Introduction
Colorectal carcinoma (CRC) is the third most common
cancer worldwide (1). Additionally,
it is the fifth leading cause of death in China and is a major
public health problem (2). The
activation of oncogenes coupled with the inactivation of tumor
suppressor genes may lead to the carcinogenesis of CRCs (3). In previous years, increasing numbers of
oncogenes and tumor suppressor genes, including Adenomatous
polyposis coli, Tumor protein 53 and KRAS proto-oncogene, GTPase
have been clearly identified as key factors in tumor formation
(4–7).
However, the identification of these authenticated targeting
molecules, which contribute to an improved understanding of the
occurrence and development of CRCs, is insufficient for the
development of a cure.
5-Hydroxytryptamine (5-HT, serotonin) was first
identified as a vasoconstrictor from the blood (8). It was subsequently characterized as a
neurotransmitter in the central nervous system (CNS) (9). It is primarily isolated in the
gastrointestinal tract, platelets, and the CNS (10). 5-HT exerts its biological function
through binding numerous cognate receptors, including the 5-HT1/5,
5-HT2, 5-HT3 and 5-HT4/6/7 subtypes (11). Generally, the serotonin receptor genes
encode G-protein-coupled serotonin receptors, with the exception of
the 5-HT3 subtype. The 5-HT3 subtype, which has 3 subunits
including 5-HT3A (also termed HTR3A), 5-HT3B (also
termed HTR3B), and HTR3C, may encode a subunit of the
ligand-gated cation channel (12–15). The
HTR3A subunit forms a functional channel as a homo-pentamer;
however, the HTR3B subunit alone does not and the underlying
cause has not yet been determined.
5-HT, as a mitogenic factor, has been suggested to
contribute to certain malignancies including breast, prostate and
bladder cancer (16–18). Tutton et al (19) demonstrated the effect of serotonin in
dimethylhydrazine-induced adenocarcinoma of the colon:
Intra-peritoneal injection of a small dose (10 mg/kg) of serotonin
resulted in an increase in the tumor cell mitotic rate. Similarly,
5HT3 and 5HT4 agonists caused significant proliferation of the
colorectal cancer HT29 cell line, while 5HT3 and 5HT4 antagonists
inhibited the cell growth (20). A
previous study also revealed that HTR3A expression was
directly associated with tumor grade in follicular lymphoma,
suggesting that HTR3A may participate in its carcinogenesis
(21). However, the precise role of
HTR3A in CRC has not been fully evaluated.
In the present study, to detect the functional role
of HTR3A in CRC, HTR3A expression was silenced in
human CRC HCT116 and SW1116 cell lines via construction of a short
hairpin RNA (shRNA) lentiviral vector. Furthermore, the effects of
HTR3A silencing on the growth and apoptosis of CRC cells
were determined by MTT, colony formation and flow cytometry
assays.
Materials and methods
Analysis of Oncomine data
In order to determine the expression of HTR3A
in human colorectal cancer, data mining using the Oncomine database
(www.oncomine.org; date of access, 03/06/2016) was
performed. The gene expression of HTR3A in cancer tissues
was compared with normal colorectal tissues, collected from a
number of datasets containing colon and colorectal data (Bittner,
colon; Gaedcke, colorectal; Hong, colorectal; Jorissen, colorectal
2; Laiho, colon; Notterman, colon; Reid, colon; Sabates-Bellver,
colon; Watanane, colon.) (22–25). An
Oncomine outlier analysis was also performed in the colorectal
cancer tissues.
Cell culture
The human embryonic kidney (HEK) 293T cell line and
CRC DLD-1, HCT116, LoVo, RKO, SW1116 and SW620 cell lines were
obtained from The Cell Bank of Type of Culture Collection of
Chinese Academy of Science (Shanghai, China). SW1116 and 293T cells
were cultured in Dulbecco's modified Eagle's medium (Hyclone; GE
Healthcare Life Sciences, Logan, UT, USA) with 10% fetal bovine
serum (FBS; Biowest, Nuaillé, France). DLD-1, HCT116, LoVo, RKO and
SW620 cells were cultured in RPMI-1640 medium (Hyclone; GE
Healthcare Life Sciences) with 10% FBS. All cells were incubated at
37°C in 5% CO2 humidified air.
