Introduction
Glioblastoma multiforme (GBM) is one of the most
common and malignant primary brain tumors (1), and is characterized by diffuse
infiltration, uncontrolled proliferation and chemoresistance. Even
following tumor resection and radio-chemotherapy, improvement in
progression-free survival (PFS) and overall survival (OS) is
limited and usually accompanied by malignant progression (2–4).
MicroRNAs (miRNA/miRs) are small, non-coding RNAs
that regulate the expression of multiple genes at the
post-transcriptional level by inducing degradation of target mRNAs
or by inhibiting translation (5,6).
Increasing evidence indicates that miRNAs regulate a variety of
tumor cell processes, including proliferation, invasion, apoptosis
and chemoresistance (7–9). miR-218 has been reported to be
downregulated in several neoplasms and serves critical roles in
tumorigenesis and metastasis. It exhibits low expression levels in
numerous tumor tissues and is associated with patient prognosis
(10–13).
Survivin, encoded by and also known as baculoviral
inhibitor of apoptosis repeat-containing 5 (BIRC5), is a member of
the inhibitor of apoptosis (IAP) family (14). Survivin is strongly expressed in a
majority of tumors and is overexpressed during embryonic
development, whereas it is absent in normal differentiated tissues
(15). Survivin regulates cell-cycle
progression, inhibits apoptosis and induces chromosomal instability
(16,17). It is also associated with poor
prognosis in ovarian cancer, breast cancer and leukemia (18,19).
miR-218 directly binds to survivin mRNA, and the miR-218/survivin
axis has been reported in a variety of tumors and regulates cancer
cells migration, invasion and lymph node metastasis (20,21). A
recent study predicted that miR-218 targets the 3′UTR of survivin
mRNA and suggested that miR-218 may play an important role in the
regulation of surviving (19).
However, the association between miR-218 and survivin mRNA in GBM
remains unknown. The present study investigated the
miR-218/survivin axis in GBM and observed the effects of miR-218
overexpression on the proliferation, migration and invasion of
glioma cells.
Materials and methods
Patients and clinical specimens
The data of 144 patients with GBM and genome RNA
sequencing were collected from the Chinese Glioma Genome Atlas
(CGGA) (http://www.cgga.org.cn). The clinical
characteristics of the patients are listed in Table I. In total, 22 patients received
radiotherapy only, 25 received temozolomide (TMZ) only, 59 were
treated with radiotherapy plus TMZ and 38 received no treatment.
Tumor specimens and adjacent tissues were obtained from six
patients (36–58 years old; 2 women and 4 men) who underwent surgery
between January 15th and February 20th, 2018) in the Department of
Neurosurgery of Bejing Tiantan Hospital (Bejing, China). The
adjacent tissues were defined as the area between the tumor and the
normal brain tissue. The present study was approved by the Ethics
Committee of The Capital Medical University (Bejing, China), and
all patients provided informed consent.
 | Table I.Clinical information for 20% of
patients with longer survival times in low-survivin expression
group. |
Table I.
Clinical information for 20% of
patients with longer survival times in low-survivin expression
group.
| Sample ID | Gender | Age, years | OS, days | TCGA subtype | IDH mutation | MGMT
methylation | RT | TMZ | TP53 mutation | EGFR mutation | ATRX mutation |
|---|
| 494 | F | 59 | 2,072 | Neural | 0 | 1 | 1 | 1 | 1 | 0 | 0 |
| 680 | M | 44 | 1,832 | Neural | 0 | 0 | 1 | 1 | 1 | 0 | 0 |
| 761 | M | 45 | 1,744 | Proneural | 1 | 1 | 1 | 0 | 0 | 1 | 0 |
| 822 | M | 46 | 1,324 | Proneural | 1 | 1 | 1 | 1 | 1 | 0 | 0 |
| 491 | F | 37 | 1,074 | Proneural | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| D37 | M | 55 | 965 | Mesenchymal | 0 | 0 | 1 | 1 | 1 | 0 | 0 |
| 1026 | M | 59 | 875 | Classical | 0 | 1 | 1 | 1 | 0 | 0 | 0 |
| D57 | M | 40 | 766 | Classical | 0 | NA | 1 | 1 | 1 | 0 | 0 |
| 802 | M | 43 | 681 | Mesenchymal | 0 | 0 | 1 | 1 | 1 | 0 | 0 |
| 1023 | F | 58 | 681 | Classical | 0 | 0 | NA | NA | 0 | 0 | 0 |
| 700 | F | 25 | 669 | Proneural | 0 | 1 | 1 | 1 | 0 | 0 | 0 |
| D35 | F | 68 | 661 | Mesenchymal | 0 | 1 | 1 | 0 | 1 | 0 | 1 |
| 710 | M | 42 | 660 | Mesenchymal | 1 | 1 | 0 | 1 | 0 | 0 | 0 |
Cell culture and transfection
Immortalized human gliocyte HEB cells (cat. no.
