Introduction
Colorectal cancer (CRC) is a cancer that originates
from parts of the large intestine (such as the colon or rectum) due
to the abnormal proliferation of tumor cells, which have the
ability to invade and migrate to other sites in the body (1,2). CRC is a
major cause of cancer-associated mortality worldwide, and is
estimated to be the third most commonly diagnosed cancer, as well
as the leading cause of cancer-associated mortality, in females and
males (3). The incidence and
mortality rates of CRC vary substantially based on different
countries and ethnicities, with the rates in developing countries
and in African-Americans being relatively higher than those in
developed countries and in Asians, which may be associated with the
disproportionately low socioeconomic status (4,5). A rapid
increase in the incidence and mortality rate of CRC in China has
been witnessed over the past few decades (6,7). In
previous years, a large number of patients with CRC were diagnosed
at an advanced stage; however, due to the widespread implementation
of colonoscopy screening and improvements in treatment (8,9), diagnosis
has now improved. However, prognostic outcomes remain poor, with a
5-year survival rate of <15% (10), as over 50% of all diagnosed cases are
likely to develop metastases, and the majority of these patients
have unresectable tumors (11,12).
Notably, major non-risk factors associated with the development of
CRC include age, gender, diet, alcohol consumption, obesity,
smoking, lack of physical exercise, diabetes and atherosclerosis,
economic status and methods of diagnosis (13–15). Furthermore, at
the cellular level, CRC is a biologically heterogeneous disease,
and the progressive accumulation of genetic changes and epigenetic
alterations may perform critical roles in inducing the pathological
changes involved in transforming normal colonic epithelium into
malignant tumors (16–18).
Proteins in the T cell immunoglobulin domain and
mucin (Tim) family are transmembrane glycoproteins that possess
common structural motifs, including immunoglobulin V, mucin,
transmembrane and cytoplasmic domains (19,20).
Members of the Tim family, which were initially recognized as
specific surface molecules of T helper (Th)1 and Th2 cells, can
provide co-stimulatory signals targeting the functional role of Th1
and Th2 cells regarding the regulation mechanism of differentiation
(21,22). The Tim family encodes eight members in
mice (Tim-1 through Tim-8), which are located at 11B1.1, and gene
loci of the three members in humans (Tim-1,-3 and −4) mapped on
chromosome 5q33.2 (23). Members of
the family are critical for Th1- and Th2-mediated immunity, and
they are implicated in various autoimmune and allergic-associated
diseases, cancer types and viral infections in humans (24–26).
Tim-3 is selectively and highly expressed by well-differentiated
Th1 cells, but not by Th2 cells (27). Following interaction with its ligand,
galectin-9, Tim-3 may induce a positive or negative stimulus for
the activation and differentiation of T cells, so exerting
immunomodulatory properties (28,29).
Similarly, Tim-3 may stimulate tumor cells to escape
immunosurveillance. Previous evidence has documented the inhibitory
effect of Tim-3 on Th1-mediated immunity (30,31).
Furthermore, Tim-3 can cause innate immune cells, including
macrophages/monocytes and natural killer (NK) cells, to inhibit
tissue damaging immune responses (32) and can release inflammatory mediators,
such as tumor necrosis factor-α and interleukin-6, to participate
in the occurrence and development of inflammation (33,34). The
mechanisms underlying Tim-3-mediated T cell activity in mediating
immune- or inflammation-associated diseases are relatively clear;
however, it remains to be clarified whether Tim-3 is expressed on
tumor cells or tissues in patients with CRC. In addition, the
mechanisms by which Tim-3 is involved in human CRC remain unclear.
In the present study, the genetic mutations and expression
activities of Tim-3, and how these affect the clinical features of
patients with CRC, were investigated in order to clarify the role
of Tim-3 in the incidence and development of CRC.
Materials and methods
Ethical statement
The present study was approved by the Ethics
Committee of The First Affiliated Hospital of Harbin Medical
University (Harbin, China). All procedures performed in the present
study involving humans were conducted in accordance with the
ethical standards of the institutional and/or national research
committee in the First Affiliated Hospital of Harbin Medical
University (Harbin, China), and with The Declaration of Helsinki
(1964) and its later amendments (35). All patients enrolled in the present
study were Chinese. The experimental objectives and details
regarding the procedures involved in the present study were
presented to all patients enrolled, and corresponding written
informed consent was provided prior to the initiation of the
experiments.
Patients
A prospective study was conducted based on the
collection and analysis of a total of 258 patients diagnosed with
CRC between December 2012 and June 2015 at The First Affiliated
Hospital of Harbin Medical University. All patients were inquired
in detail in terms of medical history and examined carefully prior
to inclusion in the present study, and the diagnosis of CRC was
determined following consideration of the physical condition of
each patient, the results of clinical laboratory tests and X-ray
imaging. Of all the patients included, 136 were male and 122 were
female, all ranging between 28 and 72 years old, with a mean age of
57.46±10.23 years. The inclusion criteria were as follows: i)
Patients were confirmed to have CRC histologically by pathologists
blinded to the clinical outcome; ii) patients did not receive any
prior preoperative treatment over the past 6 months, including
chemotherapy, radiotherapy or Traditional Chinese Medicine
treatment; iii) patients had no history of other malignant tumor
types; iv) patients were willing to be involved in the study and
could cooperate with the experimental procedures to provide
complete clinical data. The Dukes' (36) and tumor-node-metastasis (TNM) staging
systems, recommended by the Union for International Cancer Control
and the American Joint Committee on Cancer (7th Ed.) (37), were applied for the histological
pattern classification of included patients. During the same period
at the same hospital, 246 cases of normal controls from healthy
volunteers who were examined were collected. Of these, 140 were
male and 106 were female, ranging between 26 and 78 years of age
(mean, 56.50±12.00 years). The controls were confirmed to have no
history of CRC or other malignant tumor types, and could provide
complete clinical information relevant to the experiment.
