tongue trauma. The
collection of samples was performed in accordance with the policies
of the National Research Ethics Committee, and informed consent was
obtained from each patient. The clinicopathological features of the
patients are summarized in Table
I.
 | Table I.Association between the CD63
expression in the 40 tongue squamous cell carcinoma tissue
specimens and clinical characteristics. |
Table I.
Association between the CD63
expression in the 40 tongue squamous cell carcinoma tissue
specimens and clinical characteristics.
|
|
| CD63
expression |
|
|---|
|
|
|
|
|
|---|
| Characteristic | Patients, n
(%) | Low | High | P-value |
|---|
| Male | 23 (57.5) | 15 | 8 | 0.973 |
| Female | 17 (42.5) | 11 | 6 |
|
| Age, years |
|
>55 | 22 (55) | 15 | 7 | 0.744 |
|
≤55 | 18 (45) | 11 | 7 |
|
| Histological
differentiation |
|
Well | 16 (40) | 6 | 10 | 0.014 |
|
Moderate | 17 (42.5) | 14 | 3 |
|
|
Poor | 7 (17.5) | 6 | 1 |
|
| TNM stage |
|
I–II | 24 (60) | 12 | 12 | 0.02 |
|
III–IV | 16 (40) | 14 | 2 |
|
| Lymph node
metastasis |
|
N+ | 11 (27.5) | 11 | 0 | 0.007 |
|
N− | 29 (72.5) | 17 | 14 |
|
Immunohistochemistry
Immunohistochemical (IHC) analysis was performed to
investigate the expression of CD63 in different grades of human
TSCC samples using the Diaminobenzidene (DAB) Detection kit
(Streptavidin-Biotin; cat. no. SP-9000-D; OriGene Technologies,
Inc., Beijing, China) according to the manufacturer's instructions.
The tissues were fixed in 10% formalin for 60 min at room
temperature, paraffin-embedded, then deparaffinized in xylene for
10 min. The xylene was replaced twice, each time tissue sections
were soaked for 10 min at room temperature. The tissues were
transferred to anhydrous ethanol for 5 min, then 95% ethanol for 5
min, and 75% ethanol for 5 min at room temperature. Then the
tissues were rehydrated in water and rinsed in phosphate-buffered
saline (PBS; pH 7.4) for 5 min. Antigen retrieval was performed in
a pressure cooker in citrate buffer (0.01 M, pH 6.0) for 15 min,
followed by treatment with 3% hydrogen peroxide for 15 min at room
temperature and three washes with PBS for 2 min each time.
The specimens were incubated with antibody against
CD63 (cat. no. ab216130; polyclonal rabbit; 1:50 dilution; Abcam,
Cambridge, UK) for 1 h at 60°C and then washed three times with PBS
(0.01 M). The specimens were then incubated with 100 µl
streptavidin-biotin-conjugated IgG antibody (cat. no. PV-6001, goat
anti-rabbit; ready-to-use dilution; ZSGB-BIO; OriGene Technologies,
Inc.) for 20 min at room temperature and washed three times with
PBS. Streptavidin peroxidase was then applied to the specimens, and
they were incubated for 10 min at room temperature. The incubation
was followed by four rinses in PBS. Next, 30 µl of DAB Chromogen
was added to 1.5 ml of DAB substrate, which were mixed by swirling
and applied to the specimens, which were then incubated for 10 min.
The specimens were then washed four times with PBS.
The sections underwent counterstaining by Modified
Harris Hematoxylin (0.5% hematoxylin; cat. no. 6765003; Thermo
Fisher Scientific, Inc.) for 5 min at room temperature according to
the manufacturer's instructions, followed by dehydration in graded
ethanol and mounting onto coverslips. The specimens were analyzed
using a DFC310-FX light microscope (Leica Microsystems GmbH,
Wechsler, Germany). Generally, each specimen was assigned a score
according to the intensity of the staining (0, no staining; 1, weak
staining; 2, medium staining; and 3, strong staining), and the
percentage of stained cells (1, <10%; 2, 11–25%; 3, 26–50%; 4,
>50%). The final immunoreactive score was calculated as the mean
of these two scores. When evaluating the protein expression of
CD63, a score of <2.5 was defined as low and ≥2.5 as high.
Screening of effective short hairpin
RNAs (shRNAs) against CD63
For CD63 knockdown, three shRNA plasmids were
synthesized to target the sequence of CD63 mRNA (NM_001780.5) by
Shanghai GeneChem Co., Ltd. (Shanghai, China) (Table II). The 293 cells were cultured in
two 6-well plates and transfected with shRNA using Lipofectamine
2000 (Invitrogen; Thermo Fisher Scientific, Inc.) following the
transfection procedure; 48 h later, one 6-well plate of cells was
harvested and the total RNA was isolated with RNAiso Plus (cat. no.
9108; Takara Biotechnology Co., Ltd., Beijing, China) according to
the manufacturer's instructions.
 | Table II.Sequences of shRNA against CD63. |
Table II.
Sequences of shRNA against CD63.
| ID | 5′-sequence | Stem | Loop | Stem | 3′-sequence |
|---|
|
CD63-RNAi(4038–1)-a | GATCCC |
GCCTCGTGAAGAGTATCAGAA | CTCGAG |
TTCTGATACTCTTCACGAGGC | TTTTTGGAT |
|
CD63-RNAi(4038–1)-b | AGCTATCCAAAAA |
GCCTCGTGAAGAGTATCAGAA | CTCGAG |
TTCTGATACTCTTCACGAGGC | GG |
|
CD63-RNAi(4040–1)-a | GATCCC |
GCTGGCTATGTGTTTAGAGAT | CTCGAG |
ATCTCTAAACACATAGCCAGC | TTTTTGGAT |
|
CD63-RNAi(4040–1)-b | AGCTATCCAAAAA |
GCTGGCTATGTGTTTAGAGAT | CTCGAG |
ATCTCTAAACACATAGCCAGC | GG |
|
CD63-RNAi(4041–1)-a | GATCCC |
GCAAGGAGAACTATTGTCTTA | CTCGAG |
TAAGACAATAGTTCTCCTTGC | TTTTTGGAT |
|
CD63-RNAi(4041–1)-b | AGCTATCCAAAAA |
GCAAGGAGAACTATTGTCTTA | CTCGAG |
TAAGACAATAGTTCTCCTTGC | GG |
cDNA was synthesized with the PrimeScript RT reagent
kit (Takara Biotechnology Co., Ltd.). Reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
using a Thermal Cycler Dice Real Time System II with SYBR Premix Ex
Taq II (Takara Biotechnology Co., Ltd.). The specific primers used
were as follows: Human CD63 forward,
5′-CCCAAGCTTGCCACCATGGCGGTGGAAGGAGGAATGAAATG-3′ and reverse,
5′-CCGCTCGAGCATCACCTCGTAGCCACTTCTGATAC-3′; and human β-actin
forward, 5′-GCATCCACGAAACTACATTCAACTC-3′ and reverse,
5′-CACTGTGTTGGCATAGAGGTCTTTG-3′.