Lentivirus plasmids and
transfection
To knock down HTR3A expression in the colon
cancer cell lines, a lentivirus-mediated shRNA vector was
constructed. A total of 100 ng of oligonucleotides was mixed with
100 ng linearized pFH-L vector (Shanghai Hollylab Co., Ltd.,
Shanghai, China) to perform the ligation under the catalytic action
of ligase for 2 h at 25°C. The sequences of the oligonucleotides
cloned into the pFH-L vector were as follows: Control shRNA
(shCon), 5′-TTCTCCGAACGTGTCACGT-3′; human HTR3A gene shRNA
#1 [shHTR3A(S1)], 5′-CTACAGCATCACCCTGGTTAT-3′; and #2
[shHTR3A(S2)], 5′-CAAATATCCCGTACGTGTATA-3′. Lentiviruses were
generated following the co-transfection of recombinant pFH-L vector
with the pHelper plasmids including pVSVG-I and pCMV-∆R8.92
(Shanghai Hollybio Co., Ltd., China) into 293T cells using
Lipofectamine 2000® (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) according to the manufacturer's
protocol for 48 h. Then, SW1116 (40,000 cells/well) and HCT116
(50,000 cells/well) cells were plated into 6-well plates overnight
and transfected with different lentiviruses including shCon,
shHTR3A(S1) and shHTR3A(S2) vectors with a multiplicity of
infection (MOI) of 20 and 50, respectively. The infection
efficiency was monitored through observation of the expression
level of green fluorescent protein (GFP) under an Olympus CKX41
microscope (Olympus Corporation, Tokyo, Japan) with a magnification
×100 after 96 h.
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from colon cancer cell lines
using TRIzol® reagent (Life Technologies; Thermo Fisher
Scientific, Inc.), and then cDNA was synthesized using 2.0 µg Total
RNA and 1 µl Oligo dT1 (0.5 µg/µl, Shanghai Shenggong, China) by
M-MLV Reverse Transcriptase and M-MLV Reverse Transcriptase Buffer
(Promega Corporation, Madison, WI, USA) according to the
manufacturer's protocol at 40°C for 60 min. RT-qPCR was then
performed using the Bio-Rad Connect Real-Time PCR platform (CFX96
Touch™, Bio-Rad Laboratories, Inc., Hercules, CA, USA).
The solution contained 10 µl 2X SYBR premix ex Taq (Bio-Rad, USA),
150 ng cDNA and 0.8 µl forward primer and reverse primer (2.5 µM),
respectively in a total volume of 20 µl. The primers used for
amplification of human HTR3A gene were as follows: Forward,
5′-CATCTTCATTGTGCGGCTGGTG-3′; and reverse,
5′-AGTCATCAGTCTTGGTGGCTTGG-3′. As an internal standard,
β-actin was amplified using the following primers: Forward,
5′-GTGGACATCCGCAAAGAC-3′; and reverse, 5′-AAAGGGTGTAACGCAACTA-3′.
Absorbance value was read at the extension stage and the
2−ΔΔCq method was used to quantify the results (26).
Western blot analysis
The lentivirus transfected cells were lysed using
lysis buffer (100 mM Tris, 4% SDS, 10% glycerin, 200 mM NaCl, 2 mM
EDTA) to extract total protein, and the protein concentration was
measured using a bicinchoninic acid Protein Assay kit (P0012,
Beyotime Institute of Biotechnology, Haimen, China). Protein
samples (30 µg) were loaded onto 10% SDS-PAGE at 80 V for 2 h and
transferred to polyvinylidene difluoride membranes (EMD Millipore,
Billerica, MA, USA). Then, the membranes were blocked for 1 h at
25°C in 5% non-fat milk (in TBST buffer: 10 mM Tris, 100 mM NaCl,
0.1% Tween-20; pH=7.4) and incubated with primary antibodies as
follows: Rabbit anti-HTR3A (1:400 dilution; cat. no., 10443-1-AP,
ProteinTech Group, Inc., Chicago, IL, USA), rabbit
anti-Bcl-2-associated death promoter (BAD; 1:1,000 dilution; cat.
no., 10435-1-AP; ProteinTech Group, Inc.), rabbit
anti-B-cell-2-associated X protein (BAX; 1:500 dilution; cat. no.,
2774; Cell Signaling Technology, Inc., Danvers, MA, USA), rabbit
anti-Bcl-2 (1:1,000 dilution; cat. no., 2876; Cell Signaling
Technology, Inc.) and rabbit anti-GAPDH (1:100,000 dilution; cat.
no., 10494-1-AP; ProteinTech Group, Inc.) at 4°C for overnight.
Following three wash steps with TBST (10 mM Tris, 100 mM NaCl, 0.1%
Tween-20; pH=7.4), the membranes were incubated for 1 h with
anti-rabbit horse-radish peroxidase-conjugated secondary antibodies
(1:5,000 dilution; cat. no., SC-2054; Santa Cruz Biotechnology,
Inc., Dallas, TX, USA) at room temperature. The target bands were
detected with Pierce™ ECL Western Blotting Substrate (Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol.
MTT assay
The effect of HTR3A silencing on CRC cell
proliferation was assessed by MTT assay. SW1116 (2,500 cells/well)
and HCT116 (2,000 cells/well) cells infected with shCon and shHTR3A
lentiviruses were seeded in 96-wells plates. At the indicated time
points (1, 2, 3, 4 and 5 days), MTT (5 mg/ml, Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany) was added to the plate and incubated for
4 h at 37°C. Then, 100 µl acidic isopropanol (10% SDS, 5%
isopropanol and 0.01 mol/l HCl) was added to dissolve the formazan.