001398) and human GBM LN229 cells (cat. no. 001397) were obtained
from Shanghai Bioleaf Biotech Co., Ltd. The cell lines were
maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a 95%
relative humidified incubator with 5% CO2. LN229 cells
in the logarithmic growth phase were transfected with miR-218
mimics (10 µl/ml) using Lipofectamine™ 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) and cultured at 37°C in a 5%
CO2 humidified atmosphere for 6 h. Medium was then
changed and cells were left in incubator for 48 h. The sequences
transfected were as follows: Sense 5′-UUCUCCGAACGUGUCACGUTT-3′,
anti-sense 5′-ACGUGACACGUUCGGAGAATT-3′; and negative control (NC)
mimics, sense 5′-UUGUGCUUGAUCUAACCAUGU-3′, and anti-sense
5′-AUGGUUAGAUCAAGCACAAUU-3′ (Shanghai GenePharma Co., Ltd.).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
The mRNA expression levels of miR-218 and survivin
were detected in tumor specimens, adjacent tissues and the HEB and
LN229 cell lines. In addition, mRNA levels of miR-218 and survivin
were detected in the control, NC and mimics groups. The primers
were provided by Tsingke Biological Technology, Co., Ltd. and were
designed as follows: miR-218, loop primer
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACATGGTT-3′, forward
5′-TGCGCTTGTGCTTGATCTAAC-3′; U6, forward
5′-CGCTTCGGCAGCACATATAC-3′, reverse, 5′-AAATATGGAACGCTTCACGA-3′;
survivin, forward 5′-ACCACCGCATCTCTACATTCA-3′, reverse
5′-CTTTGCATGGGGTCGTCATC-3′; and β-actin, forward
5′-AGCGAGCATCCCCCAAAGTT-3′ and reverse 5′-GGGCACGAAGGCTCATCATT-3′.
U6 and β-actin were used as internal control for miR-218 and
surviving, respectively. Total RNA (1 µg) was isolated using Trizol
reagent (cat. no. 252250AX, Aidalb Biotechnologies Co., Ltd.) and
reversely transcribed to cDNA using Hiscript Reverse Transcriptase
Reagent Kit (cat. no. R101-01/02; Vazyme Ltd.) according to the
manufacturer's instructions. The PCR amplification process was
detection on an ABI 7900 or Viia7 Fast Real-Time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA,
USA). The PCR conditions for amplification were as follows:
Pre-denaturation at 95°C for 10 min, denaturation at 95°C for 30
sec, 40 cycles, annealing at 60°C for 30 sec and extension at 72°C
for 30 sec. The relative expression levels were normalized to
endogenous controls and were calculated using 2−ΔΔCq
method (22).
Cell Counting Kit-8 (CCK-8)
proliferation assay
Cell proliferation was evaluated using a CCK-8
proliferation assay (cat. no. C0037; Beyotime Institute of
Biotechnology), which is a sensitive colorimetric assay for the
determination of the number of viable cells based on the production
of formazan (23). Prior to
proliferation assays, the numbers of viable cells were equal
between the experimental and NC group. LN229 cells were seeded into
96-well plates (2,000 cells/well) and cultured at 37°C in a 5%
CO2 humidified atmosphere for 3 days. Next, 10 µl CCK-8
was added to each well and the cells were incubated at 37°C for 4
h. The absorbance value of each well was measured using a
microplate reader 450 nm (24).
Transwell migration and invasion
assays
Following transfection, an equal number of viable
cells were added to the upper chambers (3-µm pore size) of a
24-well Transwell plate. Suspensions of cells (200 µl,
5×105 cells/ml) in DMEM from different groups were added
to the upper chambers, which were pretreated with Matrigel for the
invasion assay but untreated for the migration assay. The lower
chambers contained 800 µl DMEM plus 10% FBS. After 24 h of
incubation at 37°C and 5% CO2, the upper chambers were
detached from the plates and the cells remaining on the upper
surface of the filters were removed with cotton swabs. The cells on
the lower surface were fixed with 10% methyl alcohol at 25°C for 30
min, and stained with Giemsa at 25°C for 20 min. Three high-power
fields were selected randomly to examine the invading and migrating
cells under light microscopy.
Apoptosis assay
Flow cytometry was used to analyze apoptosis in
LN229 cells transfected with miR-218 mimics or the negative control
(NC). According to the manufacturer's instructions, 48 h after
transfection, cells were stained with Annexin
V-allophycocyanin/7-amino-actinomycin D apoptosis detection kit
(cat. no. KA3807; Abnova). Apoptotic fractions were detected using
flow cytometry (AccuriC6; BD Biosciences) and CELL Quest 3.0
software.
Western blot analysis
Extraction of total protein from the tissues and
cells was achieved with RIPA Lysis Buffer (cat. no. P0013B;
Beyotime Institute of Biotechnology). Protein concentrations were
determined using a BCA protein detection kit (cat. no. P0010;
Beyotime Institute of Biotechnology). In total, 40 µg protein was
subjected to electrophoresis on 10% SDS-polyacrylamide gels and
then transferred to polyvinylidene difluoride membranes (cat. no.
IPVH00010; EMD Millipore). Membranes were blocked with 5% fat-free
milk diluted in TBST containing 0.1% Tween-20 for 2 h at 25°C.
Membranes were then incubated with a 1:800-diluted anti-survivin
antibody (cat. no. bs-0615R; BIOSS) or a 1:1,000-diluted anti-GAPDH
antibody (cat. no. AB-P-R001; Hangzhou Xianzhi Biotechnology Co.,
Ltd.) as the loading control, for 24 h at 4°C. They were then
incubated with the 1:5,000-diluted horseradish peroxidase (HRP)
labeled goat anti-rabbit secondary antibody (cat. no. BA1045;
Boster Biological Technology) for 60 min at 37°C. The immune
imprint was visualized with an enhanced chemiluminescence western
blot detection system (Bio-Rad Laboratories, Inc.). Relative
expression level of survivin was normalized to endogenous control
GAPDH ChemImager 5,500 V2.03 software.