All reactions were performed in triplicate. The data
are expressed as the mean normalized expression (MNE). The MNE is
directly proportional to the amount of RNA of the target gene
relative to the amount of RNA of the reference gene β-actin.
Analysis of relative gene expression data using the
2−ΔΔCt method (21).
The other 6-well plate of cells was harvested 48 h
after transfection, and the protein was extracted using
radioimmunoprecipitation assay (RIPA) buffer and 1%
phenylmethylsulfonyl fluoride (PMSF) (both Beyotime Institute of
Biotechnology, Haimen, China). The bicinchoninic acid (BCA) method
was used to measure the concentration of the protein with a BCA
Protein Assay kit (Aidlab Biotechnologies Co., Ltd., Beijing,
China). Equivalent amounts of protein (50 µg) were denatured in SDS
sample buffer (cat. no. S9788; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) and separated by 10% SDS-PAGE and then
transferred to a polyvinylidene difluoride (PVDF) membrane (Merck
KGaA, Darmstadt, Germany). The membrane was blocked with 1% bovine
serum albumin (BSA) (Sigma-Aldrich; Merck KGaA) for 2 h and
incubated with the following primary antibodies: rabbit polyclonal
anti-CD63 (cat. no. ab216130; 1:1,000 dilution; Abcam), mouse
polyclonal anti-β-actin (cat. no. 3700; 1:5,000 dilution; Cell
Signaling Technology, Inc., Danvers, MA, USA) overnight at 4°C, and
the signal was detected using horseradish peroxidase
(HRP)-conjugated secondary antibodies (goat anti-rabbit IgG-HRP,
cat. no. sc-2004; goat anti-mouse IgG-HRP, cat. no. sc-2005,
1:2,000 dilution; Santa Cruz Biotechnology, Inc., Dallas, TX, USA)
followed by development using a Alkaline phosphatase substrate
chromogenic kit (cat. no. PP2501; Aidlab Biotechnologies Co.,
Ltd.). The images were captured and analyzed using Omega
Lum™ G capture software (version 2.0.1027.0; Gel
Company, San Francisco, CA, USA).
Construction of the CD63
overexpression plasmid
Total RNA was extracted from TCA8113 cells using
RNAiso Plus (cat. no. 9108; Takara Biotechnology Co., Ltd.)
according to the manufacturer's instructions. cDNA was synthesized
with the PrimeScript RT reagent kit and 100 ng cDNA was used as a
template for amplifying the CD63 gene using PrimeSTAR®
HS Premix (cat. no. R040A; Takara Biotechnology Co., Ltd.)
according to the manufacturer's protocol (the primers used were
those aforementioned). The 50-µl reaction system contained 25 µl
PrimeSTAR HS Premix, 1 µl each of the forward and reverse primers
of CD63, 2 µl cDNA, and 21 µl distilled water.
The PCR thermocycling conditions were as follows:
98°C for 10 sec, 55°C for 5 sec, 72°C for 1 min and 30 cycles later
72°C for 10 min. The CD63 gene was detected by 1% agarose gel
electrophoresis and purified with an AxyPrep DNA Gel Extraction kit
(Axygen; Corning Incorporated, Corning, NY, USA) according to the
manufacturer's protocol. The purified CD63 gene was then subcloned
into the HindIII/XhoI sites of the PEGFP-N3 vector
(BioVector NTCC, Inc., Beijing, China). The recombinant plasmids
were transformed into Escherichia coli DH5α and screened by
kanamycin (100 µg/ml; Sangon Biotech Co., Ltd., Shanghai, China).
An Axyprep-96 Plasmid kit (Axygen) to purify the recombinant
plasmids from bacterial cells cultured in lysogeny broth medium
overnight. The recombinant plasmids were detected by restriction
enzyme digestion and then DNA sequencing (Sangon Biotech Co.,
Ltd.).
Transfection and selection of stable
clones
The most effective shRNA plasmid against CD63 (named
CD63-RNAi-4041) and the CD63 overexpressing plasmid (named
PEGFP-N3-CD63) were transfected into TCA8113 cells using
Lipofectamine 2000 and screened using geneticin (Gibco; Thermo
Fisher Scientific, Inc.) at 600 µg/ml. Neomycin-resistant clones
were obtained, and the CD63 expression level was detected by
western blot. An IHC assay was performed as aforementioned to
observe the expression and location of CD63 in screened cell lines.
The screened TCA8113 cells and normal cells were seeded in
glass-bottom cell culture dishes and fixed in 4% paraformaldehyde
at room temperature (Sangon Biotech Co., Ltd.) for 30 min.
Following three washes with PBS for 5 min each time,
the cells were treated with 0.1% Triton X-100 (Sigma-Aldrich) for
10 min and then washed three times with PBS. The cells were blocked
in 1% BSA for 1 h and incubated with the rabbit polyclonal
anti-CD63 antibody (1:1,000 dilution; Abcam) for 1 h at 37°C. After
being washed with PBS three times, the cells were incubated with
the fluorescein isothiocyanate-conjugated anti-R-Phycoerythrin
antibody (1:500 dilution; cat. no. ab34723; Abcam) for 30 min,
followed by three washes with PBS. The expression and location of
CD63 protein were observed using a fluorescence microscope
(magnification, ×400; FSX100; Olympus, Shanghai, China).
Wound-healing assay
The transfected cells and normal TCA8113 cells
(5×105 cells per well) were seeded in 24-well plates,
and when the cells reached a confluent state the cell layer was
scratched with a sterile 200-µl pipette tip. The medium and cell
debris was aspirated away and replaced with 1 ml of fresh RPMI 1640
medium without FBS. Images of the wounded area were captured at 0
and 24 h, using a DMI3000 B light microscope (Leica Microsystems
GmbH). The wound healing speed was calculated as the difference in
the area between 0 and 24 h divided by the height of the wound,
with the use of ImageJ1.46r software (National Institutes of
Health, Bethesda, MD, USA).