Separation of peripheral blood
mononuclear cells (PBMCs) and sample collection/processing
After 12 h of overnight fasting, peripheral venous
blood samples (10 ml) were drawn from each patient via clean
venipuncture in the early morning, and collected in
heparin-anticoagulant tubes. Collected blood was diluted (1:1) and
a 20 ml suspension was prepared. Subsequent to adding the Ficoll
separating liquid (Tianjin Haoyang Biological Products Science and
Technology Co., Ltd., Tianjin, China) to the centrifuge tube,
Ficoll density gradient centrifugation was performed (447 × g, 20
min, 18–20°C). The liquids were then divided into four layers from
top to bottom. A straw was gently inserted into the annular white
cloud layer (the second layer) to absorb the PBMCs. PBS (5:1) was
added to the PBMCs, followed by centrifugation (1,500 rpm/min, 15
min, 18–20°C). The supernatant was discarded, and the
centrifugation procedure was repeated. High purity PBMCs were
obtained subsequent to discarding the supernatant. Mononuclear cell
purity of ≥80% was achieved through density gradient centrifugation
and the adherence method (38).
In addition, intestinal samples were collected from
each patient during surgery, including CRC radical cancerous
tissues, paracancerous tissues (<2.0 cm beyond the cancer
tissue) and normal colon mucosa tissues (>5.0 cm beyond the
cancer tissue). All blood and tissue specimens were rapidly placed
into liquid nitrogen and maintained at −80°C for 10 min. All tissue
samples were prepared for the extraction of RNA and proteins for
polymerase chain reaction (PCR) amplification, reverse
transcription-quantitative PCR (RT-qPCR) and western blot analysis.
Furthermore, immunohistochemical staining was performed following
paraffin embedding and sectioning, according to the manufacturer's
protocols. All related experimental procedures were performed in
accordance with the protocol provided by the manufacturer (Boster
Bioengineering Co., Ltd., Wuhan, China). Subsequently, mouse
anti-human TIM-3 monoclonal primary antibody (dilution ratio,
1:100; cat. no. 1315, Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) was added to the sections overnight at 4°C. The samples
were then maintained at room temperature for 30 min, followed by
washing with PBS, and incubation with a rabbit anti-mouse secondary
antibody (cat. no: abs20002; Absin Biotechnology Co., Ltd.
Shanghai, China) for 30 min at 37°C. After further washing with
PBS, DAB staining was performed and the samples were mounted after
another hematoxylin counter-staining and visualized on an inverted
microscope (IX71, Olympus, Tokyo, Japan). In the course of the
experiment, PBS was used as in place of the primary antibody as a
negative control. Positive cells were defined as brown or yellow
brown granules. The pathologists and laboratory personnel involved
were blinded to the case/control status of all collected and
extracted samples.
PCR amplification
Genomic DNA was isolated from the collected whole
blood samples using the Wizard Genomic DNA Purification
Kit® (Promega Corporation, Madison, WI, USA), according
to the manufacturer's protocol. Genotyping of Tim-3
polymorphisms was performed with a PCR-restriction fragment length
polymorphism. According to the sequences of human β-actin
(GI:168480144) and Tim-3 (GI:354681988) obtained from the National
Center for Biotechnology Information GenBank (https://www.ncbi.nlm.nih.gov/), PREMIER Primer 5.0
software (PREMIER Biosoft, Palo Alto, CA, USA) was used for the
design of specific primers. The capture probe sequences were as
follows: Tim-3 forward, 5′-CCAAATCCCAGGCATAAT-3′ and
reverse, 5′-AAGCGA-CAACCCAAAGGT-3′; β-actin forward,
5′-CGAAACTACCT-TCAACTCCATC-3′ and reverse,
5′-AGTGATCTCCTTCTGCATCCT-3′. PCR was performed in a total volume of
15 µl: DNA template (0.5 µl), Taq DNA polymerase (1.25 unit;
Appligene-Oncor, Heidelberg, Germany), 10X Taq buffer (1.5 µl;
Shanghai Pharmaceutical Industries Co., Ltd., Shanghai, China), 2.5
mmol/l dNTP mix (1.0 µl), upstream/downstream primers (per 0.2 µl)
and sterile double-distilled water added up to a final volume of 15
µl. The PCR reaction was conducted under the following conditions:
Pre-degeneration at 95°C for 4 min, followed by 20 cycles of 95°C
for 30 sec, 68°C for 45 sec and 72°C for 60 sec, and then another
20 cycles of 95°C for 30 sec, 58°C for 30 sec and 72°C for 40 sec.
Following the completion of PCR, 3 µl of the PCR products were
separated and analyzed via 2% agarose gel and SDS-PAGE. PCR
products were digested overnight with XhoI and PstI
to identify the Tim-3-882C/T and 4259G/T genotypes. All PCR
products were confirmed with DNA sequencing in an automatic DNA
sequencer ABI 3100 Avant (Applied Biosystems; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), and the single nucleotide
polymorphism genotypes were auto-analyzed using Sequence Detection
System 1.4.0 software (Applied Biosystems; Thermo Fisher
Scientific, Inc.).
RT-qPCR detection
Total RNA extraction from blood samples was
performed using a TRIzol® kit (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. Total
RNA was purified with a NanoVue™ Plus spectrophotometer (GE
Healthcare, Chicago, IL, USA) at wavelengths of 260 and 280 nm. The
extracted RNA from each sample was then reverse transcribed into
cDNA via an RT-PCR assay (Qiagen GmbH, Hilden, Germany), according
to the manufacturer's protocol, followed by preservation at −20°C.