Immunohistochemical (IHC)
staining
IHC analysis of survivin expression was performed on
section fixed for 24 h at 25°C with 10% formalin and
paraffin-embedded using standard procedures. Briefly, sections
(4-µm thick) were dried for 12 h at 37°C, deparaffinized in xylene
for 10 min, rehydrated in an alcohol series, and then placed in 3%
hydrogen peroxide for 10 min to block endogenous peroxidase
activity. Sections were incubated with a 1:100 anti-survivin
antibody at 4°C for 12 h. Subsequently, sections were washed in PBS
three times for 5 min and then incubated for 50 min at 25°C with
100 µl 1:5,000-diluted HRP labeled goat anti-rabbit secondary
antibody (cat. no. BA1045; Boster Biological Technology). Finally,
immunostaining was visualized with diaminobenzidine, and sections
were counterstained with hematoxylin and eosin for 25 sec at 25°C
prior to washing again with PBS.
Fluorescence in situ hybridization
(FISH)
The expression status of miR-218 in GBM samples was
evaluated by FISH. Homo sapiens (hsa)-miR-218-5p probes
(red-labeled; Sangon Biotech Co., Ltd.)
(5′-ACATGGTTAGATCAAGCACAA-3′) directed against the mature miR-218
sequence were used. Sections (4-µm thick) were dried at 37°C for 12
h, deparaffinized in xylene for 10 min, rehydrated in an alcohol
series, washed 3 times with PBS (5 min/wash), and then digested
using proteinase K for 30 min at 37°C. After washing again 3 times
using PBS (5 min/wash), the sections were hybridized with the
probes (3 ng/ml) for 24 h at 45°C. Subsequently, the tissue
sections were washed in 5X saline sodium citrate (SSC) at 45°C for
15 min, 4X SSC at 37°C for 15 min, 2X SSC (containing 50% deionized
formamide) at 37°C for 15 min, 2X SSC (containing 20 µg/ml RNaseA)
at 37°C for 15 min and 0.5X SSC for 15 min, and then washed 3 times
for 5 min each with 0.01 mol/l PBS. Finally, sections were
counterstained with DAPI (cat. no. C1002; Beyotime Institute of
Biotechnology) for 5 min and examined with an Olympus BX53
microscope.
Statistical analysis
According to the median expression level of
survivin, the samples were divided into two groups. Each group was
assessed by the Student's t-test and χ2 test using R
language 3.2.5 (https://cran.r-project.org/bin/windows/base/old/3.2.5/)
and SPSS software 16.0 (SPSS, Inc.). OS was defined as the time
following surgery until mortality or last follow-up date. Using the
Kaplan-Meier method protract survival analysis by GraphPad Prism 7
(GraphPad Software, Inc.), the differences between the low- and
high-survivin expression groups were calculated using the log-rank
test. For continuous variables, comparisons of mean values between
multiple groups were assessed by ANOVA, and then Holm-Sidak's
multiple comparisons test was used. All data are presented as the
mean ± standard error. P<0.05 was considered to indicate a
statistically significant difference. Factors with P<0.05 in
univariate analysis were incorporated into a multivariate Cox
analysis.
Results
Survivin expression is associated with
poor prognosis in patients with GBM
Data from 144 patients with GBM and whole-genome RNA
sequencing were collected from the CGGA database and used in the
present study. The prognosis of the group with high survivin
expression was poor on Student's t-test and univariate analysis
(P<0.05) (Fig. 1A; Table III). Survivin was highly expressed
in classical subtype gliomas, while all neural subtype gliomas
exhibited low expression of survivin (P=0.013 and P<0.01,
respectively; Table II). In
multivariate Cox analysis, the expression of survivin was observed
to be associated with OS (P=0.006; Table III). In total, 81 patients with GBM
received radiation plus TMZ chemotherapy or radiation only. OS in
the radiotherapy alone group (358±82 days) was significantly
different from that in the radiation plus TMZ chemotherapy group
(885±116 days; P=0.0007 and P=0.0014, respectively) independent of
the expression levels of survivin (Fig.
1B and C).
 | Table III.Prognostic factors of overall
survival for 144 patients with glioblastoma multiforme. |
Table III.
Prognostic factors of overall
survival for 144 patients with glioblastoma multiforme.
|
| Univariate | Multivariate |
|---|
|
|
|
|
|---|
| Variable | P-value | P-value | HR | 95% CI lower | 95% CI upper |
|---|
| Sex | 0.353 |
|
|
|
|
| Age, ≤48 vs.
>48 | 0.88 |
|
|
|
|
| ATRX mutation | 0.519 |
|
|
|
|
| EGFR mutation | 0.241 |
|
|
|
|
| TERT mutation | 0.752 |
|
|
|
|
| IDH mutation | 0.069 | 0.01 | 0.204 | 0.061 | 0.686 |
| MGMT
methylation | 0.009 | 0.12 | 0.533 | 0.241 | 1.178 |
| Survivin high
expression | 0.041 | 0.006 | 2.898 | 1.355 | 6.198 |
| KPS | <0.001 | 0.005 | 0.343 | 0.162 | 0.723 |
| RT vs. RT+TMZ | <0.001 | 0.005 | 0.311 | 0.139 | 0.697 |
| Resection, GTR vs.
STR | 0.174 | 0.151 | 1.704 | 0.824 | 3.522 |
 | Table II.Clinical and molecular
characteristics of 144 patients with glioblastoma multiforme. |
Table II.