Transwell cell invasion assay
The Transwell invasion assay was performed to
examine the invasion ability of CD63-silenced and
CD63-overexpressing TCA8113 cells, using a 6.5-mm Transwell with an
8.0-µm Pore Polyester Membrane Insert (Corning Incorporated) coated
with 10 µl Matrigel (50 µl/cm2; Corning Incorporated). A
total of 1×105 cells were plated into the upper chamber
of the Transwell with 500 µl RPMI 1640 medium without FBS, and 500
µl RPMI 1640 medium with 10% FBS was added into the lower chamber.
The cells were cultured for 24 h in 5% CO2 at 37°C.
The non-invading cells in the upper side of the
filter were then gently removed with a soft cotton swab, and the
cells that had invaded to the lower side of the filter were fixed
with 4% paraformaldehyde at room temperature for 30 min and stained
with 1% crystal violet at 37°C for 15 min (Sigma-Aldrich; Merck
KGaA). The number of cells in three randomly selected fields was
counted with an Image Analysis System (version 3.3.0; Leica
Microsystems GmbH), and these numbers are expressed as the average
number of migrating cells.
Assessing expression of matrix
metalloproteinase-2 (MMP-2) and MMP-9
The stably transfected cell lines were lysed with
RIPA and 1% PMSF, and western blot analysis was performed following
BCA protein analysis, performed as aforementioned. Following 10%
SDS-PAGE (50 µg protein per lane) electrophoresis and the transfer
of the protein to a PVDF membrane, the membrane was blocked with 1%
BSA for 2 hat room temperature and incubated with the following
primary antibodies: Rabbit polyclonal anti-MMP-2 (cat. no. 40994;
1:1,000 dilution; Cell Signaling Technology, Inc.), rabbit
polyclonal anti-MMP-9 (cat. no. 2270; 1:1,000 dilution; Cell
Signaling Technology, Inc.), and mouse polyclonal anti-β-actin
(cat. no. 3700; 1:1,000 dilution; Cell Signaling Technology, Inc.)
overnight at 4°C. The signal was then detected using HRP-conjugated
secondary antibodies (goat anti-rabbit IgG-HRP; cat. no. sc-2004;
goat anti-mouse IgG-HRP, cat. no. sc-2005; 1:2,000 dilution; both
from Santa Cruz Biotechnology, Inc.) for 1 hat room temperature
followed by visualization using an Alkaline phosphatase substrate
chromogenic kit (cat. no. PP2501; Aidlab Biotechnologies Co.,
Ltd.). The images were captured and analyzed using an Omega Lum G
capture software.
Statistical analysis
Quantitative data are expressed as the mean ±
standard error of the mean. Statistical comparisons were performed
using one-way analysis of variance followed by the
Student-Newman-Keuls test. The χ2 test was used to
analyze the association between CD63 expression and
clinicopathological patient characteristics of TSCC. All
statistical analyses were performed using the SPSS Statistics 18.0
software program (SPSS, Inc., Chicago, IL, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression of CD63 in TSCC
tissues
The IHC staining revealed a significant decrease in
CD63 protein levels in the TSCC tissues compared with the normal
tongue tissues (Table III). As
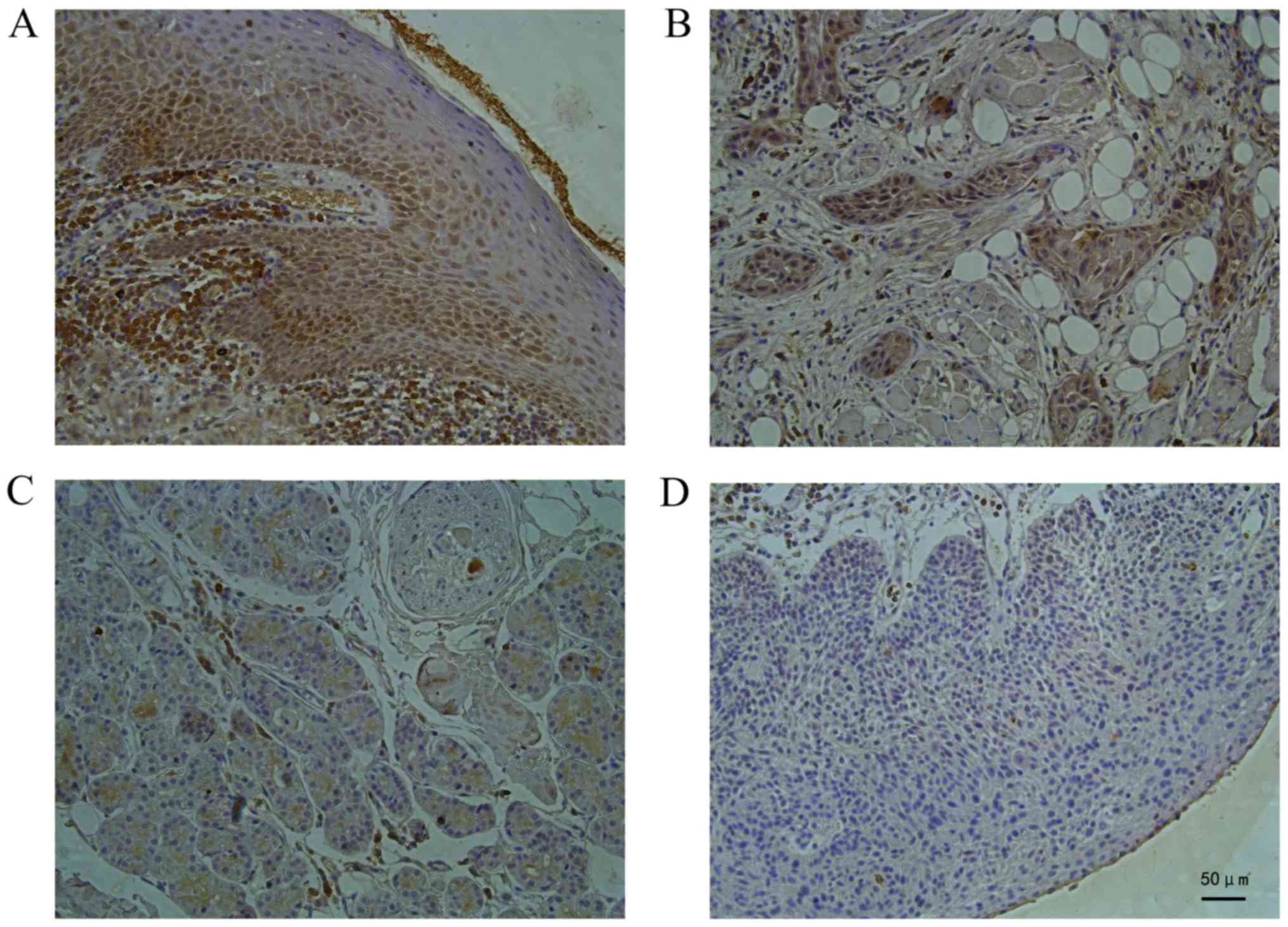
shown in Fig. 1, in the normal tongue
tissues, staining for the CD63 protein was deep
brown-yellow-colored, granular and expressed mainly in the cell
membrane and cytoplasm. In the TSCC tissue (Fig. 1A), the staining for the CD63 protein
was weaker than that in the normal tongue tissue as the
differentiation degree decreased significantly (P<0.05),
indicating a positive association between the CD63 expression level
and the histopathological differentiation of the tongue cancer
(Fig. 1B).
