Introduction
Breast cancer is a common malignancy with high
morbidity rate (1). Incidence of
breast cancer shows an increasing trend and ~1,400,000 breast
cancer patients are diagnosed each year and the mortality rate is
as high as 36% (2). In China, onset
age of breast cancer is becoming icreasingly younger (3). The survival rate of breast cancer is
closely related to the time of diagnosis; the deterioration of
breast cancer, migration of breast cancer cells and drug resistance
of cancer cells are still key factors for increase in mortality
rate of breast cancer (4).
Progression of breast cancer is related to the
imbalance between proto-oncogenes and tumor suppressor genes
(5,6).
Remodeling and spacing factor-1 (RSF-1) is a recently identified
tumor biomarker that is highly expressed in a variety of human
cancer cells, and many studies have shown that its expression level
is raised continuously in ovarian cancer (7), gastric cancer (8), colorectal carcinoma (9,10), liver
cancer (11,12) and malignancies. Overexpression of
RSF-1 is closely related to the occurrence and progression of
cancer cells. Overexpression of Rsf-1 can promote the growth and
invasion of tumor cells, which can regulate the activity of NF-κB
pathway and the expression of MMP-2, Bcl-2, p65 and ERK protein
(13). This study investigated the
effects of RSF-1 expression interference on proliferation and
apoptosis of breast cancer cells to provide a novel potential
therapeutic target for the treatment of breast cancer.
Materials and methods
Materials and reagents
Michigan cancer foundation-7 (MCF-7) and SKBR-3
breast cancer cells purchased from (Cell Bank of the Chinese
Academy of Sciences, Shanghai, China); Cell Counting Kit-8 (CCK-8),
bromodeoxyuridine (BrdU) kits, and Giemsa dye solution
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany); interferences
RSF-1 small-interfering ribonucleic acid (siRNA), scrambled control
siRNA and DharmaFECT 1 kits (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), RSF-1 human primary antibodies,
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and horseradish
peroxidase (HRP)-labeled secondary antibodies (Abcam, Cambridge,
MA, USA); Dulbecco's modified Eagles medium (DMEM) (Gibco; Thermo
Fisher Scientific, Inc.); apoptosis kits, TRIzol kits (Invitrogen;
Thermo Fisher Scientific, Inc.) and 4′,6-diamidino-2-phenylindole
(DAPI) staining kits (Nanjing Jiancheng Biotechnology Co., Ltd.,
Nanjing, China). Rabbit monoclonal RSF-1 antibody (dilution 1:500;
cat. no. ab109002), rabbit polyclonal GAPDH antibody (dilution
1:500; cat. no. ab37168) and secondary goat anti-rabbit (HRP) IgG
antibody (dilution 1:2,000; cat. no. ab6721) were all purchased
from Abcam. This study was approved by the Ethics Committee of
China Medical University (Liaoning, China).
Transfection with RSF-1 siRNA
MCF-7 cells were cultured in DMEM containing double
antibodies under standard condition and collected during
logarithmic phase. After treatment, MCF-7 breast cancer cells were
divided into two groups to be transfected with RSF-1 siRNA (RSF-1
siRNA) and negative control siRNA (NC), respectively. RSF-1
specific interference sequence: 5′-GGAAAGACAUCUCUACUAUUU-3′, and
sequence of scrambled control siRNA in NC, 5′-GAAGCAACGUAUCUUGA-3′.
Transient transfection was performed according to DharmaFECT 1
manual.
Verification of the interference
effect of RSF-1 siRNA on messenger RNA (mRNA) via reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells in each group
using TRIzol kits, and qualified total RNA samples were subjected
to reverse transcription. Total RNA was taken to synthesize
complementary deoxyribonucleic acid (cDNA) using the RT Revert Aid
First Strand cDNA synthesis kit. The specific reaction conditions
were 42°C for 15 min and 95°C for 3 min. Quantitative analysis was
carried out using the ABI 7500 fluorescence PCR amplification
instrument (Applied Biosystems; Thermo Fisher Scientific, Inc.).
PCR amplification was performed using primers listed in Table I. β-catenin was used as an endogenous
control, Ct values were processed using 2−ΔΔCq method
(14).
 | Table I.RT-qPCR primer sequences. |
Table I.