The relative expression of Tim-3 levels was measured using RT-PCR,
as described previously (39), and
the TaqMan-based qPCR reagent kit was provided by Exiqon A/S
(Vedbaek, Denmark). The primer sequences were as aforementioned.
The reaction conditions were: Pre-denaturing for 5 min at 95°C,
followed by 10 sec at 95°C and 1 min at 60°C, for 40 cycles in
total. β-actin was used as the internal control, and the
2−ΔΔCq method (40) was
applied for the detection of Tim-3 and the corresponding control.
Experiments were performed three times to stabilize the final
results.
Western blot analysis
Total protein was extracted using a Total Protein
Extraction kit (Nanjing KeyGen Biotech Co., Ltd., Nanjing, China)
in accordance with the manufacturer's protocol, and the standard
bicinchoninic acid (BCA) method was used for protein determination,
as previously reported (41).
Subsequent to calculating the amount of BCA detection reagent
according to the number of samples, 50 µg total protein was
resolved by 10% SDS-PAGE and transferred to a polyvinylidene
fluoride membrane, which was blocked for 1 h at room temperature
with 5% skimmed milk. All antibodies used in this experiment were
purchased from Abcam (Cambridge, MA, USA). Subsequently, the rabbit
anti-human AGR2 polyclonal (dilution, 1:1,000; catalog no. ab76473)
and rabbit anti-human β-actin polyclonal (dilution, 1:200; catalog
no. ab6276) primary antibodies were added and incubated overnight
at 4°C on an agitating table. Primary antibodies were purchased
from Abcam. On the second day, following washing in PBS at 37°C for
1 h (three times) and three washes with TBS-Tween-20 buffer, a
horseradish peroxidase-labeled goat anti-rabbit secondary antibody
(catalog no. ab181658; dilution, 1:10,000; Abcam) was added and
incubated at 37°C for 1 h, followed by membrane rinsing (three
times, 5 min each). Positive bands were developed and visualised by
autoradiography following the addition of a chemiluminescent
reagent (Sigma-Aldrich; Merck KGaA). Quantitative analysis
(Quantity one analysis system software; Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) was performed by estimating the gray value
ratios of the target protein Tim-3 and the reference protein
β-actin.
Statistical analysis
All data files were processed using SPSS 17.0
statistical software (SPSS, Inc., Chicago, IL, USA). Measurement
data are presented as the mean ± standard deviation and categorical
data were tested using t-test and nonparametric test. All the tests
in the present study were two-sided. Furthermore, Hardy-Weinberg
equilibrium was used to confirm the genotypes and allele
frequencies. Spearman's correlation analysis was also performed to
investigate the correlation between Tim-3 mRNA and protein
expression levels. P<0.05 was considered to indicate a
statistically significant difference.
Results
Baseline characteristics of
patients
As listed in Table I,
there were 136 males and 122 females in the case group, with a mean
age of 57.46±10.23 years. There were 246 normal controls,
consisting of 140 males and 106 females, with a mean age of
56.50±12.00 years. No evident statistical significance was detected
regarding the gender and age distribution between groups
(P>0.05), which were comparable. There were 123 cases of
patients aged <60 years, and the numbers of patients with a
detected tumor size of ≤5 and >5 cm were 131 and 127,
respectively. In addition, 125 patients had a family history of
CRC. Histological grade classification results revealed 117 cases
well-moderate differentiation and 141 cases of poor
differentiation. Lymph node metastasis and distant metastasis were
also detected and recorded; 138 patients had lymph node metastasis
and 121 patients had distant metastasis. A total of 120 patients
were TNM stage I/II and 138 patients were TNM stage III/IV.
Finally, 141 of the cases were clinical stage I or II and 117 cases
were clinical stage III or IV. Immunohistochemistry indicated that
Tim-3 staining (yellow or brown granules) was predominantly
localized in the cytoplasm and nucleus (Fig. 1).
 | Table I.Baseline characteristics of 258
patients with colorectal cancer. |
Table I.
Baseline characteristics of 258
patients with colorectal cancer.
|
| mRNA levels of
Tim-3 (n) |
|
|
|
| Variables | Low levels | High levels | Z-value | P-value |
|---|
| Total | 148 | 110 |
|
|
| Gender |
|
|
|
|
|
Male | 75 | 61 | −0.709 | 0.448 |
|
Female | 73 | 49 |
|
|
| Age |
|
|
|
|
| <60
years | 64 | 59 | −1.650 | 0.099 |
| ≥60
years | 84 | 51 |
|
|
| Tumor size |
|
|
|
|
| ≤5
cm | 66 | 65 | −2.299 | 0.022 |
| >5
cm | 82 | 45 |
|
|
| Family history |
|
|
|
|
|
With | 68 | 57 | −0.932 | 0.352 |
|
Without | 80 | 53 |
|
|
|
Differentiation |
|
|
|
|
|
Well-moderate | 59 | 58 | −2.048 | 0.041 |
|
Poor | 89 | 52 |
|
|
| TNM stages |
|
|
|
|
|
I–II | 58 | 62 | −2.730 | 0.006 |
|
III–IV | 90 | 48 |
|
|
| Lymph node
metastasis |
|
|
|
|
|
With | 88 | 50 | −2.226 | 0.026 |
|
Without | 60 | 60 |
|
|
| Distant
metastasis |
|
|
|
|
|
With | 78 | 43 | −2.163 | 0.031 |
|
Without | 70 | 67 |
|
|
| Clinical
stages |
|
|
|
|
|
I–II | 78 | 63 | −0.728 | 0.467 |
|
III–IV | 70 | 47 |
|
|
Tim-3 gene expression in CRC cases and
corresponding controls
A total of three different genotypes were detected
in the Tim-3-882C/T and 4259T/G, namely CC, CT and TT and
TT, TG and GG, respectively (Table
II). In the control group, the χ2 test revealed that
the genotypes and allele frequencies of −882C/T and 4259T/G
conformed to the Hardy-Weinberg equilibrium. The genotype
distribution and allele frequency of −882C/T and 4259T/G
polymorphisms were significantly different compared with the case
group and the control group (all P<0.05), with the exception of
the genotype distribution comparison of CC/CT between the groups
(P>0.05). Notably, the T and G alleles of −882C/T and 4259G/T
were associated with a significantly increased risk of CRC
[-882C/T: Odds ratio (OR)=1.684; 95% confidence interval (CI),
1.236–2.295; P=0.001; 4259T/G: OR=1.797; 95% CI, 1.316–2.454;
P<0.001].