 | Table III.Protein expression of CD63 in TSCC
tissues (n=40) and normal tongue tissues (n=4). |
Table III.
Protein expression of CD63 in TSCC
tissues (n=40) and normal tongue tissues (n=4).
|
|
| CD63 expression,
n |
|
|
|---|
|
|
|
|
|
|
|---|
| Histological
type | n | Low | High | Ratio, % | P-value |
|---|
| Normal tissues | 4 | 0 | 4 | 100 | 0.023 |
| TSCC tissues | 40 | 26 | 14 | 35 |
CD63 protein expression in the stage I–II TSCC
tissues was significantly higher than that in the stage III–IV TSCC
tissues (P<0.05); the expression was significantly higher in the
well- and moderately differentiated TSCC tissues compared with that
in the poorly differentiated TSCC tissues (P<0.05). Lower
expression of CD63 was significantly associated with lymph node
metastasis (P<0.01). Thus, the protein expression level of CD63
in TSCC was significantly associated with the Tumor-Node-Metastasis
stage (20), tumor differentiation
and lymph node metastasis, although it was not associated with the
age or sex of the patient (Table
I).
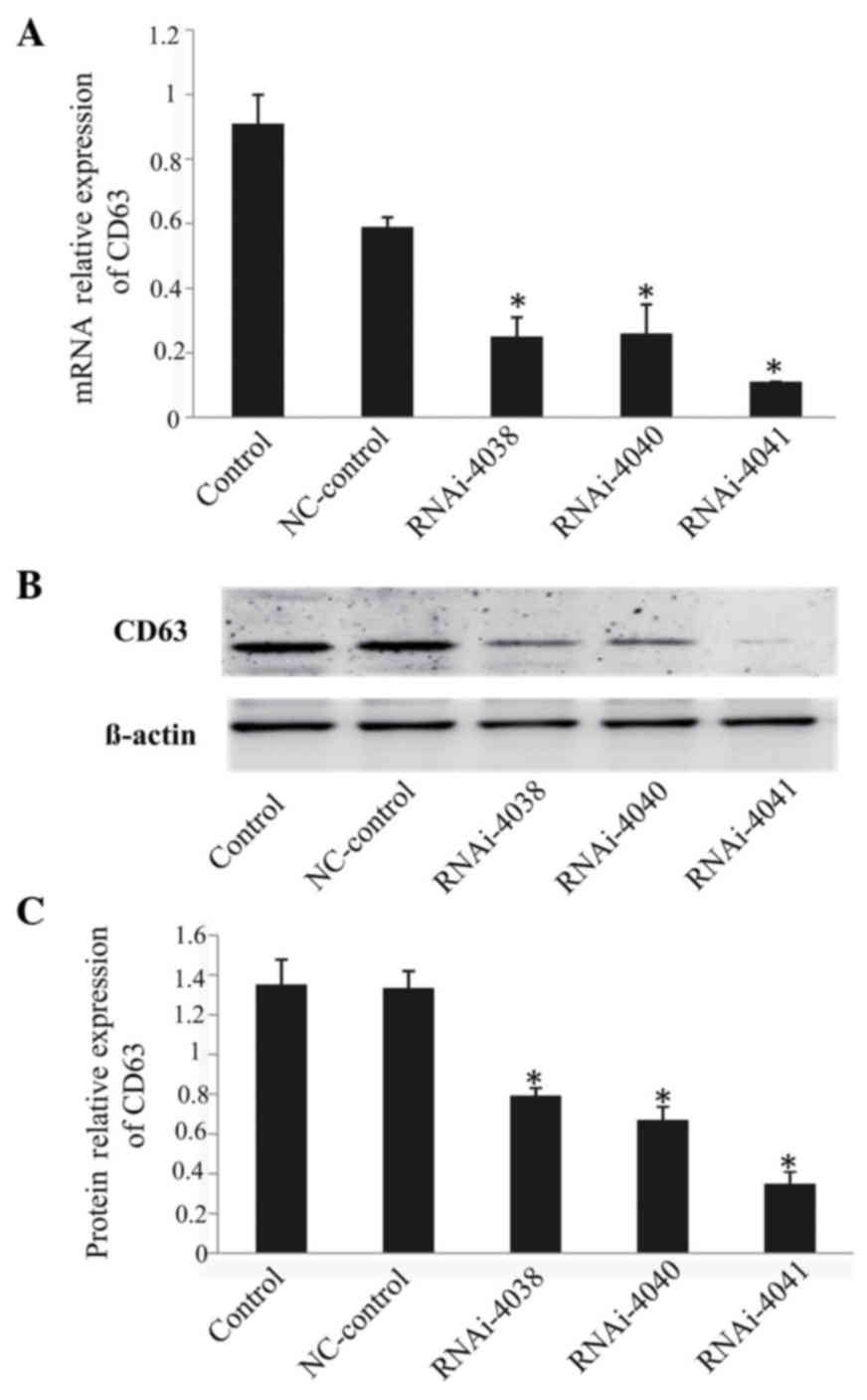
Screening of effective shRNA plasmids
against CD63
The shRNA plasmids were efficiently transfected into
293 cells with the use of Lipofectamine 2000; RT-qPCR and western
blot analysis results revealed that all three shRNA plasmids
effectively silenced the expression of CD63, with the plasmid
CD63-RNAi-4041 being the most efficient. The interference
efficiency of CD63-RNAi-4041 at the mRNA and protein levels was 88
and 74%, respectively (P<0.05; Fig.