RT-qPCR primer sequences.
| Gene | Primer sequences |
|---|
| β-catenin | F: |
5′-GCTTGGAATGAGACTGCTGA-3′ |
|
| R: |
5′-CTGGCCATATCCACCAGAGT-3′ |
| RSF-1 | F: |
5′-GATACTATGCGTCTCCAGCCAA-3′ |
|
| R: |
5′-CAACTCGTTTCGATTTCTGACAA-3′ |
Detection of RSF-1 protein expression
by western blot analysis
Cultured cells were seeded in a 6-well plate at a
density of 104/well. After 24 h, supernatant was
discarded. After transfection with RSF-1 siRNA and negative control
siRNA, cells were cultured for 72 h and total protein was
extracted. Protein concentration was measured and SDS-PAGE gel
electrophoresis was performed with 50 µg protein per lane. After
gel transfer, PVDF membranes were blocked at room temperature for 1
h, followed by incubation with rabbit monoclonal RSF-1 antibody
(dilution 1:500; cat. no. ab109002) overnight at 4°C. Membranes
were washed with Tween/Tris-buffered salt solution (TBST) and
incubated with secondary goat anti-rabbit (HRP) IgG antibody
(dilution 1:2,000; cat. no. ab6721) at room temperature for 1 h.
After washing with TTBS, color development with chromo-genic
solution and photography were performed.
Detection of cellular proliferation
inhibition rate via CCK-8 assay
Cell viability was determined by CCK-8 assay, 100 µl
of cell suspension containing 104 transfected MCF-7
cells were seeded in a 96-well plate, 20 µl of CCK-8 was added into
each well 24, 48 and 72 h later. Absorbance value (A) at 450 nm was
measured using an enzyme microplate reader. Cell proliferation
inhibition rate was calculated according to the following formula:
Cellular proliferation inhibition rate (%) = (1 -
Atest/Acontrol) × 100%.
Plate cloning formation
experiment
Cells were digested and 100 cells of each group were
inoculated into a culture dish. After 24 h, cells were transfected
with RSF-1 siRNA and scrambled sequence siRNA, respectively.
Culture medium was changed every 2 days. Cell culture was performed
for 2 weeks and was terminated when macroscopic apophyses were
found in culture dishes. After washing with phosphate-buffered
saline (PBS), fixation with 20% methanol for 15 min was performed.
Then, an appropriate amount of Giemsa solution was added and
staining was performed for 40 min. After washing and air drying,
clones were counted using a cloning counter. Cloning formation rate
(%) = (number of clones/number of inoculated cells) × 100%, and the
inhibition rate was calculated.
Detection of cell proliferation
activity via BrdU
MCF-7 cells were transfected with RSF-1 siRNA and
negative control siRNA according to instructions of BrdU stain kit.
Cells were cultured for 72 h and BrdU solution was added, followed
by addition of stationary solution for fixation, and addition of
tetramethylbenzidine (TMB) substrate. Absorbance at 450 nm was
measured using the enzyme microplate reader. Results were expressed
as relative light unit (RLU).
Detection of apoptosis of MCF-7 cells
through Annexin V (AV)/propidium iodide (PI) double staining
method
MCF-7 cells were transfected with RSF-1 and negative
control siRNAs. Cells were cultured for 72 h and digested with
trypsin. After centrifugation at 2,500 × g for 5 min at 4°C, cells
were washed twice with PBS, followed by addition of 5 µl AV and 5
µl PI. After incubation at room temperature for 15 min, flow
cytometer (Becton-Dickinson; BD Biosciences, Franklin Lakes, NJ,
USA) was used to detect apoptosis rate.
DAPI staining
MCF-7 cells were transfected with RSF-1 and negative
control siRNAs. Cells were cultured for 72 h and washed with
precooled PBS three times. DAPI solution (1 µg/ml) was added to
each well, followed by incubation at 37°C for 5 min in a 5% carbon
dioxide (CO2) incubator. After rinsing with precooled
PBS, a fluorescence microscope was used to observe and record the
results in the dark.
Statistical analysis
Statistical Product and Service Solutions (SPSS)
19.0 software (International Business Machines Corporation, New
York, NY, USA) was used for data analysis. Data were expressed as
mean ± standard deviation. Analysis of variance (ANOVA) was used
for comparison of multiple groups and the post hoc test was Dunnett
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Detection of the interference effect
of RSF-1 siRNA on RSF-1 mRNA via protein expression
Compared with control group, RSF-1 protein
expression level was significantly reduced in RSF-1 siRNA
transfected MCF-7 and SKBR-3 cells (P<0.01) (Fig. 1).
