Introduction
Melanoma is a rare type of malignant tumor in China;
however, this disease still affects a considerable number of
patients every year (1). Melanoma is
the most aggressive form of skin cancer and it has an increasing
incidence across the world (2). At
present, there are no effective treatment strategies for melanoma
in the advanced stages (3). Although
surgical resection is considered as a radical treatment for
melanoma in the early stages, survival of patients following
surgery is still poor due to high recurrence rate (4). Previous studies have demonstrated that
recurrent melanoma is closely associated with exposure to UV rays,
smoking and lack of exercise (5,6).
However, the molecular mechanisms underlying recurrent melanoma
remain to be elucidated (5,6).
In addition to messenger RNAs encoding proteins, the
human genome also transcribes a large set of non-coding RNAs, which
serve roles in physiological processes and the development of human
diseases (7). Long non-coding RNAs
(lncRNAs) are a subgroup of non-coding RNAs composed of >200
nucleotides (8). lncRNAs are central
regulators in cancer biology (9).
Long intergenic non-protein coding RNA 1638 (LINC01638) is a
recently identified lncRNA which serves an oncogenic role in
triple-negative breast cancer (10).
Preliminary microarray data revealed in the present study indicated
that LINC01638 lncRNA is upregulated in melanoma tissues compared
with healthy tissues (data not shown). The present study
demonstrated the upregulation of LINC01638 lncRNA in melanoma and
suggested that LINC01638 lncRNA may participate in the growth of
melanoma and its local recurrence following surgical resection.
Materials and methods
Patients and specimens
The current study included 40 patients with melanoma
and 23 patients with benign skin lesions diagnosed in Hunan
People's Hospital (Changsa, China) between March 2011 and March
2013. The inclusion criteria were as follows: i) Patients were
diagnosed with melanoma or benign skin lesions through pathological
examinations; ii) patients were diagnosed for the first time and no
treatment was received during the three months before the study;
iii) patients were diagnosed with American Joint Committee on
Cancer (AJCC) stage I–IIIA melanoma and did not have tumor
metastasis; iv) patients understood the experimental protocol and
were willing to participate; and v) patients complicated the whole
procedure and five-year follow-up. The exclusion criteria were as
follows: i) Patients with co-morbidities, including chronic
inflammation; ii) patients transferred to other hospitals; and iii)
patients who could not attend the follow-up or who succumbed prior
to the confirmation of recurrence. There were 10 cases of stage I,
21 cases of stage II and 9 cases of stage IIIA melanoma. A total 16
healthy patients who were willing to donate skin biopsies and blood
were included to serve as the control group. No significant
differences in age and sex were identified between the three
patient groups. Patient basic information is presented in Table I. Plasma and skin biopsy tissues were
stored in liquid nitrogen prior to use. The present study was
approved by the Ethics Committee of Hunan People's Hospital.
Written informed consent was obtained from all patients prior to
enrolment.
 | Table I.Basic information of the three groups
of patients. |
Table I.
Basic information of the three groups
of patients.
|
| Sex |
|
|
|---|
|
|
|
|
|
|---|
| Group | Male | Female | Age range
(years) | Average age
(years) |
|---|
| Melanoma | 22 | 18 | 27–66 | 46.1±5.3 |
| Benign skin
lesion | 12 | 11 | 24–67 | 45.9±6.0 |
| Control | 7 | 9 | 22–68 | 46.2±5.2 |
Treatment, blood extraction and
follow-up
Blood (5 ml) was collected on the day of admission.
All patients with melanoma were treated with surgical resection,
followed by chemotherapy. All patients were followed up for 5 years
following discharge to record recurrence. Blood was collected again
in cases of recurrence and at the end of follow-up period in cases
of no recurrence. Plasma was prepared by centrifuging blood at
1,250 × g for 15 min in EDTA tubes at room temperature.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Following RNA extraction from plasma, biopsies and
in vitro cultivated cells using TRIzol® reagent
(Invitrogen; Thermo Fisher Scienfitic, Inc., Waltham, MA, USA), a
NanoDrop™ 2000 Spectrophotometer (Thermo Fisher Scientific, Inc.,
Pittsburgh, PA, USA) was used to measure RNA concentration. To
enrich RNA concentration, RNA samples were mixed with alcohol and
glycogen (1:1,000; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany).