2).
Construction of the
CD63-overexpressing plasmid
The CD63 gene sequence was obtained by PCR, from
which a specific 717-bp band was observed (Fig. 3A). PEGFP-N3-CD63 plasmids were
successfully obtained following restriction enzyme digestion
(Fig. 3B) and DNA sequencing.
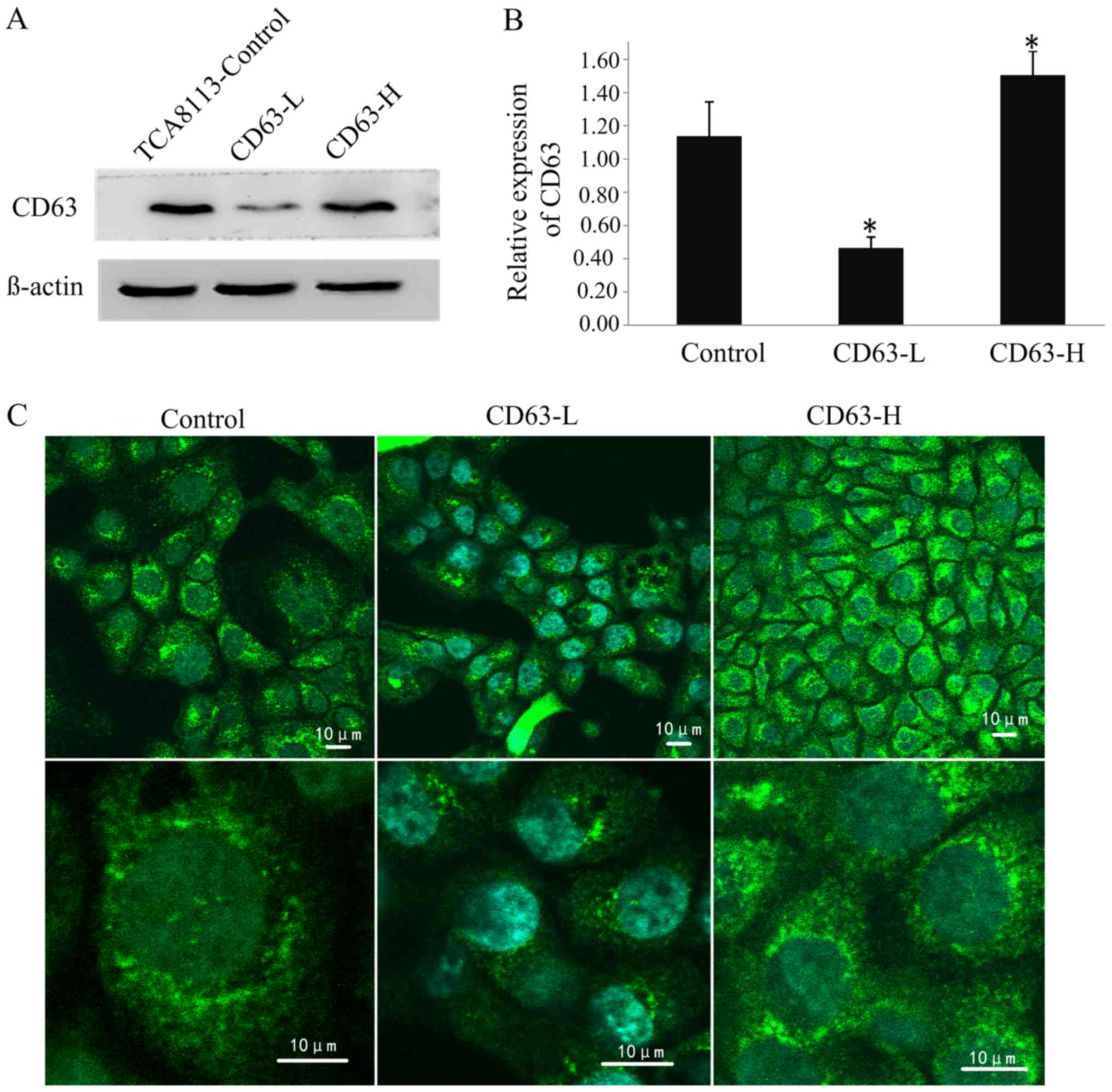
Selection of stable clones
Following screening using geneticin, stable clones
transfected with CD63-RNAi-4041 and PEGFP-N3-CD63 plasmids were
obtained. Western blotting revealed that the expression level of
CD63 in TCA8113 cells transfected with CD63-RNAi-4041 was 59% lower
than that in the controls, and the expression level of CD63 in
cells transfected with PEGFP-N3-CD63 was 32% higher than that in
the controls; these differences were significant (P<0.05;
Fig. 4A and B). The CD63-silenced
TCA8113 cell line CD63-low (CD63-L) and the CD63 overexpressing
cell line CD63-high (CD63-H).
The indirect immunofluorescence staining observed
using confocal microscopy revealed that the CD63 protein was
primarily located in the outer membrane and cytoplasm of TCA8113
cells. The fluorescent signal intensity in the CD63-H cells was the
strongest of the three cell lines, and that in the CD63-L cells was
the weakest. This difference in the fluorescence signal may be
representative of the expression level of CD63 protein (Fig. 4C).