The absorbance at 595 nm was measured quantitatively using a
microplate reader (Epoch™, BioTek Instruments,
Inc.).
Colony formation assay
The effect of HTR3A silencing on CRC cell
colony formation was measured using colony formation assay. Equal
numbers (200 cells/well) of SW1116 and HCT116 cells transfected
with shCon or shHTR3A were inoculated in 6-wells plate and cultured
continuously at 37°C for 11 and 8 days, respectively. The cell
medium (DMEM or RPMI-1640 as described above) was changed every 2
days. Then, the cell colonies were washed with PBS, fixed in 4%
paraformaldehyde for 30 min at 4°C, and stained with crystal violet
(C0121; Beyotime Institute of Biotechnology) for 20 min at 25°C.
The single colonies (>50 cells) was counted and images were
captured under a CH-2 light microscope at a magnification of ×40
(Olympus Corporation, Tokyo, Japan).
Flow cytometric analysis
The effects of HTR3A silencing on CRC cell
cycle distribution and apoptosis were examined using flow
cytometric analysis. SW1116 (80,000 cells/well) and HCT116 (120,000
cells/well) cells infected with shCon or shHTR3A were seeded in 6
cm dishes and cultured for 5 days at 37°C. For the cell cycle
analysis, the cells were harvested with centrifugation at 5,000 g
for 3 min at 25°C, washed in cold PBS and fixed in 70% ethanol for
overnight at 4°C. Then, cells were stained with 500 µl propidium
iodide solution containing RNase (C1052; Beyotime Institute of
Biotechnology) for 30 min in darkness according to the
manufacturer's protocol. For apoptosis analysis, the cells were
also harvested, washed in PBS and then stained using the Annexin
V-APC/7-AAD Apoptosis Detection kit (KGA1026, Nanjing KeyGen
Biotech Co., Ltd., Nanjing, China) according to the manufacturer's
protocol. Finally, the cells were measured using a flow cytometer
(Gallios™, Beckman Coulter, Inc., Brea, CA, USA). Cell
cycle data were analyzed using modifit software (version 5.0;
Verity Software House, ME, USA) and apoptosis data were analyzed
using FlowJo software (version 7.6; FlowJo LLC, Ashland, OR,
USA).
Statistical analysis
Data are presented as mean ± standard deviation from
three independent experiments. Statistical analysis was performed
using GraphPad Prism software (version 5.0; GraphPad Software,
Inc., La Jolla, CA, USA). An unpaired Student's t-test was used
when comparing data between two groups. For multi-group analysis,
one-way and two-way analysis of variance with Bonferroni post hoc
test was used. P<0.05 was considered to indicate a statistically
significant difference.
Results
HTR3A is efficiently silenced in CRC
cell lines
HTR3A expression levels in colorectal cancer
tissues were investigated using the publicly available Oncomine
database (www.oncomine.org) in Sep. 2017. The
results in Fig. 1 indicated that no
significant changes in HTR3A expression were observed in the
colorectal tumor samples compared with normal tissue. Considering
that cancer heterogeneity, including cellular morphology, gene
expression, metabolism, motility, proliferation and metastatic
potential was a potential challenge in proto-oncogene screening, an
Oncomine outlier analysis was performed in colorectal cancer
samples for HTR3A expression, with the outlier set at the
95th percentile. As indicated in Fig.
1, the expression of HTR3A was demonstrated to be
significantly increased in 10 databases from different studies.
following this outlier analysis. These results demonstrated that
HTR3A was overexpressed in certain colorectal cancer samples and
may be a proto-oncogene in colorectal cancer.