Clinical and molecular
characteristics of 144 patients with glioblastoma multiforme.
|
|
| Survivin
expression |
|
|---|
|
|
|
|
|
|---|
| Variables | Total | High | Low | P-value |
|---|
| Sex |
|
|
|
|
|
Female | 51 | 23 | 28 | 0.384 |
|
Male | 93 | 49 | 44 |
|
| Age, years |
|
|
|
|
|
>48 | 71 | 33 | 38 | 0.405 |
|
≤48 | 73 | 39 | 34 |
|
| IDH mutation |
|
|
|
|
|
Mutation | 39 | 20 | 19 | 0.851 |
| Wild
type | 105 | 52 | 53 |
|
| TP53 mutation |
|
|
|
|
|
Mutation | 77 | 36 | 41 | 0.404 |
| Wild
type | 67 | 36 | 31 |
|
| EGFR mutation |
|
|
|
|
|
Mutation | 39 | 19 | 20 | 0.851 |
| Wild
type | 105 | 53 | 52 |
|
| ATRX mutation |
|
|
|
|
|
Mutation | 13 | 4 | 9 | 0.146 |
| Wild
type | 131 | 68 | 63 |
|
| TERT promoter
mutation |
|
|
|
|
|
Mutation | 36 | 21 | 15 | 0.305 |
| Wild
type | 65 | 31 | 34 |
|
| MGMT
methylation |
|
|
|
|
|
Yes | 61 | 34 | 27 | 0.368 |
| No | 71 | 34 | 37 |
|
| KPS |
|
|
|
|
|
>70 | 60 | 27 | 33 | 0.317 |
|
≤70 | 36 | 20 | 16 |
|
| Resection |
|
|
|
|
|
GTR | 90 | 44 | 46 | 0.717 |
|
STR | 46 | 24 | 22 |
|
| Treatment |
|
|
|
|
| RT | 22 | 11 | 11 | 0.946 |
|
RT+TZM | 59 | 30 | 29 |
|
| TCGA subtype |
|
|
|
|
|
Proneural | 33 | 20 | 13 | 0.165 |
|
Neural | 12 | 0 | 12 | <0.001 |
|
Classical | 48 | 31 | 17 | 0.013 |
|
Mesenchymal | 51 | 21 | 30 | 0.117 |
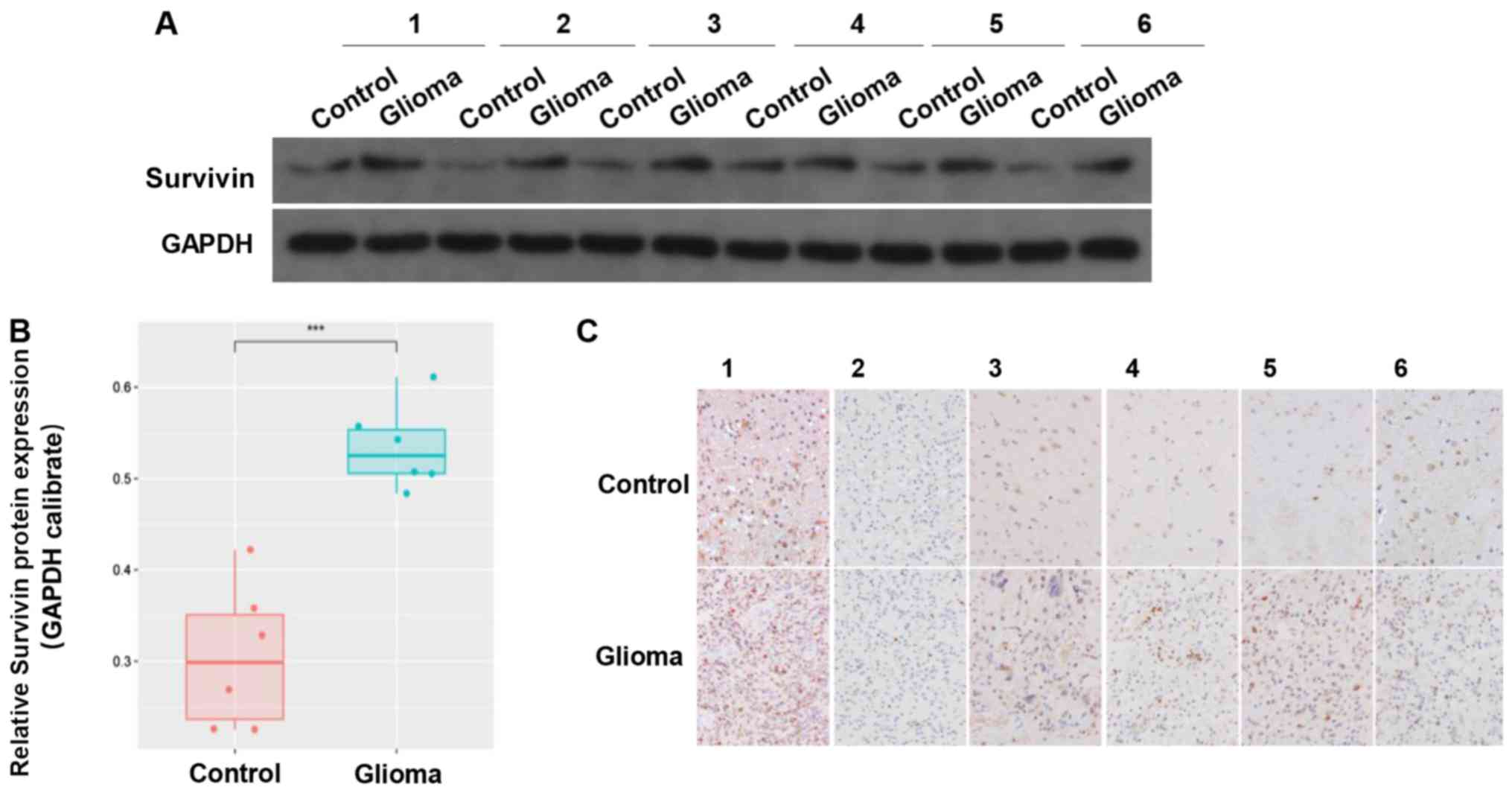
Expression of survivin is reduced in
tumor samples compared with in adjacent tissues
The clinical data of 6 cases of GBM tumor samples
were collected. The results revealed that there was higher
expression of survivin in the glioma tissues compared with in the
adjacent control tissues (Fig. 2A and
B). IHC indicated that survivin was mainly located in the
cytoplasm and nucleus (Fig. 2C).