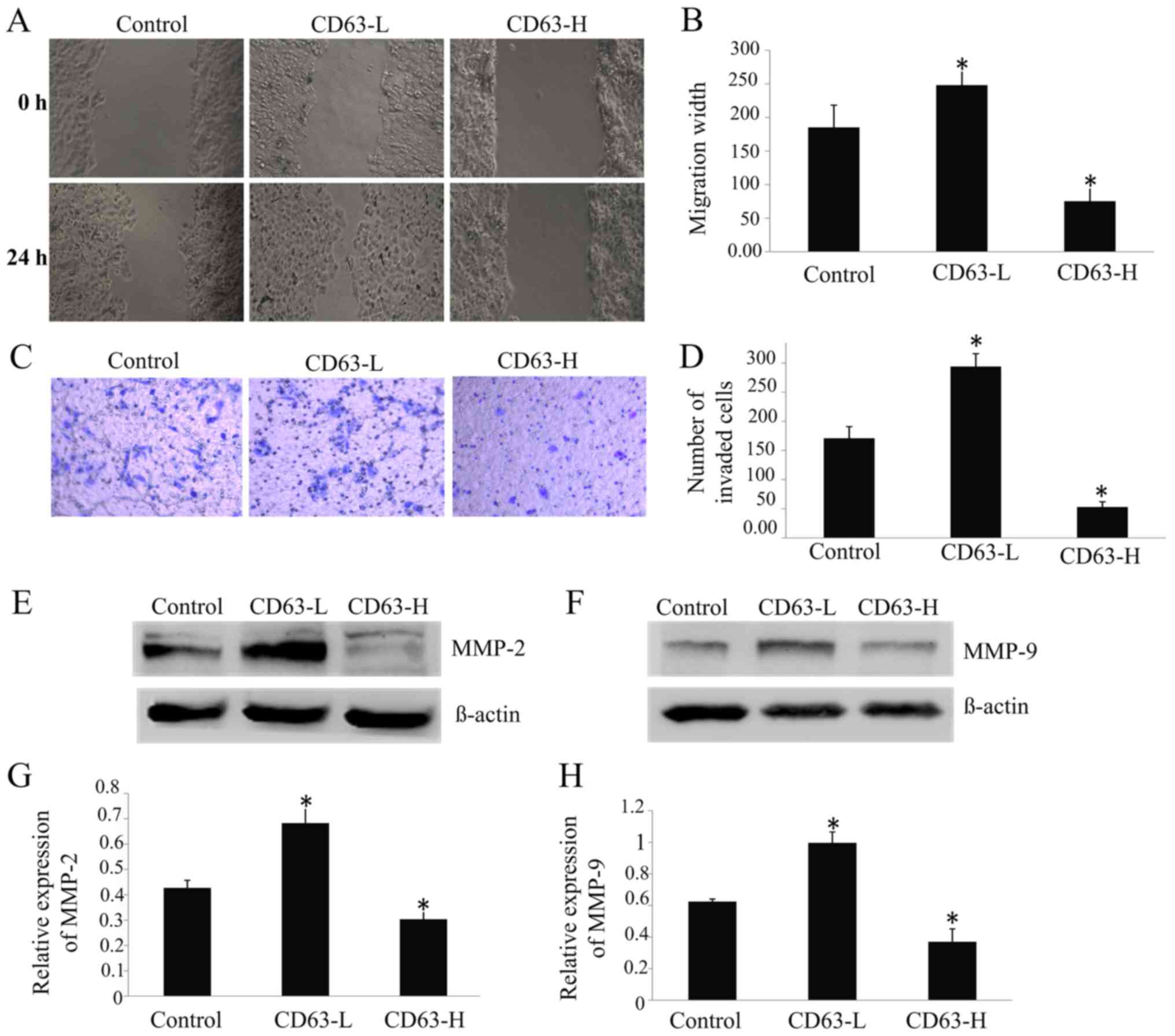
CD63 expression inhibits TCA8113 cell
migration and invasion
The CD63-L cells were able to repair the wound 73%
more rapidly than the control cells, whereas the speed of repair of
CD63-H cells was 59% slower than that of the control cells
(P<0.05; Fig. 4A and B). The
invasion assay revealed that 70% more CD63-L cells invaded through
the Matrigel-coated filters compared to the control cells, whereas
71% fewerCD63-H cells invaded compared to the control cells
(Fig. 4C and D). There was a
significant negative association between the CD63 expression level
and the migratory and invasive ability of TCA8113 cells
(P<0.05).
The results of western blotting revealed that the
expressions of MMP-2 (Fig. 4E and F)
and MMP-9 (Fig. 4G and H) were
upregulated by 60 and 61%, respectively, in CD63-L cells, but were
downregulated by 29 and 41%, respectively, in CD63-H cells,
compared with the control cells. Taken together, the findings of
the present study indicated that CD63 might serve a role in
inhibiting the migration and invasion of TSCC cells.
Discussion
TSCC is a threat to human health worldwide, and its
metastasis is believed to be one of the fundamental features that
contributes to the majority of incidences of cancer-associated
mortality in humans (1,2). However, the detailed molecular
mechanisms of TSCC remain elusive. CD63 expression is associated
with the biological behavior of solid tumors, particularly those
with metastatic potential, and it has been found to serve an
inhibitory role in the invasion and metastasis of multiple tumor
types. Kwon et al (17)
examined the expression level of CD63 in 90 cases of non-small cell
lung cancer (NSCLC) to investigate the potential of CD63 as a
prognostic biomarker for lung cancer subtypes, using tissue
microarray-based immunohistochemistry. The results of this analysis
revealed that 63.3% of the NSCLC samples were CD63-negative, and
the CD63 protein level was lower than that in normal tissue. CD63
protein negativity was significantly associated with larger tumor
size, advanced clinicopathological stage and poor patient survival
rates (P=0.008); the findings of this study (17) indicated that CD63 could be used as a
biomarker to predict the prognosis of patients with early-stages of
lung adenocarcinoma.
Woegerbauer et al (22) used the immunohistochemistry staining
of Merkel cell carcinoma specimens from 25 patients and observed
that CD63 expression was significantly associated with the
disease-free survival time of patients. In addition, a study by
Chen et al (23) demonstrated
that CD63 was strongly expressed in all normal gastric epithelium
and gastric ulcer tissues, and M0-stage gastric carcinomas
exhibited stronger expression of CD63 than M1-stage carcinomas.
Chen et al (23) also noted
that reductions in the expression of CD9, CD63 and CD82 were
indicators of the metastatic potential of gastric carcinoma cells,
and proposed that constitutive expression of CD63 may indicate that
CD63serves a direct role in human gastric carcinogenesis (23).
Similar results to those discussed in the previous
paragraph were achieved in the present study: Immunohistochemical
staining revealed that CD63 expression was downregulated in TSSC
tissues, and analysis of the clinicopathological characteristics of
the TSCC patients indicated that CD63 expression was significantly
associated with the TNM stage, tumor differentiation, and lymph
node metastasis, indicating that the expression level of CD63 may
negatively regulate the development and metastasis of TSCC.
Loss-of-function studies have benefited greatly from
the use of RNA interference techniques. The utilization of shRNAs,
which enables stable gene silencing that is reversible and provides
a method to examine the outcomes of temporary in vivo target
inhibition, assess long-term phenotypes, and conduct pool-based
forward genetic screening (24). The
293 cell line and its derivatives are commonly used as a vehicle in
cell biology studies, owing to their high transfection efficiency.