 | Table II.Distribution of the frequency and
distribution of −882C/T and 4259T/G alleles in the T cell
immunoglobulin domain and mucin-3 gene between the case and control
groups. |
Table II.
Distribution of the frequency and
distribution of −882C/T and 4259T/G alleles in the T cell
immunoglobulin domain and mucin-3 gene between the case and control
groups.
| Site | Case group
(n=258) | Control group
(n=246) | χ2 | P-value | OR (95% CI) |
|---|
| −882C/T |
|
|
|
|
|
| CC | 150 | 168 |
|
| 1.00 |
| CT | 86 | 74 | 1.844 | 0.175 | 1.302
(0.889–1.905) |
| TT | 22 | 4 | 12.024 | 0.001 | 6.160
(2.076–18.282) |
|
CT+TT | 108 | 196 | 8.680 | 0.003 | 0.617
(0.447–0.852) |
| C | 386 | 410 |
|
| 1.00 |
| T | 130 | 82 | 11.026 | 0.001 | 1.684
(1.236–2.295) |
| 4259T/G |
|
|
|
|
|
| TT | 148 | 172 |
|
| 1.00 |
| TG | 88 | 69 | 4.047 | 0.044 | 1.482
(1.009–2.177) |
| GG | 22 | 5 | 12.368 | <0.001 | 5.114
(1.889–13.839) |
|
TG+GG | 110 | 194 | 6.513 | 0.011 | 0.659
(0.478–0.908) |
| T | 384 | 413 |
|
| 1.00 |
| G | 132 | 79 | 13.804 | <0.001 | 1.797
(1.316–2.454) |
Tim-3 mRNA and protein expression in
CRC specimens and PBMCs
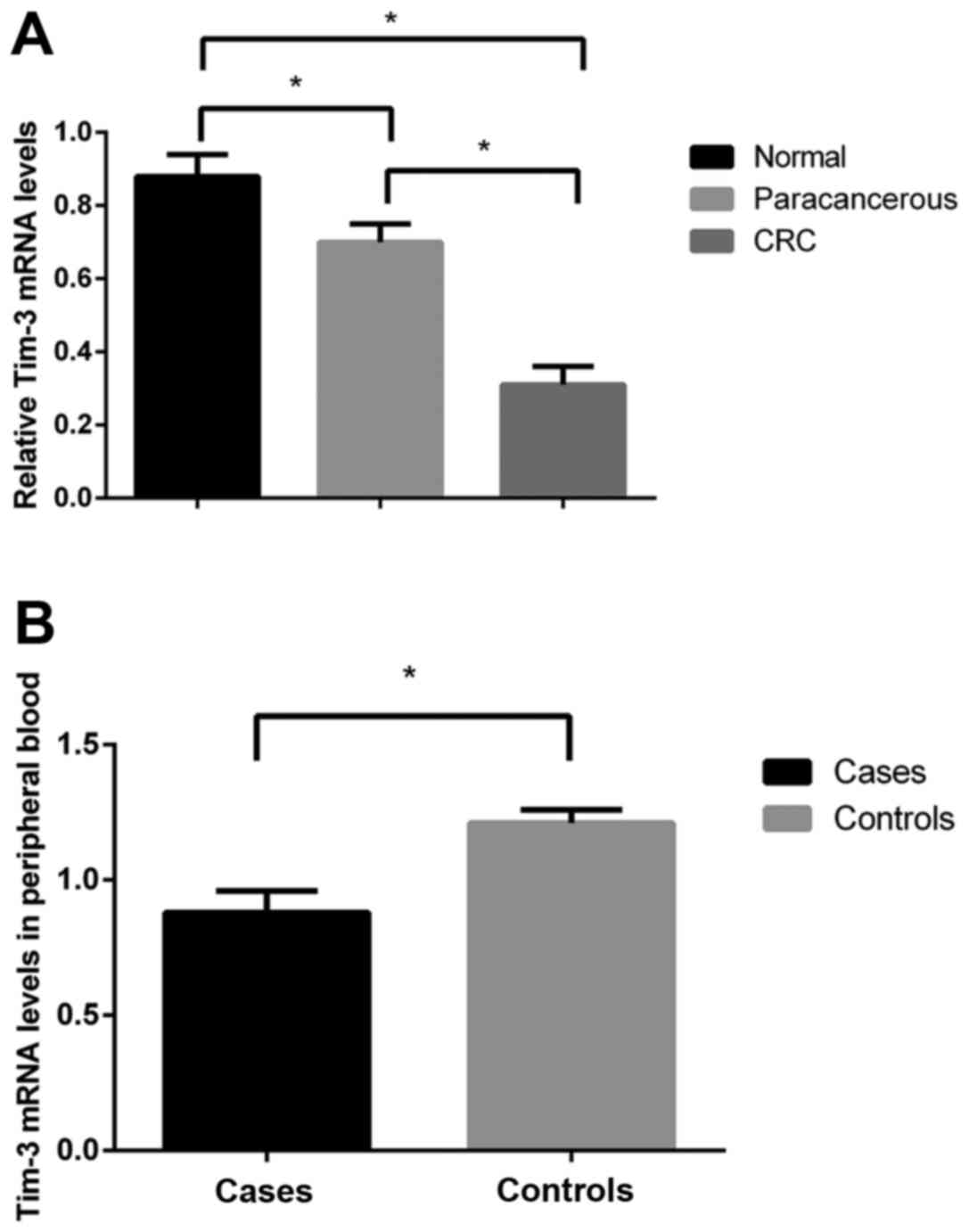
Overall, 148 cases had low Tim-3 expression levels
and 110 had high expression levels. RT-qPCR detection and western
blot analysis revealed clearly decreased levels of Tim-3 mRNA and
its protein expression in monocytes from the peripheral blood and
tissue samples from patients with CRC, as compared with the blood
samples of the normal controls, as well as with the normal colon
mucosa tissues and paracancerous tissues from patients (mRNA
levels: 0.78±0.09 vs. 1.32±0.06; protein expression: 0.34±0.05 vs.