These results suggested that survivin was highly expressed in
gliomas.
miR-218 expression in the LN229 GBM cell line is
significantly lower compared with that in the control cell line.
Compared with immortalized human gliocyte HEB cells (25,26), GBM
cells had lower miR-218 expression (Fig.
3A). The same pattern was observed in glioma and adjacent
samples from 6 patients with GBM. Compared with in the adjacent
tissues, the level of miR-218 expression in glioma was reduced
(Fig. 3B). A red-labeled
hsa-miR-218-5p probe was used for FISH. The results revealed that
miR-218-5p expression in glioma tissues was low compared with that
in adjacent tissues (Fig. 3C).
miR-218 inhibits the proliferation,
migration and invasion of GBM cells, and contributes to
apoptosis
Upon transfection with miR-218 mimics, the
expression of miR-218 in LN229 cells increased significantly
compared with that of the LN229+NC group (Fig. 3D). A CCK-8 proliferation assay
revealed that, upon transfection with miR-218 mimics, the optical
density values of LN229 cells decreased significantly compared with
the LN229+NC group, suggesting that overexpression of miR-218 can
inhibit the proliferation of LN229 cells (Fig. 3E). Transwell migration and invasion
assays revealed that miR-218 mimics can inhibit the invasion and
migration of glioma cells (Fig. 3F and
G). The results of flow cytometry demonstrated that the
apoptosis rate of LN229 cells markedly increased upon transfection
with miR-218 mimics compared with that of the LN229+NC group,
suggesting that high expression of miR-218 can promote the
apoptosis of LN229 cells (Fig.
3H).
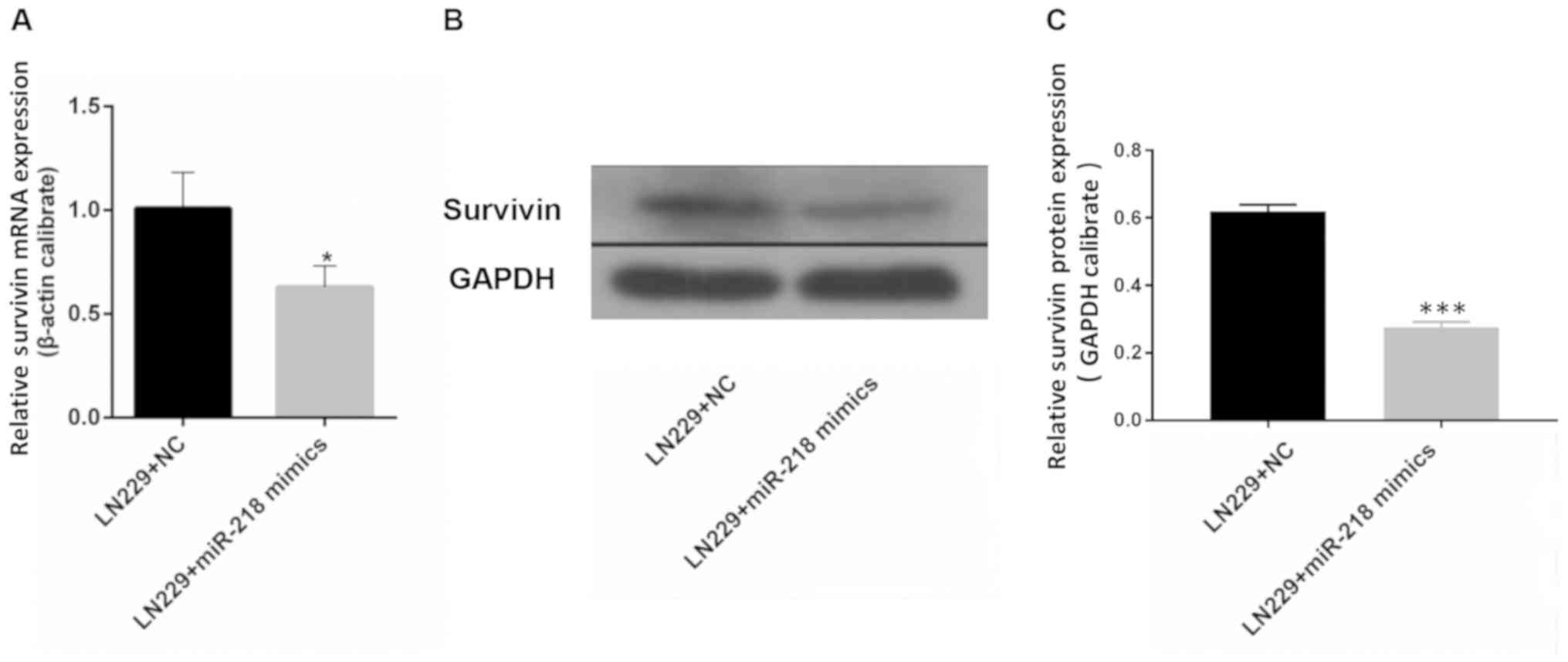
miR-218 overexpression is associated with reduced
expression of survivin. The results of RT-qPCR demonstrated that
the mRNA expression of survivin in LN229 cells was significantly
reduced upon transfection with miR-218 mimics compared with that in
the LN229+NC group (Fig. 4A).