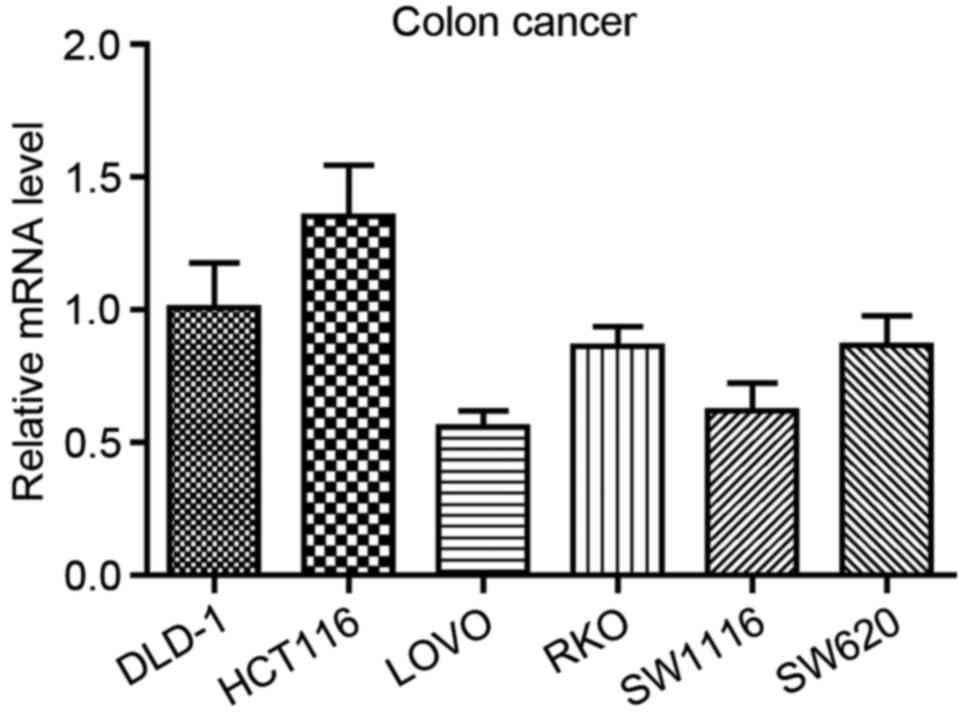
To analyze the role of HTR3A in the
occurrence and development of CRC in vitro, the endogenous
expression of HTR3A was examined in the 6 CRC DLD-1, HCT116,
LOVO, RKO, SW1116 and SW620 cell lines. As demonstrated in Fig. 2, it was identified that HTR3A
was widely expressed in CRC cell lines, and the highest expression
level was observed in HCT116 cells. Therefore, this cell line was
selected for the subsequent analyses. In addition, to demonstrate
the universality of the functional role of HTR3A in CRC
cells, SW1116 was also randomly selected. They were used to perform
the subsequent in vitro experiments through RNA
interference. The infection efficiency, as measured using a
lentivirus-mediated shRNA vector expressing GFP and observed using
fluorescence microscope, was >80% in the HCT116 cells following
infection with shHTR3A(S1) and shHTR3A(S2) groups (Fig. 3A), indicating that the off-target
effect did not occur. As the off-target effect was excluded and it
exhibited sufficient knockdown efficiency, only shHTR3A(S1) was
used in SW1116 cells to silence HTR3A expression. Additional
confirmation that HTR3A mRNA and protein levels were
significantly decreased in HCT116 and SW1116 cells infected with
shHTR3A was obtained by RT-qPCR and western blot analysis
(P<0.01; Fig. 3B and C).
Collectively, these results indicated that the lentivirus-mediated
shRNA may silence HTR3A expression successfully in CRC cell
lines in vitro.
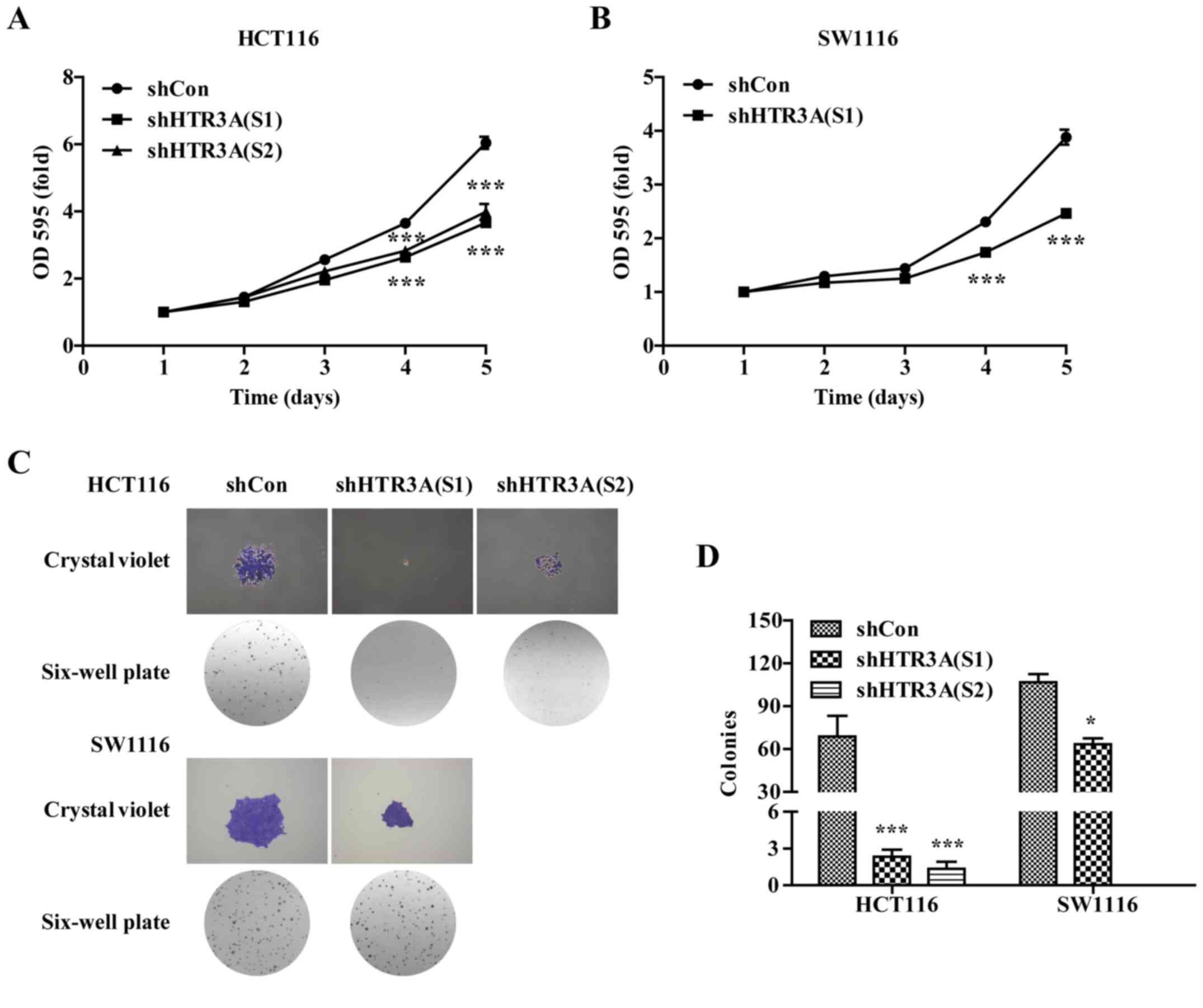
HTR3A silencing inhibits CRC cell
growth
The effect of HTR3A silencing on CRC cell
growth was explored by MTT and colony formation assays. As
indicated in Fig. 4A and B, the
proliferative ability of the CRC cells was significantly decreased
in HTR3A knockdown group compared with those in the shCon group
(P<0.001). Concurrently, it was identified that the size and
number of colonies formed in the shHTR3A (S1) or shHTR3A (S2)
groups was decreased compared with those in shCon group in the
HCT116 and SW1116 cells (Fig. 4C).
The quantitative analysis results confirmed that the number of
colonies was significantly decreased in the shHTR3A(S1) and shHTR3A
(S2) groups in HCT116 cells (Fig. 4D;
