Introduction
Hypopharyngeal carcinoma, one of the most common
types of head and neck squamous cell carcinoma, results in
substantial morbidities and mortalities annually. The majority of
patients with hypopharyngeal carcinoma are frequently observed to
be at an advanced stage, with lymph node metastasis at the initial
diagnosis. The predominant therapy for hypopharyngeal carcinoma
remains confined to surgery with additional treatments of
radiotherapy and chemotherapy (1).
Although surgical techniques and anticancer agents have advanced,
the overall survival rates have not significantly improved during
the last two decades (2,3). Therefore, identification of the
associated target factors and the potential mechanism of
hypopharyngeal carcinoma metastasis and proliferation are important
for the survival of patients.
The epithelial cell adhesion molecule (EpCAM), also
known as CD326, a 39–42-kDa type I trans-membrane glycoprotein,
consists of an extracellular domain, a single transmembrane domain
and a short 26-amino acid intracellular domain (EpICD) (4). EpCAM expression is observed at the
basolateral membrane of the majority of epithelium, with the
exception of squamous epithelium (5,6).
Furthermore, EpCAM is overexpressed in the majority of human
epithelial carcinomas, including breast, colorectal, prostate,
hepatic and head and neck carcinomas, and its overexpression in
these cancers is associated with proliferation and neoplastic
transformation (7–11). Although EpCAM is regarded as a
Ca2+-independent homophilic cell-cell adhesion molecule,
its intercellular adhesive activity is extremely weak (12,13).
EpCAM predominantly contributes to proliferation and metastasis by
regulating E-cadherin mediated-adhesion and β-catenin signaling
(14,15). EpCAM has previously been regarded as
an additional marker for the identification of cancer-initiating
stem cells (16). Cancer stem cells
exhibiting a high level of EpCAM expression are more tumorigenic
and malignant than those exhibiting low expression levels (17). Therefore, based on the radiation and
drug resistance of cancer stem cells, targeting EpCAM may present a
promising approach for cancer therapy.
However, few studies have analyzed the effect of
EpCAM in hypopharyngeal carcinomas and thus, knowledge regarding
the role of EpCAM in the process of carcinogenesis, tumor
progression and metastasis requires further elucidation. In the
present study, the expression of EpCAM in hypopharyngeal carcinoma
was examined. Furthermore, the EpCAM small interfering RNA (siRNA)
was employed to downregulate EpCAM in hypopharyngeal carcinoma FaDu
cells for studying the role of EpCAM and its mechanism. The present
study also attempted to clarify whether EpCAM may be regarded as a
potential candidate for hypopharyngeal carcinoma therapy.
Materials and methods
Tissue sections and
immunohistochemistry
A total of 40 hypopharyngeal carcinoma tissue
samples were obtained from patients with hypopharyngeal carcinoma
at the Shandong University Affiliated Provincial Hospital (Jinan,
China). For controls, samples of normal squamous epithelium tissue
were obtained from the non-cancerous regions of these patients. No
patients had previously received pre-operative chemotherapy or
radiotherapy. Patient information, including age, gender and
tumor-node-metastasis stage, were obtained from the surgical and
pathological records, and written informed consent was obtained
from all patients. The study was approved by the ethics committee
of Shandong University (Jinan, China).
The paraffin-embedded tissue sections were hydrated
in xylene (Guangcheng Chemical Reagent Co., Ltd., Tianjin, China)
and a graded alcohol series. Antigen retrieval was performed in a
water bath at 95°C for 20 min with citric acid buffer (Beijing
Zhongshan Golden Bridge Biotechnology Co., Ltd., Beijing, China),
and endogenous peroxidase activity was blocked with 3%
H2O2. Next, the tissue sections were
incubated with goat serum (Beijing Zhongshan Golden Bridge
Biotechnology Co., Ltd.) for 45 min and stained with rabbit
anti-human EpCAM antibody (Abcam, Cambridge, MA, USA) at 4°C
overnight. For the negative control, an equal amount of
phosphate-buffered saline (PBS) was used instead of the primary
antibody. Subsequent to washing the tissue section with PBS three
times, the biotin-labeled goat anti-rabbit IgG primary antibody
(IgG/Bio, Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd.)
was detected following incubation with a secondary antibody
(horseradish peroxidase-labeled streptavidin; S-A/HRP, Beijing
Zhongshan Golden Bridge Biotechnology Co., Ltd.) for 30 min at
37°C. 3,3′-Diamino-benzidine tetrahydrochloride solution (Beijing
Zhongshan Golden Bridge Biotechnology Co., Ltd.) was used to
visualize positive staining, and hematoxylin was used to
counterstain the nucleoli.