Western blotting revealed a similar outcome: Expression of survivin
in LN229 cells was lower following transfection with miR-218 mimics
compared with that in the LN229+NC group (Fig. 4B and C).
Discussion
Survivin is a member of the IAP protein family, and
is encoded by the BIRC5 gene. Survivin is associated with
inhibition of apoptosis and regulation of cell-cycle progression
(17) and acts as an inhibitor of
programmed cell death by binding to caspase-3/7 in the G2/M phase
(27). The expression of survivin is
higher during fetal development and in the majority of tumors,
whereas it is completely absent in adults and in normal cells
(17,28). Overexpression of survivin has been
confirmed in breast, lung, ovarian, prostate and colon cancer
(21,29). Expression of survivin has also been
associated with poor prognosis and chemotherapy resistance
(21,30,31). A
previous study have demonstrated that high expression of survivin
may increase chemotherapy resistance, and that survivin inhibition
represents an approach to overcoming drug resistance (32). However, no relevant genome atlas
studies have been reported in GBM. Survivin is also a potential
target of miR-218, which is involved in the proliferative,
migratory and invasive behavior of various tumors (20,21). Our
previous study, which included 220 gliomas from the CGGA database,
demonstrated that the expression of survivin is associated with
tumor grade, and that it may be a novel prognostic factor in
gliomas (10). Using whole-genome
RNA sequencing data of 144 patients with GBM in the present study,
it was observed that patients with high expression of survivin had
shorter OS times than those with low expression. However, patients
who received radiation plus TMZ chemotherapy had a better prognosis
compared with patients receiving radiation only, regardless of
their survivin expression levels. This may be due to the limited
number of cases included in the present study and the heterogeneity
of gliomas. Cheng et al (13)
demonstrated that miR-218 has a significantly decreased expression
level in gliomas, which was also associated with poor PFS and OS.
It was also reported that miR-218 may regulate tumor progression
(33–36). HEB cells are not fully ‘normal’
astrocytes, as the cell line has been immortalized, which may
reduce the differences in proliferative ability between astrocytes
and GBM cells (25,26). Therefore, patient samples were used
in the present study to verify the results. The current study
indicated that the expression of miR-218 in the LN229 cell line was
significantly lower than that in HEB cells, which was also
validated in clinical samples. In vitro, the results of the
present study confirmed that miR-218 could inhibit the
proliferation, migration and invasion of GBM cells, and contribute
to their apoptosis. As indicated by Gao and Jin (37), miR-218 can influence cancer-related
processes by targeting a series of genes, including survivin.
However, the role of the miR-218/survivin axis in GBM remains
unknown. Compared with previous studies, the present study explored
the association between miR-218 and survivin in GBM, and
demonstrated that the expression of survivin in LN229 cells
decreased significantly upon transfection with miR-218 mimics.
Combined with luciferase assay results from previous studies, these
observations suggest that miR-218 may directly target surviving
(19–21,38).
These results indicate that survivin may serve as a potential
predictive biomarker targeting miR-218.
However, there are several limitations in the
present study. LN229 is a typical glioma cell line, and when
compared with other GBM cells, there are no significant differences
in adherence and proliferation capacity (39–41).
However, in the current study, only one GBM cell line was used and
there was no verification using animal models. Furthermore, the
sample size was limited. In addition, the association between
survivin protein expression and patient survival is unknown. Thus,
multiple cell lines, animal models, and larger multicenter sites
for patient recruitment will be considered in future studies. In
the laboratory, a stably overexpressing cell line will be set up
and used to determine relative proliferation and migration.
Patients or their relatives will be followed up to obtain extra
prognostic information. Associated pathways will also be
investigated, such as SLIT2-ROBO1 pathway. Furthermore, survivin
has been reportedly associated with mitotic catastrophe (16,17),
which should be further explored in glioma. Cell cycle analysis
will also be performed to assess the impact of miR-218
overexpression on LN229 cells. Experiments using a caspase
inhibitor, such as z-VAD-fmk, to identify the type of cell death
are required to confirm the involvement of the apoptotic cascade
(42). A luciferase assay will be
used to demonstrate that miR-218 directly targets survivin. These
remaining questions will be addressed in future studies.
Acknowledgements
The authors would like to thank Ms. Hua Huang from
the Beijing Neurosurgical Institute (Beijing, China) for technical
support with experiments and her contribution to the CGGA
database.
Funding
The present study was supported by the Natural
Science Foundation of Beijing (Beijing, China; grant no. 7152052),
the Natural Science Foundation of China Capital Medical University,
(Beijing, China; grant no. PYZ2017145), and the Miaopu Project of
Beijing Tiantan Hospital (Beijing, China; grant no. 2017MP05).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the Chinese Glioma Genome Atlas
(CGGA) (http://www.cgga.org.cn).
Authors' contributions
XT, PY and KW performed most of the experiments and
drafted the manuscript. YL and XL helped with data analysis. XS, RH
and KZ helped with tumor samples collection, designed figures and
revised the manuscript. JW designed the study and revised the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
The Capital Medical University (Beijing, China). All patients
provided informed consent.
Patient consent for publication
Written informed consent was obtained from each
patient involved in this study.
Competing interests
The authors declare that they have no conflicts of
interest.