Following incubation at −80°C for 30 min, the mixture was
centrifuged at 4°C for 30 min at 12,000 × g. RNA samples with an
A260/A280 ratio of 1.8–2.0 were subjected to reverse transcription
using SuperScript IV Reverse Transcriptase (Thermo Fisher
Scientific, Inc.) to synthesize cDNA at 55°C for 30 min and 75°C
for 10 min. All PCR reactions were prepared using SYBR®
Green Real-Time PCR Master Mixes (Thermo Fisher Scientific, Inc.)
and the following primers: 5′-AATACATCAGCACTGTTGCCTTT-3′ (forward)
5′-CTCCATACATACATCTCCAAAAAGT-3′ (reverse) for LINC01638 lncRNA;
5′-GACCTCTATGCCAACACAGT-3′ (forward) and 5′-AGTACTTGCGCTCAGGAGGA-3′
(reverse) for β-actin. The following thermocycling conditions were
used for the PCR: 95°C for 1 min; 40 cycles of 95°C for 12 sec and
58.5°C for 40 sec. Data normalization was performed using the
2−ΔΔCq method (11) with
β-actin as the internal reference gene. PCR products were sequenced
to ensure the correct products were obtained.
Cell lines and cell culture
A normal human skin cell line CCD-1059Sk and two
human melanoma cell lines C32 and SK-MEL-28 were obtained from the
American Type Culture Collection (ATCC; Manassas, VA, USA). The
cells were cultured with Eagle's Minimum Essential Medium with 10%
fetal bovine serum (Sangon Biotech Co., Ltd., Shanghai, China) at
37°C with 5% CO2. Vectors containing full length
LINC01638 lncRNA, short hairpin RNA (shRNA)
(5′-GGCCCTCCTGCTGATGAGAGAC-3′) and negative control shRNA
(5′UUCUCCGAACGUGUCACGUGGC-3) were purchased from GeneCopoeia Inc.
(Rockville, MD, USA). LINC01638 expression vector (15 nM) and
shRNAs (20 nM) were transfected into cells using
Lipofectamine® 3000 according to the manufactuer's
protocol (Thermo Fisher Scientific, Inc.). Transfection with empty
vectors or negative control shRNA was used as negative control.
Untransfected cells were used as control cells. LINC01638 lncRNA
expression was detected by RT-qPCR 12 h following transfection.
Cell proliferation assay
Following transfection (24 h later), a cell
proliferation assay was performed to determine the proliferation in
cells with an LINC01638 overexpression rate >200% or knockdown
rate <50%. The cell counting kit-8 (CCK-8; Sigma-Aldrich; Merck
KGaA) was used to detect cell proliferation. Briefly, cells were
harvested, and cell suspensions were prepared with a density of
4×104 cells/ml. Each well of a 96-well plate was filled
with 0.1 ml cell suspension. Cells were cultured at 37°C in a 5%
CO2 incubator, followed by addition of 10 µl CCK-8
solution after 24, 48, 72 and 96 h. Cells were cultured for
additional 4 h and optical density values were measured using a
microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA)
at a wavelength of 450 nm.
Statistical analysis
GraphPad Prism software (version 6; GraphPad
Software Inc., La Jolla, CA, USA) was used for all statistical
analyses. Data are presented as the mean ± standard deviation.
Comparisons between two groups were performed by unpaired t- test.
Comparisons between two time points within the same group were
performed by paired t-test. Comparisons among multiple groups were
performed by one way analysis of variance followed by a Tukey test.
Associations between LINC01638 lncRNA and clinicopathological data
of patients with melanoma were analyzed by the χ2 test.
Receiver operating characteristic (ROC) curve analysis was
performed to evaluate the diagnostic value of LINC01638 for
melanoma. P<0.05 was considered to indicate a statistically
significant difference.
Results
LINC01638 lncRNA is significantly
upregulated in patients with melanoma
Expression of LINC01638 lncRNA in skin biopsies and
plasma samples of patients with melanoma, patients with benign skin
lesions and healthy controls was quantified using RT-qPCR.
Expression of LINC01638 lncRNA in skin biopsies was significantly
upregulated in patients with melanoma compared with the other two
patient groups (P<0.05; Fig. 1A).
In addition, plasma levels of LINC01638 lncRNA were significantly
increased in patients with melanoma compared with the other two
patient groups (P<0.05; Fig. 1B).