P<0.05), and shHTR3A (S1) group in the SW1116 group compared
with shCon groups (Fig. 4D;
P<0.05). Together, the data indicated that CRC cell growth was
inhibited following HTR3A knockdown in vitro.
HTR3A silencing induces CRC cell cycle
arrest
In light of the effect of HTR3A silencing on
CRC cell growth, whether HTR3A depletion also affected the
cell-cycle progression was investigated. Flow cytometric analysis
indicated that HTR3A silencing significantly increased the
percentage of cells in S phase in the shHTR3A(S1) and shHTR3A(S2)
groups [shHTR3A(S1) group, 31.98±0.92 vs. 23.43±0.77%, P<0.01;
shHTR3A(S2) group, 28.69±1.13 vs. 23.43±0.77%, P<0.001],
accompanied with decreased G2/M phase [shHTR3A(S1)
group, 19.70±0.83% vs. 21.70±0.95%, P<0.05; shHTR3A(S2) group,
13.03±0.57% vs. 21.70±0.95%, P<0.001] compared with those in the
shCon group in HCT116 cells (Fig. 5A and
B). Additionally, in the SW1116 cells, an increase in the
percentage of cells in S phase in the shHTR3A(S1) group (27.51±0.12
vs. 32.50±0.64%; P<0.01) was observed, but a decrease in
G2/M phase cells [24.68±0.10 vs. 17.14±0.77%; P<0.01
in shHTR3A(S1) group] in comparison with the shCon group was
indicated (Fig. 5A and C). These
results suggested that HTR3A knockdown may damage cell
growth through arresting cell cycle in CRC cells.
Knockdown of HTR3A induces apoptosis
by regulating apoptosis-associated molecules
Next, the effect of HTR3A silencing on
apoptosis was examined to initially explore how HTR3A was
involved in CRC cell growth. As expected, HTR3A knockdown
led to an increased proportion of early apoptotic cells [Annexin
V+/7AAD-shHTR3A(S1) group, 33.04±0.78%; shHTR3A(S2) group,
78.85±0.55%] and late apoptotic cells [Annexin V+/7AAD+ shHTR3A(S1)
group, 9.65±0.72%; shHTR3A(S2) group, 15.64±0.66%] compared with
the shCon group (Annexin V+/7AAD-, 22.05±0.96%, P<0.001; Annexin
V+/7AAD+, 4.45±1.06%, P<0.001) in HCT116 cells (Fig. 6A and B). Accordingly, the expression
of apoptosis-associated proteins including BAD, BAX, and Bcl-2 were
also detected, and it was identified that the expression levels of
BAD and BAX were upregulated while Bcl-2 was downregulated in
HTR3A-knockdown cells (Fig.