Glossary
Abbreviations
Abbreviations:
|
GBM
|
glioblastoma multiforme
|
|
PFS
|
progression-free survival
|
|
OS
|
overall survival
|
|
miR-218
|
microRNA-218
|
|
IAP
|
inhibitor of apoptosis
|
|
BIRC5
|
baculoviral inhibitor of apoptosis
repeat-containing 5
|
|
miRNAs/miRs
|
microRNAs
|
|
CGGA
|
Chinese Glioma Genome Atlas
|
|
TMZ
|
temozolomide
|
|
DMEM
|
Dulbecco's modified Eagle's medium
|
|
FBS
|
fetal bovine serum
|
|
NC
|
negative control
|
|
CCK-8
|
Cell Counting Kit-8
|
|
IHC
|
immunohistochemistry
|
|
FISH
|
fluorescence in situ
hybridization
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
SSC
|
saline sodium citrate
|
References
|
1
|
Weller M, van den Bent M, Hopkins K, Tonn
JC, Stupp R, Falini A, Cohen-Jonathan-Moyal E, Frappaz D,
Henriksson R, Balana C, et al: EANO guideline for the diagnosis and
treatment of anaplastic gliomas and glioblastoma. Lancet Oncol.
15:e395–e403. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Thakkar JP, Dolecek TA, Horbinski C,
Ostrom QT, Lightner DD, Barnholtz-Sloan JS and Villano JL:
Epidemiologic and molecular prognostic review of glioblastoma.
Cancer Epidemiol Biomarkers Prev. 23:1985–1996. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ostrom QT, Gittleman H, Stetson L, Virk SM
and Barnholtz-Sloan JS: Epidemiology of gliomas. Cancer Treat Res.
163:1–14. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Omuro A and DeAngelis LM: Glioblastoma and
other malignant gliomas: A clinical review. JAMA. 310:1842–1850.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li N, Wang L, Tan G, Guo Z, Liu L, Yang M
and He J: MicroRNA-218 inhibits proliferation and invasion in
ovarian cancer by targeting Runx2. Oncotarget. 8:91530–91541.
2017.PubMed/NCBI
|
|
6
|
Shi D and Zhang Y, Lu R and Zhang Y: The
long non-coding RNA MALAT1 interacted with miR-218 modulates
choriocarcinoma growth by targeting Fbxw8. Biomed Pharmacother.
97:543–550. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mohr AM and Mott JL: Overview of microRNA
biology. Semin Liver Dis. 35:3–11. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hammond SM: An overview of microRNAs. Adv
Drug Deliv Rev. 87:3–14. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen P, Zhao Y and Li Y: MiR-218 inhibits
migration and invasion of lung cancer cell by regulating Robo1
expression. Zhongguo Fei Ai Za Zhi. 20:452–458. 2017.(In Chinese).
PubMed/NCBI
|
|
10
|
Bao ZS, Li MY, Wang JY, Zhang CB, Wang HJ,
Yan W, Liu YW, Zhang W, Chen L and Jiang T: Prognostic value of a
nine-gene signature in glioma patients based on mRNA expression
profiling. CNS Neurosci Ther. 20:112–118. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen W, Yu Q, Chen B, Lu X and Li Q: The
prognostic value of a seven-microRNA classifier as a novel
biomarker for the prediction and detection of recurrence in glioma
patients. Oncotarget. 7:53392–53413. 2016.PubMed/NCBI
|
|
12
|
Yang Y, Ding L, Hu Q, Xia J, Sun J, Wang
X, Xiong H, Gurbani D, Li L, Liu Y and Liu A: MicroRNA-218
functions as a tumor suppressor in lung cancer by targeting
IL-6/STAT3 and negatively correlates with poor prognosis. Mol
Cancer. 16:1412017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cheng MW, Wang LL and Hu GY: Expression of
microRNA-218 and its clinicopathological and prognostic
significance in human glioma cases. Asian Pac J Cancer Prev.
16:1839–1843. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mobahat M, Narendran A and Riabowol K:
Survivin as a preferential target for cancer therapy. Int J Mol
Sci. 15:2494–2516. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jaskoll T, Chen H, Min Zhou Y, Wu D and
Melnick M: Developmental expression of survivin during embryonic
submandibular salivary gland development. BMC Dev Biol. 1:52001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Conde M, Michen S, Wiedemuth R, Klink B,
Schröck E, Schackert G and Temme A: Chromosomal instability induced
by increased BIRC5/Survivin levels affects tumorigenicity of glioma
cells. BMC Cancer. 17:8892017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sheng L, Wan B, Feng P, Sun J, Rigo F,
Bennett CF, Akerman M, Krainer AR and Hua Y: Downregulation of
Survivin contributes to cell-cycle arrest during postnatal cardiac
development in a severe spinal muscular atrophy mouse model. Hum
Mol Genet. 27:486–498. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yoshikawa K, Shimada M, Higashijima J,
Nakao T, Nishi M, Takasu C, Kashihara H, Eto S and Bando Y: Ki-67
and survivin as predictive factors for rectal cancer treated with
preoperative chemoradiotherapy. Anticancer Res. 38:1735–1739.
2018.PubMed/NCBI
|
|
19
|
Li PL, Zhang X, Wang LL, Du LT, Yang YM,
Li J and Wang CX: MicroRNA-218 is a prognostic indicator in
colorectal cancer and enhances 5-fluorouracil-induced apoptosis by
targeting BIRC5. Carcinogenesis. 36:1484–1493. 2015.PubMed/NCBI
|
|
20
|
Kogo R, How C, Chaudary N, Bruce J, Shi W,
Hill RP, Zahedi P, Yip KW and Liu FF: The microRNA-218~Survivin
axis regulates migration, invasion, and lymph node metastasis in
cervical cancer. Oncotarget. 6:1090–1100. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hu Y, Xu K and Yagüe E: miR-218 targets
survivin and regulates resistance to chemotherapeutics in breast
cancer. Breast Cancer Res Treat. 151:269–280. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ishiyama M, Miyazono Y, Sasamoto K, Ohkura
Y and Ueno K: A highly water-soluble disulfonated tetrazolium salt
as a chromogenic indicator for NADH as well as cell viability.