0.73±0.06 vs. 0.88±0.07; all P<0.05), respectively. Furthermore,
in a comparison of Tim-3 protein expression levels between the
paracancerous and the normal colon mucosa tissues, the former had
lower expression levels when compared with the latter (P<0.05)
(Figs. 2 and 3).
Association between Tim-3
polymorphisms and mRNA/protein expression
A significant positive correlation was also observed
between Tim-3 mRNA and protein expression levels, with a
Spearman's correlation coefficient of 0.315 (P=0.029). In the
polymorphism group, a significant negative correlation was observed
between Tim-3 polymorphisms and the mRNA expression levels,
with a Spearman's correlation coefficient of −0.635
(P<0.001).
Association between Tim-3 mRNA levels
and CRC clinicopathological parameters
The associations between Tim-3 mRNA levels and the
clinicopathological parameters of CRC are presented in Table I. Tim-3 mRNA levels were revealed to
be significantly associated with tumor size, differentiation, TNM
stage, lymph node metastasis and distant metastasis (all
P<0.05). However, no significant associations were observed
regarding other clinicopathological parameters, including gender,
age, family history and clinical stage (all P>0.05). These
results suggest that significantly lower Tim-3 mRNA levels could be
detected in patients with a tumor size >5 cm, poor
differentiation degree, higher TNM stage (stage III and IV), and
with lymph node and distant metastasis.
Discussion
Immune homeostasis is an equilibrium state formed
and maintained by the body's immune system interacting with the
environment and components of the immune system, and by the
activation of T lymphocytes and the polarization of Th1/Th2, immune
cells are involved in the regulation of immune homeostasis
(42,43). The Tim gene family, mainly expressed
on the surface of immune cells, has a regulatory effect in tumor
immune surveillance and immune escape (44). A number of previous studies have
demonstrated that once the microenvironment of immune homeostasis
is damaged, it may promote the occurrence of immunological injury
in disease progression, including sepsis, inflammatory bowel
disease, tuberculosis and various malignant tumors (45–48). The
Tim-3 protein performs critical roles in regulating immune function
subsequent to binding with its ligands, which may be regarded as a
novel target for human cancer treatment (29,49,50).
Prior evidence has revealed the expression of Tim-3
in numerous types of malignant tumors, including renal cell
carcinoma and melanoma (51,52); however, few studies have focused on
detecting the expression of Tim-3 in CRC. In the present study,
immunohistochemical detection revealed Tim-3 expression in the
cytoplasm and nuclei of normal gastric mucosa and gastric carcinoma
tissues, confirming that Tim-3 is expressed in patients with CRC
and that it may be critical for the development of CRC. To clarify
the role of Tim-3 in CRC, PCR amplification, RT-qPCR and western
blot analyses were conducted to investigate the roles of its
associated genes and proteins. Firstly, PCR amplification verified
that polymorphisms of Tim-3 were detectable in patients with
CRC, and that Tim-3 mRNA was detectable via RT-qPCR. The results
indicated that the T and G alleles of −882C/T and 4259G/T may be
risk factors for the development of CRC. Secondly, Tim-3 mRNA and
protein expression of the incorporated tissues were detected by
RT-qPCR and western blot analysis, respectively, and the protein
expression levels results were concordant with those of the Tim-3
mRNA levels. Therefore, it was hypothesized that Tim-3
polymorphisms associated with low expression levels of Tim-3 may be
strongly associated with CRC. This hypothesis was verified by
correlation analysis, wherein a negative association between
Tim-3 polymorphisms and mRNA levels was identified,
indicating that Tim-3 polymorphisms may decrease the
expression levels of Tim-3, thus affecting the biological function
of this protein, and be involved in the development of CRC.