By contrast, there were no significant differences in expression of
LINC01638 lncRNA in skin biopsies (Fig.
1A) and plasma (Fig. 1B) between
patients with benign skin lesions and healthy controls. Slightly
increased LINC01638 lncRNA expression in both biopsies and plasma
was observed with increasing clinical and tumor stages (no
statistical significance; data not shown).
Upregulation of LINC01638 lncRNA
distinguishes patients with melanoma from the other patient
groups
Diagnostic values of LINC01638 lncRNA for melanoma
were analyzed by receiver operating characteristic curve analysis.
For LINC01638 lncRNA expression in skin biopsies compared with
healthy controls as references, the area under the curve (AUC) was
0.8734, with a 95% confidence interval (CI) of 0.7699–0.9700 and
standard error of 0.05281 (P<0.0001; Fig. 2A); with patients with benign skin
lesions as references, the AUC was 0.8370, with a 95% CI of
0.7380–0.9359 and standard error of 0.05049 (P<0.0001; Fig. 2B). For LINC01638 lncRNA expression in
plasma, with healthy controls as references, the AUC was 0.8391,
with a 95% CI of 0.7287–0.9494 and standard error of 0.05269
(P<0.0001; Fig. 2C); with
patients with benign skin lesions as references, the AUC was
0.8136, with a 95% CI of 0.7097–0.9174 and standard error of
0.05298 (P<0.0001; Fig. 2D).
LINC01638 lncRNA expression is
significantly associated with tumor size
Patients were divided into high and low expression
groups according to the median expression level of LINC01638
lncRNA. The association between LINC01638 lncRNA and
clinicopathological data of patients with melanoma was analyzed
using χ2 test. Results revealed that LINC01638 lncRNA
expression in skin biopsies (Table
II) and plasma (Table III) was
significantly associated with tumor size but not age, sex or
patients' smoking and drinking habits.
 | Table II.Association between long intergenic
non-protein coding RNA 1638 long non-coding RNA expression in skin
biopsies and clinicopathological data of patients with
melanoma. |
Table II.
Association between long intergenic
non-protein coding RNA 1638 long non-coding RNA expression in skin
biopsies and clinicopathological data of patients with
melanoma.
| Variable | Group | Number of cases | High-expression | Low-expression | χ2 | P-value |
|---|
| Age (years) | >45 | 21 | 13 | 8 | 2.51 | 0.11 |
|
| <45 | 19 | 7 | 12 |
|
|
| Sex | Male | 22 | 10 | 12 | 0.40 | 0.52 |
|
| Female | 18 | 10 | 8 |
|
|
| Primary tumor
diameter | >4 mm | 18 | 13 | 5 | 7.03 | 0.03 |
|
| 2–4 mm | 13 | 5 | 8 |
|
|
|
| <2 mm | 9 | 2 | 7 |
|
|
| Drinking | Yes | 17 | 7 | 10 | 0.92 | 0.34 |
|
| No | 23 | 13 | 10 |
|
|
| Smoking | Yes | 16 | 7 | 9 | 0.42 | 0.52 |
|
| No | 24 | 13 | 11 |
|
|
 | Table III.Association between long intergenic
non-protein coding RNA 1638 long non-coding RNA expression in
plasma and clinicopathological data of patients with melanoma. |
Table III.
Association between long intergenic
non-protein coding RNA 1638 long non-coding RNA expression in
plasma and clinicopathological data of patients with melanoma.