Talanta. 44:1299–1305. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun Y, Zhang Y and Chi P: Pirfenidone
suppresses TGF-β1-induced human intestinal fibroblasts activities
by regulating proliferation and apoptosis via the inhibition of the
Smad and PI3K/AKT signaling pathway. Mol Med Rep. 18:3907–3913.
2018.PubMed/NCBI
|
|
25
|
Dai Z, Wu J, Chen F, Cheng Q, Zhang M,
Wang Y, Guo Y and Song T: CXCL5 promotes the proliferation and
migration of glioma cells in autocrine- and paracrine-dependent
manners. Oncol Rep. 36:3303–3310. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang T, Shao Y, Chu TY, Huang HS, Liou
YL, Li Q and Zhou H: MiR-135a and MRP1 play pivotal roles in the
selective lethality of phenethyl isothiocyanate to malignant glioma
cells. Am J Cancer Res. 6:957–972. 2016.PubMed/NCBI
|
|
27
|
Lechler P, Renkawitz T, Campean V,
Balakrishnan S, Tingart M, Grifka J and Schaumburger J: The
antiapoptotic gene survivin is highly expressed in human
chondrosarcoma and promotes drug resistance in chondrosarcoma cells
in vitro. BMC Cancer. 11:1202011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao G, Wang Q, Gu Q, Qiang W, Wei JJ,
Dong P, Watari H, Li W and Yue J: Lentiviral CRISPR/Cas9 nickase
vector mediated BIRC5 editing inhibits epithelial to mesenchymal
transition in ovarian cancer cells. Oncotarget. 8:94666–94680.
2017.PubMed/NCBI
|
|
29
|
Slattery ML, Mullany LE, Sakoda LC, Wolff
RK, Samowitz WS and Herrick JS: Dysregulated genes and miRNAs in
the apoptosis pathway in colorectal cancer patients. Apoptosis.
23:237–250. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jingjing L, Wangyue W, Qiaoqiao X and
Jietong Y: MiR-218 increases sensitivity to cisplatin in esophageal
cancer cells via targeting survivin expression. Open Med (Wars).
11:31–35. 2016.PubMed/NCBI
|
|
31
|
Zarogoulidis P, Petanidis S, Kioseoglou E,
Domvri K, Anestakis D and Zarogoulidis K: MiR-205 and miR-218
expression is associated with carboplatin chemoresistance and
regulation of apoptosis via Mcl-1 and Survivin in lung cancer
cells. Cell Signal. 27:1576–1588. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zheng HC: The molecular mechanisms of
chemoresistance in cancers. Oncotarget. 8:59950–59964.
2017.PubMed/NCBI
|
|
33
|
Tu Y, Gao X, Li G, Fu H, Cui D, Liu H, Jin
W and Zhang Y: MicroRNA-218 inhibits glioma invasion, migration,
proliferation, and cancer stem-like cell self-renewal by targeting
the polycomb group gene Bmi1. Cancer Res. 73:6046–6055. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gu JJ, Gao GZ and Zhang SM: miR-218
inhibits the migration and invasion of glioma U87 cells through the
Slit2-Robo1 pathway. Oncol Lett. 9:1561–1566. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xia H, Yan Y, Hu M, Wang Y, Wang Y, Dai Y,
Chen J, Di G, Chen X and Jiang X: MiR-218 sensitizes glioma cells
to apoptosis and inhibits tumorigenicity by regulating
ECOP-mediated suppression of NF-κB activity. Neuro Oncol.
15:413–422. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xu J, Xu W and Zhu J: Propofol suppresses
proliferation and invasion of glioma cells by upregulating
microRNA-218 expression. Mol Med Rep. 12:4815–4820. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gao X and Jin W: The emerging role of
tumor-suppressive microRNA-218 in targeting glioblastoma stemness.
Cancer Lett. 353:25–31. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Alajez NM, Lenarduzzi M, Ito E, Hui AB,
Shi W, Bruce J, Yue S, Huang SH, Xu W, Waldron J, et al: MiR-218
suppresses nasopharyngeal cancer progression through downregulation
of survivin and the SLIT2-ROBO1 pathway. Cancer Res. 71:2381–2391.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yang X, Zhang C, Guo T, Feng Y, Liu Q,
Chen Y and Zhang Q: Reduced expression of microRNA206 regulates
cell proliferation via cyclinD2 in gliomas. Mol Med Rep.
11:3295–3300. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang H, Wang Y, Bao Z, Zhang C, Liu Y, Cai
J and Jiang C: Hypomethylated Rab27b is a progression-associated
prognostic biomarker of glioma regulating MMP-9 to promote
invasion. Oncol Rep. 34:1503–1509. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu Y, Tang K, Yan W, Wang Y, You G, Kang
C, Jiang T and Zhang W: Identifying Ki-67 specific miRNA-mRNA
interactions in malignant astrocytomas. Neurosci Lett. 546:36–41.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhou GS, Song LJ and Yang B:
Isoliquiritigenin inhibits proliferation and induces apoptosis of
U87 human glioma cells in vitro. Mol Med Rep. 7:531–536.
2013. View Article : Google Scholar : PubMed/NCBI
|