Cell line and cell culture
The human hypopharyngeal carcinoma FaDu cell line
was obtained from the American Type Culture Collection (Manassas,
VA, USA). The FaDu cells were cultured in Dulbecco’s modified
Eagle’s medium (DMEM; Gibco-BRL, Carlsbad, CA, USA) containing 10%
fetal bovine serum (FBS; Gibco-BRL), 100 U/ml penicillin and 100
mg/ml streptomycin at 37°C in a humidified atmosphere of 5%
CO2.
RNA interference
All siRNAs were designed by GeneChem (Shanghai,
China), with sequences as follows: EpCAM siRNA forward,
5′-CGTAAACTGCTTTGTGAATdTdT-3′ and reverse,
5′-ATTCACAAAGCAGTTTACGdTdT-3′; and scrambled (SCR) siRNA forward,
5′-ACGUGACACGUUCGGAGA AdTdT-3′ and reverse, 5′-UUCUCCGAACGUGUCACG
UdTdT-3′.
The FaDu cells were transfected with Lipofectamine
2000 (Invitrogen Life Technologies, Carlsbad, CA, USA) according to
the manufacturer’s instructions. Briefly, 3.0×104 cells
were plated in six-well plates (Costar, Cambridge, MA, USA) and
cultured at 37°C for ~24 h until 70–80% confluent. siRNA and
Lipofectamine 2000 were then diluted in 300 μl Opti-MEM reduced
serum media (Gibco-BRL). Next, the solutions were mixed together
and incubated for 20 min at room temperature. The cells were then
washed twice with Opti-MEM and the mixture solution was transferred
to the six-well plates. Following incubation for 8 h, the mixed
solution was discarded and 2 ml of 10% FBS-DMEM was added to each
well of the six-well plates. The cells were then incubated at 37°C
for ~72 h and lysed with 0.25% trypsin-EDTA (Gibco-BRL) for
subsequent assays.
RNA isolation and quantitative polymerase
chain reaction (qPCR)
Total RNA was isolated using TRIzol reagent
(Invitrogen Life Technologies). In accordance with the
manufacturer’s instructions, the first-strand of cDNA was
synthesized using the PrimeScript first-strand cDNA synthesis kit
[Takara Biotechnology (Dalian) Co., Ltd., Dalian, China] in a
reaction mixture with a final volume of 20 μl, containing 1 μg
total RNA, 4 μl 5× PrimeScript buffer, 1 μl deoxynucleotide
triphosphate mixture, 1 μl Oligo(dT) primer, 0.5 μl PrimeScript
RTase, 0.5 μl RNase inhibitor and RNase-free water. The reverse
transcription reaction was performed under the following
conditions: 42°C for 15 min, followed by a termination step at 95°C
for 2 min. The qPCR analyses were performed using an Eppendorf
Mastercycler (Eppendorf, Westbury, NY, USA). The standard reaction
volume was 25 μl, containing 1 μl QuantiTect SYBR Green PCR Master
Mix [Takara Biotechnology (Dalian) Co., Ltd], 2 μl cDNA template
and 0.25 μM forward and reverse primers. The initial PCR step was
as follows: 2 min at 50°C, followed by a 15 min hold at 95°C. This
was followed by 40 cycles, consisting of a 15-sec denaturation step
at 95°C, a 20-sec annealing/extension step at 59°C, and a 72°C
incubation step for 20 sec. All reactions were performed in
triplicate. Following normalization to the GAPDH gene, the
expression levels for each target gene were analyzed using the
comparative threshold cycle (CT) method. The 2−ΔΔct was
calculated to determine the relativity using the following formula:
ΔΔct = Δct(experimental group) - Δct(control group). The Δct values
were calculated using the following formula: Δct = ct(target gene)
- ct(GAPDH). The EpCAM and GAPDH primers were designed by Takara
Biotechnology (Dalian) Co., Ltd., and the primer sequences used
were as follows: EpCAM forward, 5′-GAATGG CAAAGTATGAGAAGGCTGA-3′
and reverse, 5′-TCCCAC GCACACACATTTGTAA-3′; and GAPDH forward,
5′-CAA GGTCATCCCTGACACTTG-3′ and reverse, 5′-GTCCAC
CACCCTGTTGCTGTAG-3′.
Protein extraction and western blot
analysis
To obtain the total cell lysates, the cells were
rinsed twice with ice-cold PBS and lysed in 300 μl of hot (100°C)
10 mM EDTA containing 1% sodium dodecyl sulfate (SDS). The
detergent-soluble (not anchored to the cytoskeleton) and
detergent-insoluble (anchored to the cytoskeleton) proteins were
extracted using the technique reported by Osta et al
(7), with minor revisions. Briefly,
the cells were rinsed three times with cold PBS and 300 μl cold
extraction buffer, which was composed of 50 mM Tris-HCl (pH 7.0),
50 mM NaCl, 3 mM MgCl2, 0.5% Triton X-100, 300 mM
sucrose and 1% protease inhibitor mixture (Sigma-Aldrich, St.