| Variable | Group | Number of cases | High-expression | Low-expression | χ2 | P-value |
|---|
| Age (years) | >45 | 21 | 13 | 8 | 2.51 | 0.11 |
|
| <45 | 19 | 7 | 12 |
|
|
| Sex | Male | 22 | 9 | 13 | 1.60 | 0.20 |
|
| Female | 18 | 11 | 7 |
|
|
| Primary tumor
diameter (mm) | >4 | 18 | 13 | 5 | 6.48 | 0.04 |
|
| 2–4 | 13 | 4 | 9 |
|
|
|
| <2 | 9 | 3 | 6 |
|
|
| Drinking | Yes | 17 | 8 | 9 | 0.10 | 0.75 |
|
| No | 23 | 12 | 11 |
|
|
| Smoking | Yes | 16 | 6 | 10 | 1.67 | 0.20 |
|
| No | 24 | 14 | 10 |
|
|
LINC01638 lncRNA expression is further
upregulated in patients with local recurrence but not in patients
without recurrence during follow-up
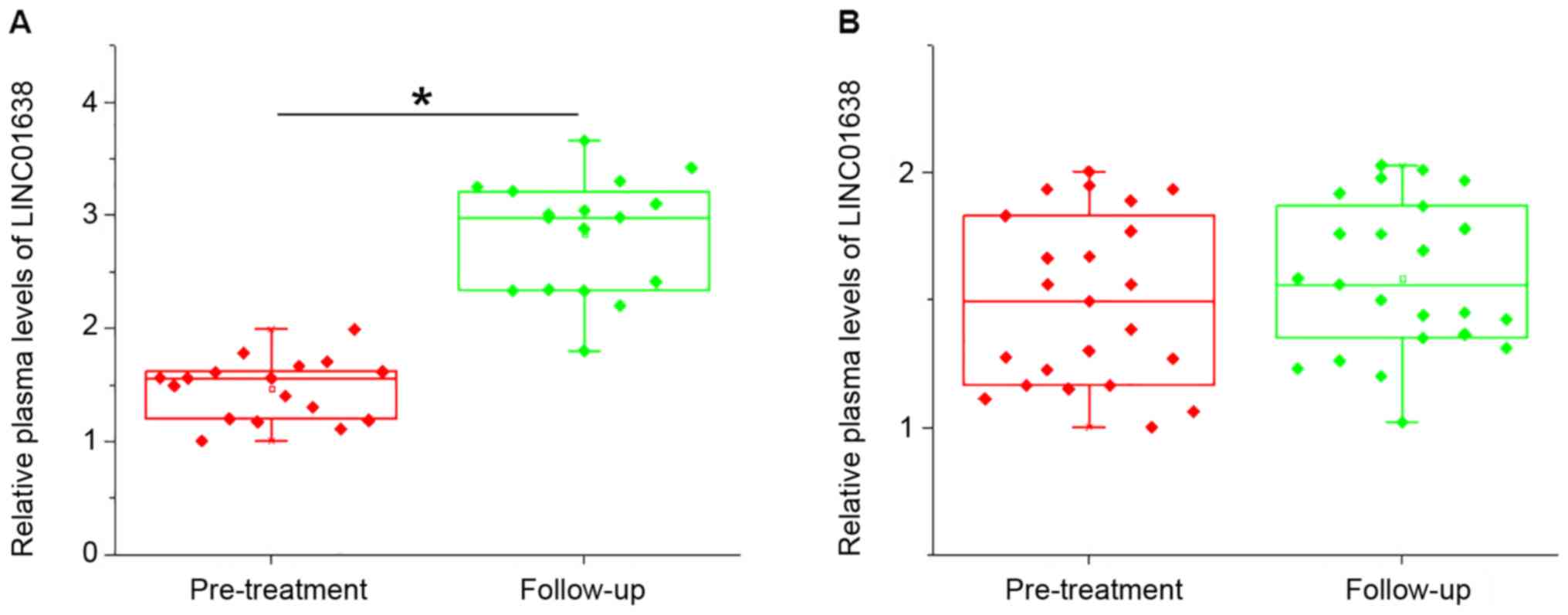
During the five year follow-up, local recurrence
(cancer grows in the same region it first started) was observed in
17 cases and no distant recurrence was observed. Compared with
pre-treatment levels, LINC01638 lncRNA expression was further
upregulated in patients with local recurrence (P<0.05; Fig. 3A), but not in patients without
recurrence (Fig. 3B). Hazard ratios
of tumor stages and LINC01638 lncRNA levels for local recurrence
were 1.78 (mean, range, 1.07–2.91) and 2.20 (mean, range,
1.11–4.30), respectively.
LINC01638 lncRNA overexpression
promotes, while knockdown inhibits cancer cell proliferation
The significant association between LINC01638 lncRNA
overexpression and tumor size suggested the involvement of
LINC01638 lncRNA in the growth of melanoma. In order to test this
further, LINC01638 lncRNA expression vectors and shRNAs were
transfected into cancer cells and cell proliferation was analyzed
using the CCK-8 assay. Following transfection, LINC01638 lncRNA
overexpression (Fig. 4A) and
knockdown (Fig. 4B) were achieved in
two human melanoma cell lines C32 and SK-MEL-28 and a normal skin
cell line CCD-1059Sk (P<0.05). Compared with control cells and
negative control cells, LINC01638 lncRNA overexpression
significantly promoted (Fig. 5A),
while LINC01638 lncRNA knockdown significantly inhibited (Fig. 5B) cell proliferation of the two human
melanoma cell lines C32 and SK-MEL-28 (P<0.05), but not cells of
the normal skin cell line CCD-1059Sk.