A possible mechanism associated with the
aforementioned hypothesis may be that Tim-3 expressed on tumor
cells has diverse tumorigenic activities contributing to
tumor-initiating and tumor-promoting activities. Previous studies
also demonstrated that Tim-3 signals may promote a significant
increase in interferon-γ (IFN-γ), and that suppressed expression of
Tim-3 could inhibit the role of IFN-γ in antitumor immunity
(53,54). However, the mechanisms by which Tim-3
regulates immune factors, cytokines and other parameters must be
identified and verified in future studies. In the long-term process
of tumor formation, tumor cells and the body's immune cells
interact and mutually adapt with each other steadily, and the
functions of immune cells change from the monitoring to the removal
of tumor cells (55,56), to interact and support each other, and
finally to promote tumor immune tolerance, angiogenesis, invasion
and metastasis through the release of cytokines, vascular
endothelial growth factor and matrix degrading enzymes (57). Thus, the inhibition of Tim-3
expression using antagonists, or the interference of relevant
signal pathways, may be a possible method for effecting tumor
suppression. Recently, Gao et al (58) proposed that the Tim-3-574G/T
polymorphism is associated with an increased risk of digestive
system cancer and other cancer (such as hepatocellular cancer and
renal cell carcinoma) in the Chinese Han population. da Silva et
al (52) also revealed that Tim-3
may be considered as a NK cell exhaustion marker in advanced
melanoma, and supported the use of Tim-3-targeted therapies to
restore the antitumor immunity in patients with advanced melanoma
(52). It is therefore essential that
treatment with anti-Tim-3 suppress its tumorigenic effects, which
requires confirmation in in vitro and in vivo
settings. The present study revealed that clinicopathological
parameters, including tumor size, differentiation degree, TNM
stage, lymph node metastasis and distant metastasis, may be
effected by Tim-3 and thus responsible for the development of CRC.
However, Tim-3 expression was not determined to be associated with
gender, age, family history or clinical stage, which may be
accepted only cautiously due to the restricted inclusion of samples
and different backgrounds, including lifestyle, economic status and
family history. With regard to the aforementioned results, and in
combination with those of previous studies, the results suggested
that patients with a tumor size >5 cm, poor differentiation
degree, higher TNM stage (stage III and IV), and with lymph node
and distant metastasis may have a significantly increased risk of
CRC compared with patients without these characteristics, which in
turn strengthened the importance of monitoring patients
clinicopathological parameters in screening for CRC.
The present study had several limitations. Firstly,
an in-depth study of the underlying mechanisms of the role of Tim-3
in the incidence and development of CRC should be conducted, in
order to improve systematic understanding of this topic. Secondly,
the present study used a relatively small sample size, which may
have an effect on the wider clinical applications. Thirdly, the
mechanism by which decreased Tim-3 expression promotes CRC cell
invasion and metastasis requires further study. Finally, the
results of the current study require confirmation through in
vivo and in vitro experimental models with long-term
follow-up in cases and trials to verify the potential effect of
targeted therapies based on Tim-3 in CRC, and in other types of
malignant tumors. The present study hypothesized that the
inhibition of Tim-3 with a specific antagonist may have an
anticancer effect on the development of CRC.
In summary, Tim-3 expression is primarily located in
in the cytoplasm and nucleus, genetic changes of Tim-3 in
the blood and tissue samples are expressed as polymorphisms, and
decreased mRNA and protein expression may be partially responsible
for the incidence and development of CRC. Reduced Tim-3 expression,
induced by genetic polymorphisms of Tim-3, may promote CRC
invasion and metastasis.
References
|
1
|
Richter J, Rudeck S, Kretz AL, Kramer K,
Just S, Henne-Bruns D, Hillenbrand A, Leithauser F, Lemke J and
Knippschild U: Decreased CK1δ expression predicts prolonged
survival in colorectal cancer patients. Tumour Biol. 27:8731–8739.
2016. View Article : Google Scholar
|
|
2
|
Day LW and Velayos F: Colorectal cancer of
the elderly. Curr Treat Options Gastroenterol. 12:269–282. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Burada F, Nicoli ER, Ciurea ME, Uscatu DC,
Ioana M and Gheonea DI: Autophagy in colorectal cancer: An
important switch from physiology to pathology. World J Gastrointest
Oncol. 7:271–284. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pibiri F, Kittles RA, Sandler RS, Keku TO,
Kupfer SS, Xicola RM, Llor X and Ellis NA: Genetic variation in
vitamin D-related genes and risk of colorectal cancer in African
Americans. Cancer Causes Control. 25:561–570. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kumar K, Brim H, Giardiello F, Smoot DT,
Nouraie M, Lee EL and Ashktorab H: Distinct BRAF (V600E) and KRAS
mutations in high microsatellite instability sporadic colorectal
cancer in African Americans. Clin Cancer Res. 15:1155–1161. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sun K, Deng HJ, Lei ST, Dong JQ and Li GX:
miRNA-338-3p suppresses cell growth of human colorectal carcinoma
by targeting smoothened. World J Gastroenterol. 19:2197–2207. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sun K, Wang W, Zeng JJ, Wu CT, Lei ST and
Li GX: MicroRNA-221 inhibits CDKN1C/p57 expression in human
colorectal carcinoma. Acta Pharmacol Sin. 32:375–384. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Triantafillidis JK, Vagianos C and
Malgarinos G: Colonoscopy in colorectal cancer screening: Current
aspects. Indian J Surg Oncol. 6:237–250. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Malkomes P, Lunger I, Luetticke A,
Oppermann E, Haetscher N, Serve H, Holzer K, Bechstein WO and
Rieger MA: Selective AKT inhibition by MK-2206 represses colorectal
cancer-initiating stem cells. Ann Surg Oncol. 23:2849–2857. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nedrebø BS, Søreide K, Eriksen MT, Kvaløy
JT, Søreide JA and Kørner H: Excess mortality after curative
surgery for colorectal cancer changes over time and differs for
patients with colon versus rectal cancer. Acta Oncol. 52:933–940.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
van Gestel YR, de Hingh IH, van Herk-Sukel
MP, van Erning FN, Beerepoot LV, Wijsman JH, Slooter GD, Rutten HJ,
Creemers GJ and Lemmens VE: Patterns of metachronous metastases
after curative treatment of colorectal cancer. Cancer Epidemiol.