6C). Taken together, these results demonstrated that
HTR3A knockdown accelerated CRC cell apoptosis by the
regulation of partial Bcl-2 family protein expression, including
BAD, BAX and BCL-2 protein.
 | Figure 6.HTR3A depletion induces
colorectal cancer cell apoptosis by regulating apoptosis-associated
molecules. (A) Representative flow cytometry data of apoptosis in
HCT116 cells following HTR3A silencing. (B) Statistical
analysis of cell apoptosis. (C) Apoptosis-associated proteins
including BAD, BAX and Bcl-2 were measured in HCT116 cells
following HTR3A silencing by western blot analysis. GAPDH
was used as an internal reference. ***P<0.001 vs. shCon.
HTR3A, 5-hydroxytryptamine receptor 3A; sh, short hairpin;
Con, control; BAD, BAX and Bcl-2, B-cell lymphoma 2; BAD,
Bcl-2-associated death promotor; BAX, B-cell-1-associated X
protein; early apoptosis, Annexin V+/7AAD-; late apoptosis, Annexin
V+/7AAD+; viable cells, Annexin V-/7AAD-; necrotic cells, Annexin
V-/7AAD+. |
Discussion
HTR3A is considered to be involved in various
biological processes, including heart arrhythmias, organismal
energy homeostasis, including inhibited thermogenesis through Htr3
in BAT and increased lipogenesis, and interneuron migration
(27–29). Additionally, it has been revealed that
HTR3A is present in large B cell lymphomas and its agonists may
promote growth of CRC cells, which implies that HTR3A may
contribute to tumor development (30,31). In
the present study, the potential role of HTR3A in CRC was first
demonstrated by silencing HTR3A expression, and it was demonstrated
that HTR3A regulated the growth of CRC cells in vitro by
controlling partial Bcl-2 family protein expression, including BAD,
BAX and BCL-2 protein.
The involvement of sustaining uncontrolled
proliferation is one of most fundamental traits of cancer cells
(30). In the present study, the
effect of HTR3A on the proliferation of CRC cells in
vitro was examined. Using MTT and colony formation assays, it
was identified that HTR3A silencing was able to inhibit the
growth in CRC HCT116 and SW1116 cells. The cell cycle controls the
transition from quiescence to proliferation in cells (31,32).
Notably, knockdown of HTR3A arrested the cell cycle at S
phase in HCT116 cells, but at G2/M phase in SW1116
cells.
It was suggested that 5-HT, as the ligand of
HTR3A, may decrease apoptosis rate through 5-HT1B receptors
and 5-HT transporters in pulmonary artery smooth muscle cells
(33). Concurrently, 5-HT1B receptor
antagonists exerted anti-mitogenic and apoptotic effects on the CRC
HT29 cell line (34). Additionally,
the HTR3A antagonist tropisetron was able to mediate
apoptosis in the breast cancer MCF-7 cell line (34,35). An
additional HTR3A antagonist, Y25130 hydrochloride, was also
demonstrated to induce apoptosis in the HT29 cell line (36). These results indicate that the
inhibition of 5HT3A-induced apoptosis may be a widespread
phenomenon in cancer cells. Consistent with the previous results,
the present study identified that HTR3A depletion
accelerated cell apoptosis in CRC cells HCT116 and altered the
expression of apoptosis-associated proteins including BAD, BAX and
Bcl-2. Cell apoptosis is important for maintaining homeostasis, and
impaired apoptosis is widely considered to be a pivotal process in
oncogenesis. It has been established that the Bcl-2 family proteins
mediate cell apoptosis primarily through the involvement of
apoptosis-associated signaling pathways (37). Among these proteins, BAD and BAX are
pro-apoptotic regulators (38);
however, Bcl-2 is primarily a regulator against apoptosis (39). However, the exact mechanisms of
HTR3A in CRC require additional investigation.
HTR3A has been demonstrated to induce emesis
during chemotherapy and radiotherapy administered during cancer
treatment (40). It was revealed that
HTR3A antagonists exhibited anti-emetic effects during
chemotherapy and radiotherapy (40).
At present, HTR3A antagonists remain the primary drugs used
in clinical settings to treat emesis. Considering the fact that
dysregulation of HTR3A in CRC may induce apoptosis, it is
hypothesized that HTR3A antagonists may enhance the
efficiency in CRC treatment. Additional studies are required to
demonstrate this hypothesis and determine whether the combined use
of HTR3A antagonists and medicine for CRC is an improved
choice compared with current strategies for CRC treatment.