Discussion
The function of LINC01638 lncRNA has only been
characterized in triple-negative breast cancer, while its
involvement in other diseases is unknown (10). The results obtained in the current
study suggested that LINC01638 lncRNA is upregulated in melanoma
and may participate in the growth of melanoma as well as its local
recurrence following surgical resection.
In spite of efforts to treat patients with melanoma
at advanced stages, the survival rates of patients with metastatic
melanoma are poor (12,13). At present, early diagnosis followed
by proper treatment remain key for increasing the survival rate of
patients with melanoma (14).
LINC01638 lncRNA has been reported to be upregulated in
triple-negative cancer tissues compared with adjacent healthy
tissues (10). In the present study,
the involvement of LINC01638 lncRNA in the early stages of melanoma
was studied in patients with melanoma at AJCC stage I–IIIA.
Compared with healthy controls, upregulation of LINC01638 lncRNA in
biopsies and plasma was observed in patients with melanoma but not
in patients with benign skin cancer. Overexpression of LINC01638
lncRNA in biopsies and plasma effectively distinguished patients
with melanoma from patients with benign skin cancer and healthy
controls. In addition, expression of LINC01638 lncRNA was not
affected by age, sex and smoking and drinking habits, which are
factors that affect the expression of certain lncRNAs (15,16).
This indicated that the overexpression of LINC01638 lncRNA may be
consistent across patients with melanoma with different
backgrounds. Therefore, detecting the expression of LINC01638
lncRNA may aid in the early diagnosis of melanoma.
In the current study, the expression of LINC01638
lncRNA was quantified in skin biopsies of patients with melanoma,
patients with benign skin lesion and healthy controls. Skin biopsy
is an invasive technique with low patient acceptability, and
therefore, a small number of samples was included in the current
study. The development of human diseases may be accompanied by
changes of blood substances, and the detection of these changes may
provide guidance for the treatment of melanoma (17,18).
Circulating lncRNAs, including Pvt1 oncogene (19) and maternally expressed 3 (20) have demonstrated diagnostic value for
melanoma. In the current study, the diagnostic value of plasma
LINC01638 lncNRA was comparable to that of LINC01638 lncNRA
expression in skin biopsies. Therefore, the detection of plasma
LINC01638 lncNRA may allow the diagnosis of melanoma in cases where
skin biopsy is not applicable.
Accurate prediction of tumor recurrence is required
for the prevention and treatment of melanoma (21,22). In
the present study, patients with local recurrence exhibited
significantly increased plasma level of LINC01638 lncNRA during
follow-up compared with patients with no recurrence, indicating
that LINC01638 lncNRA overexpression may participate in the local
recurrence of melanoma. Although the results obtained in the
current study suggest that LINC01638 lncNRA enhances the
proliferation of melanoma cells in vitro, which may promote
local recurrence, the molecular mechanism underlying recurrence
remains unknown. Future studies are required to elucidate the
molecular mechanisms by which LINC01638 lncNRA results in the
recurrence of melanoma. In the current study, LINC01638 lncRNA
overexpression promoted melanoma cell proliferation, but failed to
significantly affect cell migration and invasion. Therefore,
LINC01638 lncRNA may specifically participate in the growth of
melanoma.
In conclusion, LINC01638 lncRNA is overexpressed in
patients with melanoma. Upregulation of LINC01638 lncRNA is likely
to be associated with the local recurrence of melanoma following
surgical resection.
Acknowledgements
Not applicable.
Funding
This study was supported by Hunan Provincial
People's Hospital 2007 Renshu Fund (Changsha, China).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WX and AY performed all the experiments, analyzed
all data. AY drafted the manuscript. Both authors read and approved
the final manuscript.
Ethics approval and consent to
participate
This study received ethical approval from the Hunan
People's Hospital (Changsha, China) Ethics Committee. All
participants signed informed consent.