To identify the function of CD63 in the cell line TCA8113, three
shRNA plasmids against CD63 gene were designed and constructed in
the present study. Lipofectamine 2000 was used to transfect the
shRNA plasmids into 293 cells, and the interference efficiency was
evaluated using RT-qPCR and western blotting. The results confirmed
that all three shRNA plasmids were able to greatly reduce the
expression of CD63, with the plasmid CD63-RNAi-4041 was the most
efficient in this regard. The interference values of CD63-RNAi-4041
at the mRNA and protein levels were 88 and 74%, respectively
(P<0.05). However, Stepanenko and Dmitrenko (25) revealed that 293 cells are tumorigenic,
whereas acute changes to expression of the cancer-associated genes
aggravate tumorigenicity by promoting chromosomal instability. Even
the transfection of a stable empty vector can alter the karyotype
and phenotype (25). In the present
study, in order to exclude the cell karyotype and phenotype changes
in 293 cells, the study further examined the interfering efficiency
of the shRNA plasmids in TCA8113 cells. Therefore, any controversy
from the usage of 293 cells could be dismissed.
A CD63-overexpressing plasmid (PEGFP-N3-CD63) was
constructed using a PCR-based method. CD63-RNAi-4041 and
PEGFP-N3-CD63 plasmids were transfected into TCA8113 cells, and
G418 was used to screen stable cell lines. Stable TCA8113 cell
lines were eventually obtained, and the CD63 expression level was
detected by western blotting. Thus, CD63-silenced and
CD63-overexpressing TCA8113 cells were generated, which were termed
CD63-L and CD63-H, respectively.
Tumor growth and spread is a multistage process.
First, normal cells undergo genetic changes that alter their
phenotypes and enable their ability to spread and colonize, even to
distant sites. A number of factors regulate tumor growth and
spread, and interactions between the tumor and its microenvironment
provide protein products that are crucial to each step of tumor
progression (26). In addition, the
expression of proteolytic enzymes is associated with metastatic
phenotypes. For example, the MMPs, a family of degradative enzymes
associated with malignancy, are involved in the degradation of the
extracellular matrix, including the basement membrane, which is a
specialized matrix composed of type IV collagen, laminin, entactin,
proteoglycans and glycosaminoglycans (27). The ubiquitously present basement
membrane serves as a barrier between tissue compartments, and if
the integrity of the basement membrane is disrupted (which happens
in invasive tumors), the disruption allows the tumor to spread
locally and distantly (28,29).
MMP-2 and MMP-9 serve notable roles in the
degradation of the basement membrane. MMP-2 and MMP-9 are closely
associated with tumor progression in human oral squamous cell
carcinoma, and several studies indicate that these gelatinases are
localized to the advancing tumor front, and have been implicated in
metastatic dissemination (30–34). In
the present study, western blot analysis of the expression of MMP-2
and MMP-9 in CD63-silenced and CD63-overexpressing TCA8113 cells
revealed that when the expression of CD63 was silenced in CD63-L
cells, the expressions of MMP-2 and MMP-9 were increased by 60 and
61%, respectively, whereas the expression of these proteins were
reduced by 29 and 41%, respectively, in CD63-H cells. Considering
that the changes in expression of MMP-2 and MMP-9 may alter the
biological behavior of TCA8113 cells, wound-healing and Transwell
invasion assays were used to measure the migratory and invasive
ability of TCA8113 cells.
In the wound-healing assay, the wound-healing
ability of the control cells was 73% lower than that of the CD63-L
cells, but 59% higher than that of the CD63-H cells. In the
Transwell invasion assay, TCA8113 cells degraded the Matrigel
matrix and passed through the 8.0-µm-pore membrane through cellular
plasticity. The knockdown of CD63 enhanced the invasive ability of
TCA8113 cells.
In conclusion, the findings of the present study
indicate that CD63 may serve an inhibitory role in the malignancy
and lymph node metastasis of TSCC, and may have potential
applications in the prediction of prognosis and gene therapy for
TSCC patients.
Acknowledgements
The authors would like to thank Professor Rongjian
Su and Professor Cuifen Bao of Jinzhou Medical University for their
technical guidance. The authors acknowledge the Pathology
Department of the First Affiliated Hospital of Jinzhou Medical
University for providing human tongue and TSCC tissue
specimens.
Funding
The present study was supported by grants from the
National Nature Science Foundation of China (no. 81201285); the
National Nature Science Foundation of Liaoning Province (no.
20170540398); the Excellent Talents Project in Colleges and
Universities of the Liaoning Province Foundation (no. LJQ2015067),
and the Quanmin Oral Graduate Sci-tech Innovation Foundation, the
President Fund of Jinzhou Medical University (project no.
QM2014003).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WZ and XL conceived and designed the experiments.
WHL performed the experiments and wrote the paper. XLZ and MLH
analyzed the data.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committees of Jinzhou Medical University (approval nos. 20140005
and 20171105). Written informed consent was obtained from all
patients.
Consent for publication
Written informed consent for the publication of data
was obtained from all patients.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sano D and Myers JN: Metastasis of
squamous cell carcinoma of the oral tongue. Cancer Metastasis Rev.
26:645–662. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Neville BW and Day TA: Oral cancer and
precancerous lesions. CA Cancer J Clin. 52:195–215. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ren W, Lian P, Cheng L, Du P, Guan X, Wang
H, Ding L, Gao Z, Huang X, Xiao F, et al: FHL1 inhibits the growth
of tongue squamous cell carcinoma cells via G1/S cell cycle arrest.
Mol Med Rep. 12:3958–3964. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu Z, He Q, Ding X, Zhao T, Zhao L and
Wang A: SOD2 is a C-myc target gene that promotes the migration and
invasion of tongue squamous cell carcinoma involving cancer
stem-like cells. Int J Biochem Cell Biol. 60:139–146. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jia LF, Gan YH and Yu GY: Relationships
between microRNA expressions and prognosis in patients with tongue
squamous cell carcinoma and the mechanisms microRNA regulating
tongue squamous cell carcinoma biological behavior. Beijing Da Xue
Xue Bao Yi Xue Ban. 48:5–9. 2016.(In Chinese). PubMed/NCBI
|
|
8
|
Hotta H, Ross AH, Huebner K, Isobe M,
Wendeborn S, Chao MV, Ricciardi RP, Tsujimoto Y, Croce CM and
Koprowski H: Molecular cloning and characterization of an antigen
associated with early stages of melanoma tumor progression. Cancer
Res. 48:2955–2962. 1988.PubMed/NCBI
|
|
9
|
Maecker HT, Todd SC and Levy S: The
tetraspanin superfamily: Molecular facilitators. FASEB J.