Effect of RSF-1 siRNA on the
proliferation of MCF-7 and SKBR-3 cells
CCK-8 assay was performed to detect cell
proliferation of MCF-7 and SKBR-3 cells at 48 h after transfection.
The results showed that compared with the control group, cell
proliferation rate of MCF-7 and SKBR-3 cells was significantly
reduced after RSF-1 siRNA transfection (P<0.01) (Fig. 2).
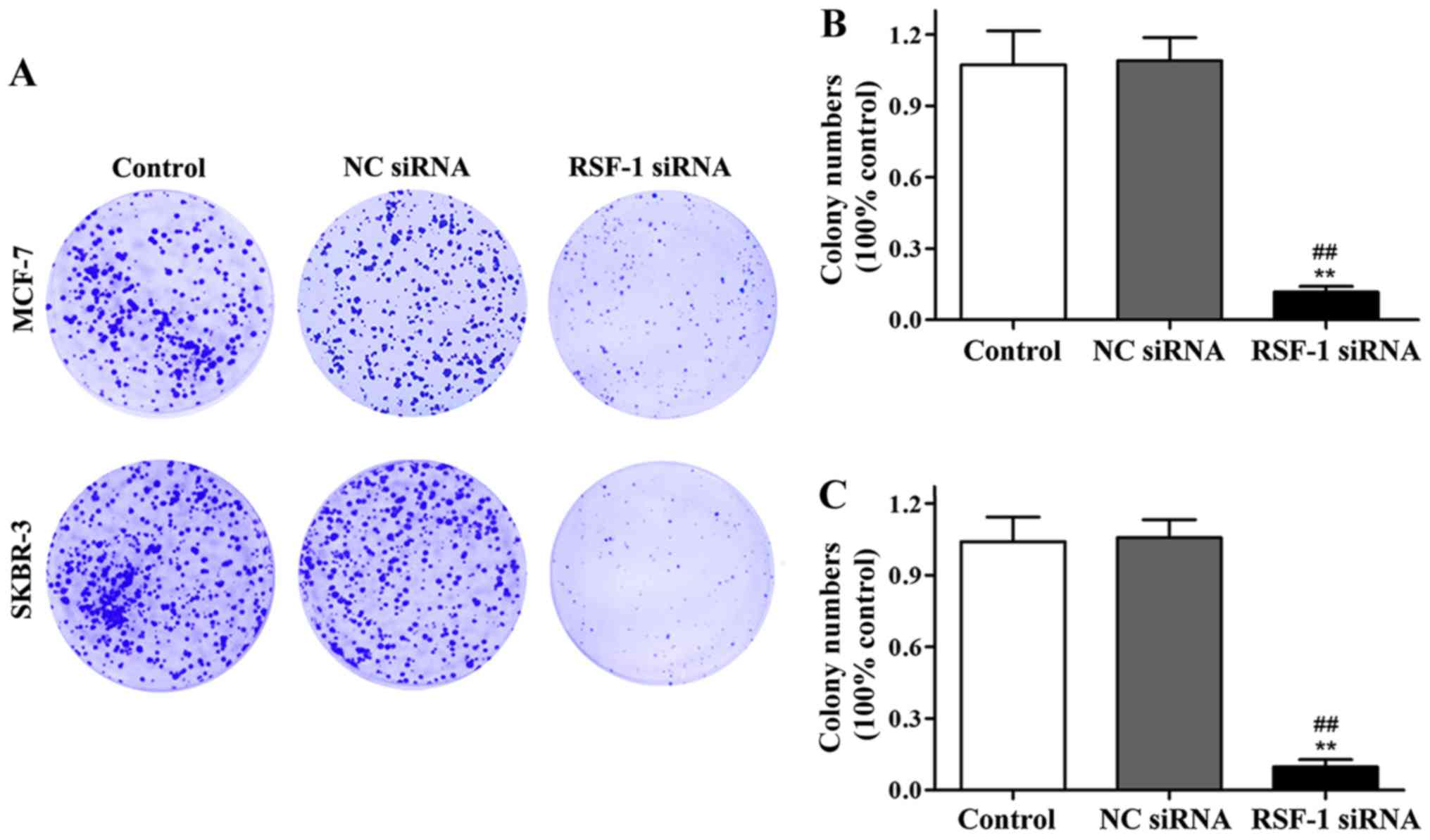
Effect of RSF-1 siRNA on the clone
formation ability of MCF-7 and SKBR-3 cells
Effect of RSF-1 siRNA on clonal formation of MCF-7
and SKBR-3 cells was detected by cell clone formation assay.