Patient consent for publication
The study followed the tenets of the Declaration of
Helsinki, and informed written consent was obtained from all
patients and controls.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W: Cancer statistics: Updated cancer
burden in China. Chin J Res. 27:12015.
|
|
2
|
Karimkhani C, Green AC, Nijsten T,
Weinstock MA, Dellavalle RP, Naghavi M and Fitzmaurice C: The
global burden of melanoma: Results from the global burden of
disease study 2015. Br J Dermatol. 177:134–140. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tsao H, Chin L, Garraway LA and Fisher DE:
Melanoma: From mutations to medicine. Genes Dev. 26:1131–1155.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Farma JM, Kulkarni N and Hsu C: Surgical
management of primary and recurrent melanoma. Surg Oncol Clin N Am.
24:239–247. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hocker T and Tsao H: Ultraviolet radiation
and melanoma: A systematic review and analysis of reported sequence
variants. Hum Mutat. 28:578–588. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Silverberg JI and Ratner D: Associations
of non-melanoma skin cancer and melanoma, extra-cutaneous cancers
and smoking in adults: A US population-based study. J Eur Acad
Dermatol Venereol. 29:1389–1397. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Luo L, Tang H, Ling L, Li N, Jia X, Zhang
Z, Wang X, Shi L, Yin J, Qiu N, et al: LINC01638 lncRNA activates
MTDH-Twist1 signaling by preventing SPOP-mediated c-Myc degradation
in triple-negative breast cancer. Oncogene. 37:6166–6179. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hodi FS, O'Day SJ, McDermott DF, Weber RW,
Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel
JC, et al: Improved survival with ipilimumab in patients with
metastatic melanoma. N Engl J Med. 363:711–723. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schadendorf D, Hodi FS, Robert C, Weber
JS, Margolin K, Hamid O, Patt D, Chen TT, Berman DM and Wolchok JD:
Pooled analysis of long-term survival data from phase II and phase
III trials of ipilimumab in unresectable or metastatic melanoma. J
Clin Oncol. 33:1889–1894. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
American Academy of Dermatology Ad Hoc
Task Force for the ABCDEs of Melanoma, ; Tsao H, Olazagasti JM,
Cordoro KM, Brewer JD, Taylor SC, Bordeaux JS, Chren MM, Sober AJ,
Tegeler C, et al: Early detection of melanoma: Reviewing the
ABCDEs. J Am Acad Dermatol. 72:717–723. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Grammatikakis I, Panda AC, Abdelmohsen K
and Gorospe M: Long noncoding RNAs(lncRNAs) and the molecular
hallmarks of aging. Aging (Albany NY). 6:992–1009. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang J, Qiu M, Xu Y, Li M, Dong G, Mao Q,
Yin R and Xu L: Long noncoding RNA CCAT2 correlates with smoking in
esophageal squamous cell carcinoma. Tumour Biol. 36:5523–5528.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zeng L, Liu J, Wang Y, Wang L, Weng S,
Tang Y, Zheng C, Cheng Q, Chen S and Yang GY: MicroRNA-210 as a
novel blood biomarker in acute cerebral ischemia. Front Biosci
(Elite Ed). 3:1265–1272. 2011.PubMed/NCBI
|
|
18
|
Shehadeh LA, Yu K, Wang L, Guevara A,
Singer C, Vance J and Papapetropoulos S: SRRM2, a potential blood
biomarker revealing high alternative splicing in Parkinson's
disease. PLoS One. 5:e91042010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen X, Gao G, Liu S, Yu L, Yan D, Yao X,
Sun W, Han D and Dong H: Long noncoding RNA PVT1 as a novel
diagnostic biomarker and therapeutic target for melanoma. Biomed
Res Int. 2017:70385792017.PubMed/NCBI
|
|
20
|
Long J and Pi X: lncRNA-MEG3 suppresses
the proliferation and invasion of melanoma by regulating CYLD
expression mediated by sponging miR-499-5p. Biomed Res Int.
2018:20865642018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mallidi S, Watanabe K, Timerman D,
Schoenfeld D and Hasan T: Prediction of tumor recurrence and
therapy monitoring using ultrasound-guided photoacoustic imaging.
Theranostics. 5:289–301. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mattonen SA, Palma DA, Haasbeek CJ, Senan
S and Ward AD: Early prediction of tumor recurrence based on CT
texture changes after stereotactic ablative radiotherapy (SABR) for
lung cancer. Med Phys. 41:0335022014. View Article : Google Scholar : PubMed/NCBI
|