11:428–442. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pols MS and Klumperman J: Trafficking and
function of the tetraspaninCD63. Exp Cell Res. 315:1584–1592. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kobayashi T, Vischer UM, Rosnoblet C,
Lebrand C, Lindsay M, Parton RG, Kruithof EK and Gruenberg J: The
tetraspanin CD63/lamp3 cycles between endocytic and secretory
compartments in human endothelial cells. Mol Biol Cell.
11:1829–1843. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kang M, Ryu J, Lee D, Lee MS, Kim HJ, Nam
SH, Song HE, Choi J, Lee GH, Kim TY, et al: Correlations between
transmembrane 4 L6 family member 5 (TM4SF5), CD151, and CD63 in
liver fibrotic phenotypes and hepatic migration and invasive
capacities. PLoS One. 9:e1028172014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hunziker W and Geuze HJ: Intracellular
trafficking of lysosomal membrane proteins. Bioessays. 18:379–389.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Atkinson B, Ernst CS, Ghrist BF, Herlyn M,
Blaszczyk M, Ross AH, Herlyn D, Steplewski Z and Koprowski H:
Identification of melanoma-associated antigens using fixed tissue
screening of antibodies. Cancer Res. 44:2577–2581. 1984.PubMed/NCBI
|
|
15
|
Jang HI and Lee H: A decrease in the
expression of CD63 tetraspanin protein elevates invasive potential
of human melanoma cells. Exp Mol Med. 35:317–323. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Radford KJ, Thorne RF and Hersey P:
Regulation of tumor cell motility and migration by CD63 in a human
melanoma cell line. J Immunol. 158:3353–3358. 1997.PubMed/NCBI
|
|
17
|
Kwon MS, Shin SH, Yim SH, Lee KY, Kang HM,
Kim TM and Chung YJ: CD63 as a biomarker for predicting the
clinical outcomes in adenocarcinoma of lung. Lung Cancer. 57:46–53.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sauer G, Kurzeder C, Grundmann R,
Kreienberg R, Zeillinger R and Deissler H: Expression of
tetraspanin adaptor proteins below defined threshold values is
associated with in vitro invasiveness of mammary carcinoma
cells. Oncol Rep. 10:405–410. 2003.PubMed/NCBI
|
|
19
|
Sordat I, Decraene C, Silvestre T,
Petermann O, Auffray C, Piétu G and Sordat B: Complementary DNA
arrays identify CD63 tetraspanin and alpha3 integrin chain as
differentially expressed in low and high metastatic human colon
carcinoma cells. Lab Invest. 82:1715–1724. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shear M: The aggressive nature of the
odontogenic keratocyst: Is it a benign cystic neoplasm? Part 3.
Immunocytochemistry of cytokeratin and other epithelial cell
markers. Oral Oncol. 38:407–415. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Woegerbauer M, Thurnher D, Houben R,
Pammer J, Kloimstein P, Heiduschka G, Petzelbauer P and Erovic BM:
Expression of the tetraspanins CD9, CD37, CD63, and CD151 in Merkel
cell carcinoma: Strong evidence for a posttranscriptional
fine-tuning of CD9 gene expression. Mod Pathol. 23:751–762. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen Z, Gu S, Trojanowicz B, Liu N, Zhu G,
Dralle H and Hoang-Vu C: Down-regulation of TM4SF is associated
with the metastatic potential of gastric carcinoma TM4SF members in
gastric carcinoma. World J Surg Oncol. 9:432011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fellmann C and Lowe SW: Stable RNA
interference rules for silencing. Nat Cell Biol. 16:10–18. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Stepanenko AA and Dmitrenko VV: HEK293 in
cell biology and cancer research: Phenotype, karyotype,
tumorigenicity, and stress-induced genome-phenotype evolution.
Gene. 569:182–190. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nelson AR, Fingleton B, Rothenberg ML and
Matrisian LM: Matrix metalloproteinases: Biologic activity and
clinical implications. J Clin Oncol. 18:1135–1149. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yurchenco PD and Schittny JC: Molecular
architecture of basement membranes. FASEB J. 4:1577–1590. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Barsky SH, Siegal GP, Jannotta F and
Liotta LA: Loss of basement membrane components by invasive tumors
but not by their benign counterparts. Lab Invest. 49:140–147.
1983.PubMed/NCBI
|
|
29
|
Liotta LA, Tryggvason K, Garbisa S, Hart
I, Foltz CM and Shafie S: Metastatic potential correlates with
enzymatic degradation of basement membrane collagen. Nature.
284:67–68. 1980. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kusukawa J, Sasaguri Y, Shima I, Kameyama
T and Morimatsu M: Expression of matrix metalloproteinase-2 related
to lymph node metastasis of oral squamous cell carcinoma. A
clinicopathologic study. Am J Clin Pathol. 99:18–23. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kawamata H, Uchida D, Hamano H,
Kimura-Yanagawa T, Nakashiro KI, Hino S, Omotehara F, Yoshida H and
Sato M: Active-MMP2 in cancer cell nests of oral cancer patients:
Correlation with lymph node metastasis. Int J Oncol. 13:699–704.
1998.PubMed/NCBI
|
|
32
|
Ikebe T, Shinohara M, Takeuchi H, Beppu M,
Kurahara S, Nakamura S and Shirasuna K: Gelatinolytic activity of
matrix metalloproteinase in tumor tissues correlates with the
invasiveness of oral cancer. Clin Exp Metastasis. 17:315–323. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hong SD, Hong SP, Lee JI and Lim CY:
Expression of matrix metalloproteinase-2 and −9 in oral squamous
cell carcinomas with regard to the metastatic potential. Oral
Oncol. 36:207–213. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yorioka CW, Coletta RD, Alves F, Nishimoto
IN, Kowalski LP and Graner E: Matrix metalloproteinase-2 and −9
activities correlate with the disease-free survival of oral
squamous cell carcinoma patients. Int J Oncol. 20:189–194.
2002.PubMed/NCBI
|