Compared with control group, the number of colonies formed in
transfection group was significantly reduced (P<0.01) (Fig. 3).
Annexin V/PI staining to detect the
effect of RSF-1 siRNA on cell apoptosis
AV/PI staining was used to examine the effect of
RSF-1 siRNA on cell apoptosis. As shown in Fig. 4, compared with control group, RSF-1
siRNA transfection significantly increased apoptosis rate of MCF-7
and SKBR-3 cells (P<0.01).
Western blot analysis of the effect of
RSF-1 siRNA on nuclear factor-κB (NF-κB) and its downstream
signaling pathways
As shown in Fig. 5,
compared with control group, Helenalin and Rsf-1 siRNA transfection
significantly reduced the expression levels of p65, Bcl-2 and XIAP
proteins, but showed no significant effects on expression of
p65.
Discussion
Breast cancer seriously affects women's health.
Onset age of breast cancer is becoming increasingly younger, and
its incidence rate ranks first among all malignancies in urban
women (15). Therefore,
identification of novel targets for the treatment of breast cancer
is needed.
RSF-1 can bind to hSNF2H to form RSF. When RSF-1
binds to hSNF2H, RSF-1 assumes the role of a histone chaperone, and
the RSF complex alters chromosome structure through
ATPase-dependent chromatin remodeling processes (16). This process can influence gene
expression, cell cycle progression, and DNA replication. The above
changes in biochemical processes play a very important role in
maintaining the normal cell cycle, cell death, cell apoptosis, cell
senescence, and other cellular processes (17). RSF-1 is a protein with cancer
promoting function. It was found that RSF-1 can promote the
proliferation of tumor cells through interaction with cyclin. It
has been reported that RSF-1 plays a key role in the recombination
repair and non-homologous recombination repair of ovarian cancer
cells (7). Chromosomal breakage may
lead to apoptosis, but repair induced by RSF-1 can maintain the
survival and proliferation of damaged cells, but this repair can
only improve the survival and proliferation of mutant cells and
does not restore cells to normal condition (18). RSF-1 can improve tumor cell viability
by repairing abnormal chromosomes, and it can also influence tumor
cell resistance to chemotherapeutic drugs by regulating NF-κB
signaling pathway (19).
In this study, endogenous RSF-1 expression was
interfered by siRNA interference. MCF-7 and SKBR-3 cells were
cultured in vitro and divided into blank control group,
negative siRNA control group (NC) and RSF-1 siRNA group. The
expression of RSF protein after interference was detected by
western blot analysis. Results showed that expression level of
RSF-1 protein in RSF-1 siRNA transfected cells was significantly
reduced. Cell proliferation assays showed that RSF-1 siRNA
significantly reduced the proliferation ability and clone formation
ability of MCF-7 and SKBR-3 cells compared to the blank control
group. AV/PI double staining results showed that compared with the
blank control group, RSF-1 siRNA significantly increased the
apoptosis rate of MCF-7 and SKBR-3 cells. Helenalin and Rsf-1 siRNA
significantly reduced the expression levels of p-p65, Bcl-2 and
XIAP proteins, indicating that Rsf-1 can regulate the expression of
NF-κB and its downstream signaling pathway related genes. It has
been reported that breast cancer tissues with high expression level
of RSF-1 are usually derived from young patients (20,21).