In summary, the data of the present study
demonstrated that the dysregulation of HTR3A primarily
participated in the proliferation, cell cycle progression and
apoptosis process of CRC cells. Additional in-depth studies of the
mechanism underlying these observations are required to demonstrate
whether HTR3A may be used as a novel target for CRC
therapy.
Acknowledgements
Not applicable.
Funding
The present study was supported by Program for
Clinical Research and Innovation of Renji Hospital, School of
Medicine, Shanghai Jiao Tong University (grant no. PYZY16-007).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JC performed the vector construction experiments and
drafted the manuscript. JT participated in the research design,
reviewed the literature and examined the data. ZW participated in
the qPCR and western blot experiments. JL participated in the
cellular function experiments. CZ participated in the data analysis
and figure formatting.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
May M: Statistics: Attacking an epidemic.
Nature. 509:50–51. 2014. View
Article : Google Scholar
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhou D, Yang L, Zheng L, Ge W, Li D, Zhang
Y, Hu X, Gao Z, Xu J, Huang Y, et al: Exome capture sequencing of
adenoma reveals Genetic alterations in multiple cellular pathways
at the early stage of colorectal tumorigenesis. PLoS One.
8:e533102013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Korinek V, Barker N, Morin PJ, vanWichen
D, de Weger R, Kinzler KW, Vogelstein B and Clevers H: Constitutive
transcriptional activation by a β-catenin-Tcf complex in
APC−/− colon carcinoma. Science. 275:1784–1787. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Morin PJ, Sparks AB, Korinek V, Barker N,
Clevers H, Vogelstein B and Kinzler KW: Activation of
beta-catenin-Tcf signaling in colon cancer by mutations in
beta-catenin or APC. Science. 275:1787–1790. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Baker SJ, Markowitz S, Fearon ER, Willson
JK and Vogelstein B: Suppression of human colorectal carcinoma cell
growth by wild-type p53. Science. 249:912–915. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Aoki Y, Niihori T, Narumi Y, Kure S and
Matsubara Y: The RAS/MAPK syndromes: Novel roles of the RAS pathway
in human genetic disorders. Hum Muta. 29:992–1006. 2008. View Article : Google Scholar
|
|
8
|
Young LW, Darios ES and Watts SW: An
immunohistochemical analysis of SERT in the blood-brain barrier of
the male rat brain. Histochem Cell Biol. 144:321–329. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Stuart JN, Ebaugh JD, Copes AL, Hatcher
NG, Gillette R and Sweedler JV: Systemic serotonin sulfate in
opisthobranch mollusks. J Neurochem. 90:734–742. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Oh KH, Nam Y, Jeong JH, Kim IK and Sohn
UD: The effect of DA-9701 on 5-hydroxytryptamine-induced
contraction of feline esophageal smooth muscle cells. Molecules.
19:5135–5149. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shyu KG: Serotonin 5-HT2B
receptor in cardiac fibroblast contributes to cardiac hypertrophy:
A new therapeutic target for heart failure? Circ Res. 104:1–3.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Davies PA, Pistis M, Hanna MC, Peters JA,
Lambert JJ, Hales TG and Kirkness EF: The 5-HT3B subunit
is a major determinant of serotonin-receptor function. Nature.
397:359–363. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Amirabad Mohammadi L, Ahangari G and
Deilami Derakhshan G: Significant changes of 5-Hydroxytriptamine 3A
receptor gene expression in peripheral blood mononuclear cells of
allergic asthmatic patients. Iran J Allergy Asthma Immunol.
13:33–39. 2014.PubMed/NCBI
|
|
14
|
Lummis SC: 5-HT3 receptors. J
Biol Chem. 287:40239–40245. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jansen M, Bali M and Akabas MH: Modular
design of Cys-loop ligand-gated ion channels: Functional
5-HT3 and GABA rho 1 receptors lacking the large
cytoplasmic M3M4 loop. J Gen Physiol. 131:137–146. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dizeyi N, Bjartell A, Nilsson E, Hansson
J, Gadaleanu V, Cross N and Abrahamsson PA: Expression of serotonin
receptors and role of serotonin in human prostate cancer tissue and
cell lines. Prostate. 59:328–336. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Siddiqui EJ, Shabbir MA, Mikhailidis DP,
Mumtaz FH and Thompson CS: The effect of serotonin and serotonin
antagonists on bladder cancer cell proliferation. BJU Int.
97:634–639. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sonier B, Arseneault M, Lavigne C,
Ouellette RJ and Vaillancourt C: The 5-HT2A serotoninergic receptor
is expressed in the MCF-7 human breast cancer cell line and reveals
a mitogenic effect of serotonin. Biochem Biophys Res Commun.
343:1053–1059. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tutton PJ and Barkla DH: The influence of
serotonin on the mitotic rate in the colonic crypt epithelium and
in colonic adenocarcinoma in rats. Clin Exp Pharmacol Physiol.