38:448–454. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Matsumoto T, Hasegawa S, Matsumoto S,
Horimatsu T, Okoshi K, Yamada M, Kawada K and Sakai Y: Overcoming
the challenges of primary tumor management in patients with
metastatic colorectal cancer unresectable for cure and an
asymptomatic primary tumor. Dis Colon Rectum. 57:679–686. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Vissers PA, Thong MS, Pouwer F, den
Oudsten BL, Nieuwenhuijzen GA and van de Poll-Franse LV: The
individual and combined effect of colorectal cancer and diabetes on
health-related quality of life and sexual functioning: Results from
the PROFILES registry. Support Care Cancer. 22:3071–3079. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jeon JY and Meyerhardt JA: Can we change
the past for colorectal cancer patients and how do we move forward?
Cancer. 120:1450–1452. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kontou N, Psaltopoulou T, Soupos N,
Polychronopoulos E, Xinopoulos D, Linos A and Panagiotakos DB:
Metabolic syndrome and colorectal cancer: The protective role of
Mediterranean diet-a case-control study. Angiology. 63:390–396.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Okugawa Y, Grady WM and Goel A: Epigenetic
alterations in colorectal cancer: Emerging Biomarkers.
Gastroenterology. 149:1204–1225.e12. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mundade R, Imperiale TF, Prabhu L, Loehrer
PJ and Lu T: Genetic pathways, prevention, and treatment of
sporadic colorectal cancer. Oncoscience. 1:400–406. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zoratto F, Rossi L, Verrico M, Papa A,
Basso E, Zullo A, Tomao L, Romiti A, Lo Russo G and Tomao S: Focus
on genetic and epigenetic events of colorectal cancer pathogenesis:
Implications for molecular diagnosis. Tumour Biol. 35:6195–6206.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hu T, Wu Z, Vervelde L, Rothwell L, Hume
DA and Kaiser P: Functional annotation of the T-cell immunoglobulin
mucin family in birds. Immunology. 148:287–303. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu G, Zheng K, Lu X and Wang J, Chai Y and
Wang J: Association between polymorphisms in the promoter region of
T cell immunoglobulin and mucin domain-3 and myasthenia
gravis-associated thymoma. Oncol Lett. 9:1470–1474. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Angiari S and Constantin G: Regulation of
T cell trafficking by the T cell immunoglobulin and mucin domain 1
glycoprotein. Trends Mol Med. 20:675–684. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ju Y, Hou N, Meng J, Wang X, Zhang X, Zhao
D, Liu Y, Zhu F, Zhang L, Sun W, et al: T cell immunoglobulin- and
mucin-domain-containing molecule-3 (Tim-3) mediates natural killer
cell suppression in chronic hepatitis B. J Hepatol. 52:322–329.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ding Q, Yeung M, Camirand G, Zeng Q, Akiba
H, Yagita H, Chalasani G, Sayegh MH, Najafian N and Rothstein DM:
Regulatory B cells are identified by expression of TIM-1 and can be
induced through TIM-1 ligation to promote tolerance in mice. J Clin
Invest. 121:3645–3656. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xiao S, Brooks CR, Sobel RA and Kuchroo
VK: Tim-1 is essential for induction and maintenance of IL-10 in
regulatory B cells and their regulation of tissue inflammation. J
Immunol. 194:1602–1608. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xu J, Jiang P and Liu J: Pooled-analysis
of the association between TIM-1 5383_5397 insertion/deletion
polymorphism and asthma susceptibility. Mol Biol Rep. 41:7825–7831.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li M, Ablan SD, Miao C, Zheng YM, Fuller
MS, Rennert PD, Maury W, Johnson MC, Freed EO and Liu SL:
TIM-family proteins inhibit HIV-1 release. Proc Natl Acad Sci USA.
111:E3699–E3707. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chou FC, Kuo CC, Chen HY, Chen HH and
Sytwu HK: DNA demethylation of the TIM-3 promoter is critical for
its stable expression on T cells. Genes Immun. 17:179–186. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cai XZ, Huang WY, Qiao Y, Chen Y, Du SY,
Chen D, Yu S, Liu N, Dou LY and Jiang Y: Downregulation of TIM-3
mRNA expression in peripheral blood mononuclear cells from patients
with systemic lupus erythematosus. Braz J Med Biol Res. 48:77–82.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shan NN, Hu Y, Hou M, Gao J, Wang X, Liu X
and Li Y: Decreased Tim-3 and its correlation with Th1 cells in
patients with immune thrombocytopenia. Thromb Res. 133:52–56. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lee MJ, Woo MY, Heo YM, Kim JS, Kwon MH,
Kim K and Park S: The inhibition of the T-cell immunoglobulin and
mucin domain 3 (Tim3) pathway enhances the efficacy of tumor
vaccine. Biochem Biophys Res Commun. 402:88–93. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dardalhon V, Anderson AC, Karman J, Apetoh
L, Chandwaskar R, Lee DH, Cornejo M, Nishi N, Yamauchi A, Quintana
FJ, et al: Tim-3/galectin-9 pathway: Regulation of Th1 immunity
through promotion of CD11b+Ly-6G+ myeloid cells. J Immunol.
185:1383–1392. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Roth CG, Garner K, Eyck ST, Boyiadzis M,
Kane LP and Craig FE: TIM3 expression by leukemic and non-leukemic
myeloblasts. Cytometry B Clin Cytom. 84:167–172. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Moller-Hackbarth K, Dewitz C, Schweigert
O, Trad A, Garbers C, Rose-John S and Scheller J: A disintegrin and
metalloprotease (ADAM) 10 and ADAM17 are major sheddases of T cell
immunoglobulin and mucin domain 3 (Tim-3). J Biol Chem.