In conclusion, this study showed that endogenous
RSF-1 expression interference can significantly inhibit
proliferation of human breast cancer cells and induce their
apoptosis. RSF-1 may be a potential therapeutic target for breast
cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YL helped with transfection with RSF-1 siRNA. YL and
JG performed PCR. LF and XZ were responsible for western blot
analysis and CCK-8 assay. EW and QL contributed to plate cloning
formation experiment. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
China Medical University (Liaoning, China) and informed consents
were signed by the patients or the guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hu Z, Mao JH, Curtis C, Huang G, Gu S,
Heiser L, Lenburg ME, Korkola JE, Bayani N, Samarajiwa S, et al:
Genome co-amplification upregulates a mitotic gene network activity
that predicts outcome and response to mitotic protein inhibitors in
breast cancer. Breast Cancer Res. 18:702016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen WQ, Zeng HM, Zheng RS, Zhang SW and
He J: Cancer incidence and mortality in China, 2007. Chin J Cancer
Res. 24:1–8. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Macià F, Porta M, Murta-Nascimento C,
Servitja S, Guxens M, Burón A, Tusquets I, Albanell J and Castells
X: Factors affecting 5- and 10-year survival of women with breast
cancer: An analysis based on a public general hospital in
Barcelona. Cancer Epidemiol. 36:554–559. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jones C, Ford E, Gillett C, Ryder K,
Merrett S, Reis-Filho JS, Fulford LG, Hanby A and Lakhani SR:
Molecular cytogenetic identification of subgroups of grade III
invasive ductal breast carcinomas with different clinical outcomes.
Clin Cancer Res. 10:5988–5997. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bange J, Zwick E and Ullrich A: Molecular
targets for breast cancer therapy and prevention. Nat Med.
7:548–552. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shamay M, Barak O and Shaul Y: HBXAP, a
novel PHD-finger protein, possesses transcription repression
activity. Genomics. 79:523–529. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hu BS, Yu HF, Zhao G and Zha TZ: High
RSF-1 expression correlates with poor prognosis in patients with
gastric adenocarcinoma. Int J Clin Exp Pathol. 5:668–673.
2012.PubMed/NCBI
|
|
9
|
Liu S, Dong Q and Wang E: Rsf-1
overexpression correlates with poor prognosis and cell
proliferation in colon cancer. Tumour Biol. 33:1485–1491. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lin CY, Tian YF, Wu LC, Chen LT, Lin LC,
Hsing CH, Lee SW, Sheu MJ, Lee HH and Wang YH: Rsf-1 expression in
rectal cancer: with special emphasis on the independent prognostic
value after neoadjuvant chemoradiation. J Clinl Pathol. 65:6872012.
View Article : Google Scholar
|
|
11
|
Xie C, Fu L, Xie L, Liu N and Li Q: Rsf-1
overexpression serves as a prognostic marker in human
hepatocellular carcinoma. Tumour Biol. 35:7595–7601. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang X, Xu H, Gao Q, Jiang H and Li Q:
The expression and significance of Rsf-1 in hepatocellular
carcinoma. J Moder Oncol. 1:98–100. 2016.
|
|
13
|
Li Q, Dong Q and Wang E: Rsf-1 is
overexpressed in non-small cell lung cancers and regulates cyclin
D1 expression and ERK activity. Biochem Biophys Res Commun.
420:6–10. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chan SW, Lim CJ, Guo K, Ng CP, Lee I,
Hunziker W, Zeng Q and Hong W: A role for TAZ in migration,
invasion, and tumorigenesis of breast cancer cells. Cancer Res.
68:2592–2598. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sheu JJ, Choi JH, Yildiz I, Tsai FJ, Shaul
Y, Wang TL and Shih IeM: The roles of human sucrose nonfermenting
protein 2 homologue in the tumor-promoting functions of Rsf-1.
Cancer Res. 68:4050–4057. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Loyola A, Huang JY, LeRoy G, Hu S, Wang
YH, Donnelly RJ, Lane WS, Lee SC and Reinberg D: Functional
analysis of the subunits of the chromatin assembly factor RSF. Mol
Cell Biol. 23:6759–6768. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Flanagan JF and Peterson CL: A role for
the yeast SWI/SNF complex in DNA replication. Nucleic Acids Res.
27:2022–2028. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cosma MP, Tanaka T and Nasmyth K: Ordered
recruitment of transcription and chromatin remodeling factors to a
cell cycle- and developmentally regulated promoter. Cell.
97:299–311. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hausheer FH and Yarbro JW: Diagnosis and
treatment of malignant pleural effusion. Semin Oncol. 12:54–75.
1985.PubMed/NCBI
|
|
21
|
Martínez-Moragón E, Aparicio J, Sanchis J,
Menéndez R, Cruz Rogado M and Sanchis F: Malignant pleural
effusion: Prognostic factors for survival and response to chemical
pleurodesis in a series of 120 cases. Respiration. 65:108–113.
1998. View Article : Google Scholar : PubMed/NCBI
|