5:91–94. 1978. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ataee R, Ajdary S, Rezayat M, Shokrgozar
MA, Shahriari S and Zarrindast MR: Study of 5HT3 and HT4 receptor
expression in HT29 cell line and human colon adenocarcinoma
tissues. Arch Iran Med. 13:120–125. 2010.PubMed/NCBI
|
|
21
|
Rinaldi A, Chiaravalli AM, Mian M, Zucca
E, Tibiletti MG, Capella C and Bertoni F: Serotonin receptor 3A
expression in normal and neoplastic B cells. Pathobiology.
77:129–135. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gaedcke J, Grade M, Jung K, Camps J, Jo P,
Emons G, Gehoff A, Sax U, Schirmer M, Becker H, et al: Mutated KRAS
results in overexpression of DUSP4, a MAP-kinase phosphatase, and
SMYD3, a histone methyltransferase, in rectal carcinomas. Genes
Chromosomes Cancer. 49:1024–1034. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hong Y, Downey T, Eu KW, Koh PK and Cheah
PY: A ‘metastasis-prone’ signature for early-stage mismatch-repair
proficient sporadic colorectal cancer patients and its implications
for possible therapeutics. Clin Exp Metastasis. 27:83–90. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Notterman DA, Alon U, Sierk AJ and Levine
AJ: Transcriptional gene expression profiles of colorectal adenoma,
adenocarcinoma, and normal tissue examined by oligonucleotide
arrays. Cancer Res. 61:3124–3130. 2001.PubMed/NCBI
|
|
25
|
Sabates-Bellver J, van der Flier LG, de
Palo M, Cattaneo E, Maake C, Rehrauer H, Laczko E, Kurowski MA,
Bujnicki JM, Menigatti M, et al: Transcriptome profile of human
colorectal adenomas. Mol Cancer Res. 5:1263–1275. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Park H, Oh CM, Park J, Park H, Cui S, Kim
HS, Namkung J, Park SK, Pak HN, Lee MH, et al: Deletion of the
serotonin receptor type 3A in mice leads to sudden cardiac death
during pregnancy. Circ J. 79:1807–1815. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Oh CM, Namkung J, Go Y, Shong KE, Kim K,
Kim H, Park BY, Lee HW, Jeon YH, Song J, et al: Regulation of
systemic energy homeostasis by serotonin in adipose tissues. Nat
Commun. 6:67942015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Murthy S, Niquille M, Hurni N, Limoni G,
Frazer S, Chameau P, van Hooft JA, Vitalis T and Dayer A: Serotonin
receptor 3A controls interneuron migration into the neocortex. Nat
Commun. 5:55242014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hanahan D and Weinberg R: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Malumbres M and Barbacid M: Cell cycle,
CDKs and cancer: A changing paradigm. Nat Rev Cancer. 9:153–166.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kastan MB and Bartek J: Cell-cycle
checkpoints and cancer. Nature. 432:316–323. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu Y, Tian HY, Yan XL, Fan FL, Wang WP,
Han JL, Zhang JB, Ma Q, Meng Y and Wei F: Serotonin inhibits
apoptosis of pulmonary artery smooth muscle cell by pERK1/2 and PDK
through 5-HT 1B receptors and 5-HT transporters. Cardiovasc Pathol.
22:451–457. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ataee R, Ajdary S, Zarrindast M, Rezayat M
and Hayatbakhsh MR: Anti-mitogenic and apoptotic effects of 5-HT1B
receptor antagonist on HT29 colorectal cancer cell line. J Cancer
Res Clin Oncol. 136:1461–1469. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hejazi SH, Ahangari G and Deezagi A:
Alternative view point against breast cancer based on selective
serotonin receptors 5HTR3A and 5HTR2A antagonists that can mediate
apoptosis in MCF-7 cell line. Curr Drug Discov Technol. 12:240–249.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ataee R, Ajdary S, Zarrindast M, Rezayat
M, Shokrgozar MA and Ataee A: Y25130 hydrochloride, a selective
5HT3 receptor antagonist has potent antimitogenic and apoptotic
effect on HT29 colorectal cancer cell line. Eur J Cancer Prev.
19:138–143. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cory S, Huang DC and Adams JM: The Bcl-2
family: Roles in cell survival and oncogenesis. Oncogene.
22:8590–8607. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Story M and Kodym R: Signal transduction
during apoptosis; implications for cancer therapy. Front Biosci.
3:d365–d375. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cory S and Adams JM: The Bcl2 family:
Regulators of the cellular life-or-death switch. Nat Rev Cancer.
2:647–656. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
40
|
Johnston KD, Lu Z and Rudd JA: Looking
beyond 5-HT3 receptors: A review of the wider role of
serotonin in the pharmacology of nausea and vomiting. Eur J
Pharmacol. 722:13–25. 2014. View Article : Google Scholar : PubMed/NCBI
|