288:34529–34544. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhao D, Hou N, Cui M, Liu Y, Liang X,
Zhuang X, Zhang Y, Zhang L, Yin D, Gao L, et al: Increased T cell
immunoglobulin and mucin domain 3 positively correlate with
systemic IL-17 and TNF-α level in the acute phase of ischemic
stroke. J Clin Immunol. 31:719–727. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
M PN: World medical association publishes
the revised declaration of Helsinki. Natl Med J India.
27:562014.PubMed/NCBI
|
|
36
|
Ali R, Toh HC and Chia WK: ASCOLT Trial
Investigators: The utility of Aspirin in Dukes C and High Risk
Dukes B Colorectal cancer-the ASCOLT study: Study protocol for a
randomized controlled trial. Trials. 12:2612011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hu H, Krasinskas A and Willis J:
Perspectives on current tumor-node-metastasis (TNM) staging of
cancers of the colon and rectum. Semin Oncol. 38:500–510. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ma B, Qu P, Zhang XM, Zhang YF, Hu PZ, Ge
W, Si SY, Huang Y, Li X and Sui YF: Culturing dendritic cells from
the peripheral blood with successive adherence method and observing
the utralmicrostructure of cells. J Mod Oncol. 2007.
|
|
39
|
Forlenza M, Kaiser T, Savelkoul HF and
Wiegertjes GF: The use of real-time quantitative PCR for the
analysis of cytokine mRNA levels. Methods Mol Biol. 820:7–23. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sun Y, Shen S, Liu X, Tang H, Wang Z, Yu
Z, Li X and Wu M: MiR-429 inhibits cells growth and invasion and
regulates EMT-related marker genes by targeting Onecut2 in
colorectal carcinoma. Mol Cell Biochem. 390:19–30. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jayachandran R and Pieters J: Regulation
of immune cell homeostasis and function by coronin 1. Int
Immunopharmacol. 28:825–828. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sharma R, Kapila R, Dass G and Kapila S:
Improvement in Th1/Th2 immune homeostasis, antioxidative status and
resistance to pathogenic E. coli on consumption of probiotic
Lactobacillus rhamnosus fermented milk in aging mice. Age (Dordr).
36:96862014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Freeman GJ, Casasnovas JM, Umetsu DT and
DeKruyff RH: TIM genes: A family of cell surface phosphatidylserine
receptors that regulate innate and adaptive immunity. Immunol Rev.
235:172–189. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang L, Ai Y and Tsung A: Clinical
application: Restoration of immune homeostasis by autophagy as a
potential therapeutic target in sepsis. Exp Ther Med. 11:1159–1167.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Rossi M and Bot A: The Th17 cell
population and the immune homeostasis of the gastrointestinal
tract. Int Rev Immunol. 32:471–474. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Takeuchi Y and Nishikawa H: Roles of
regulatory T cells in cancer immunity. Int Immunol. 28:401–409.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Cabrera R, Ararat M, Xu Y, Brusko T,
Wasserfall C, Atkinson MA, Chang LJ, Liu C and Nelson DR: Immune
modulation of effector CD4+ and regulatory T cell function by
sorafenib in patients with hepatocellular carcinoma. Cancer Immunol
Immunother. 62:737–746. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Liu Y, Gao LF, Liang XH and Ma CH: Role of
Tim-3 in hepatitis B virus infection: An overview. World J
Gastroenterol. 22:2294–2303. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang L, Zhao C, Peng Q, Shi J and Gu G:
Expression levels of CD28, CTLA-4, PD-1 and Tim-3 as novel
indicators of T-cell immune function in patients with chronic
hepatitis B virus infection. Biomed Rep. 2:270–274. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Cai C, Xu YF, Wu ZJ, Dong Q, Li MY, Olson
JC, Rabinowitz YM, Wang LH and Sun Y: Tim-3 expression represents
dysfunctional tumor infiltrating T cells in renal cell carcinoma.
World J Urol. 34:561–567. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
da Silva IP, Gallois A, Jimenez-Baranda S,
Khan S, Anderson AC, Kuchroo VK, Osman I and Bhardwaj N: Reversal
of NK-cell exhaustion in advanced melanoma by Tim-3 blockade.
Cancer Immunol Res. 2:410–422. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Cheng YQ, Ren JP, Zhao J, Wang JM, Zhou Y,
Li GY, Moorman JP and Yao ZQ: MicroRNA-155 regulates
interferon-gamma production in natural killer cells via Tim-3
signalling in chronic hepatitis C virus infection. Immunology.
145:485–497. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Fu X, Wu B, Huang B, Zheng H, Huang S, Gan
Y, Shen J, Lun ZR and Lu F: The correlation of Tim-3 and IFN-γ
expressions in mice infected with Toxoplasma gondii during
gestation. Parasitol Res. 114:125–132. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Charwat V, Rothbauer M, Tedde SF, Hayden
O, Bosch JJ, Muellner P, Hainberger R and Ertl P: Monitoring
dynamic interactions of tumor cells with tissue and immune cells in
a lab-on-a-chip. Anal Chem. 85:11471–11478. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Bose T and Trimper S: Noise-assisted
interactions of tumor and immune cells. Phys Rev E Stat Nonlin Soft
Matter Phys. 84:0219272011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Mazer B: Is there a place for B cells as
regulators of immune tolerance in allergic diseases? Clin Exp
Allergy. 44:469–471. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Gao X, Yang J, He Y and Zhang J:
Quantitative assessment of TIM-3 polymorphisms and cancer risk in
Chinese Han population. Oncotarget. 7:35768–35775. 2016.PubMed/NCBI
|