Louis, MO, USA). Next, the cells were agitated for 60 min at 4°C
and centrifuged in the Sigma 3K15 ultracentrifuge (JinanSigma
Zentrifugen GmbH, Osterode, Germany) for 1 h at 10,000 × g and 4°C.
The supernatant was collected and the pellet was lysed with 300 μl
of hot (100°C) 1% SDS/10 mM EDTA and then incubated at 100°C for 10
min. The nuclear protein was obtained using the Nuclear Extract kit
(Active Motif, Tokyo, Japan) according to the manufacturer’s
instructions. The protein concentration was then determined by the
Enhanced Bicinchoninic Acid Protein Assay kit (Beyotime, Shanghai,
China).
The total, soluble, insoluble and nuclear proteins
with denaturing conditions were loaded for SDS-polyacrylamide gel
electrophoresis (5% stacking gel and 8% separating gel), followed
by separation at 80 V for ~30 min and then 120 V for ~90 min. The
proteins were subsequently transferred to a polyvinylidene
difluoride membrane. Following blocking with 5% skimmed
milk/Tris-buffered saline-Tween (TBST) for 1 h at room temperature,
the membranes were incubated with the primary antibodies, rabbit
anti-human EpCAM (1:2,000), rat anti-human E-cadherin (1:2,000),
rabbit anti-human β-catenin (1:5,000) and mouse anti-human β-actin
(1:2,000), which were diluted in 3% skimmed milk/TBST overnight at
4°C and then washed three times with TBST for 5 min separately.
Subsequently, the membrane was incubated with secondary antibodies,
and the signals were visualized by electrochemiluminescence using
an LAS-4000 image reader (Fujifilm, Tokoyo, Japan). The EpCAM,
E-cadherin, and β-catenin primary antibodies were purchased from
Abcam, whereas the β-actin primary antibody and all secondary
antibodies were purchased from Beijing Zhongshan Golden Bridge
Biotechnology Co., Ltd.
In vitro invasion/migration assays
Cell invasion assays were performed using Transwell™
chambers (Costar, Cambridge, MA, USA). Briefly, subsequent to
coating the filter with 50 μl Matrigel (BD Biosciences, NY, USA),
which had been diluted by DMEM at 1:6 overnight at 37°C, the upper
chamber of the 24-well Transwell plates were filled with 100 μl
serum-free DMEM containing 1×105 cells/ml. The lower
chamber was filled with 0.5 ml DMEM containing 10% FBS as a
chemical attractant. Following incubation for 24 h at 37°C in a 5%
atmosphere, non-invading cells were removed by scrubbing with a
cotton swab. The filters were then fixed with methanol and stained
with crystal violet for 15 min. The number of cells that penetrated
the filter was quantified under a microscope (magnification, ×200).
To assess migration, cell migration assays were performed under the
same conditions as the Transwell invasion assays without
Matrigel-coated Transwell chambers. All experiments were performed
in triplicate and repeated three times.
Cell proliferation assay
Briefly, the cells were plated in 96-well plates at
a concentration of 5×103 cells per well and incubated
for 24, 48 or 72 h following the addition of siRNA. At each
time-point, the cells were incubated with 0.5 mg/ml
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT;
Sigma-Aldrich). After 4 h, the medium was replaced with 100 μl
dimethyl sulfoxide (Sigma-Aldrich) and vortexed for 10 min. The
absorbance was then recorded at a wavelength of 570 nm using Thermo
Multiskan MK3 (Thermo Fisher Scientific Inc, Waltham, MA, USA).
In vitro colony formation assay
The in vitro colony formation assay was
performed to measure oncogenic potential. The control, SCR siRNA
and EpCAM siRNA-treated FaDu cells were suspended in 10% FBS-DMEM,
then plated in 6 wells at 500 cells/well with 2 ml DMEM
supplemented with 10% FBS. The number of colonies were counted on
the tenth day.
Statistical analysis
Data are presented as the mean ± standard deviation.
Statistical calculations were performed using SPSS version 13.0
(SPSS, Inc., Chicago, IL, USA). The χ2 test, one-way
analysis of variance and least significance divergence were applied
to analyze the data. P<0.05 was considered to indicate a
statistically significant difference, and all tests were
two-tailed.
Results
EpCAM overexpression in primary
hypopharyngeal carcinoma
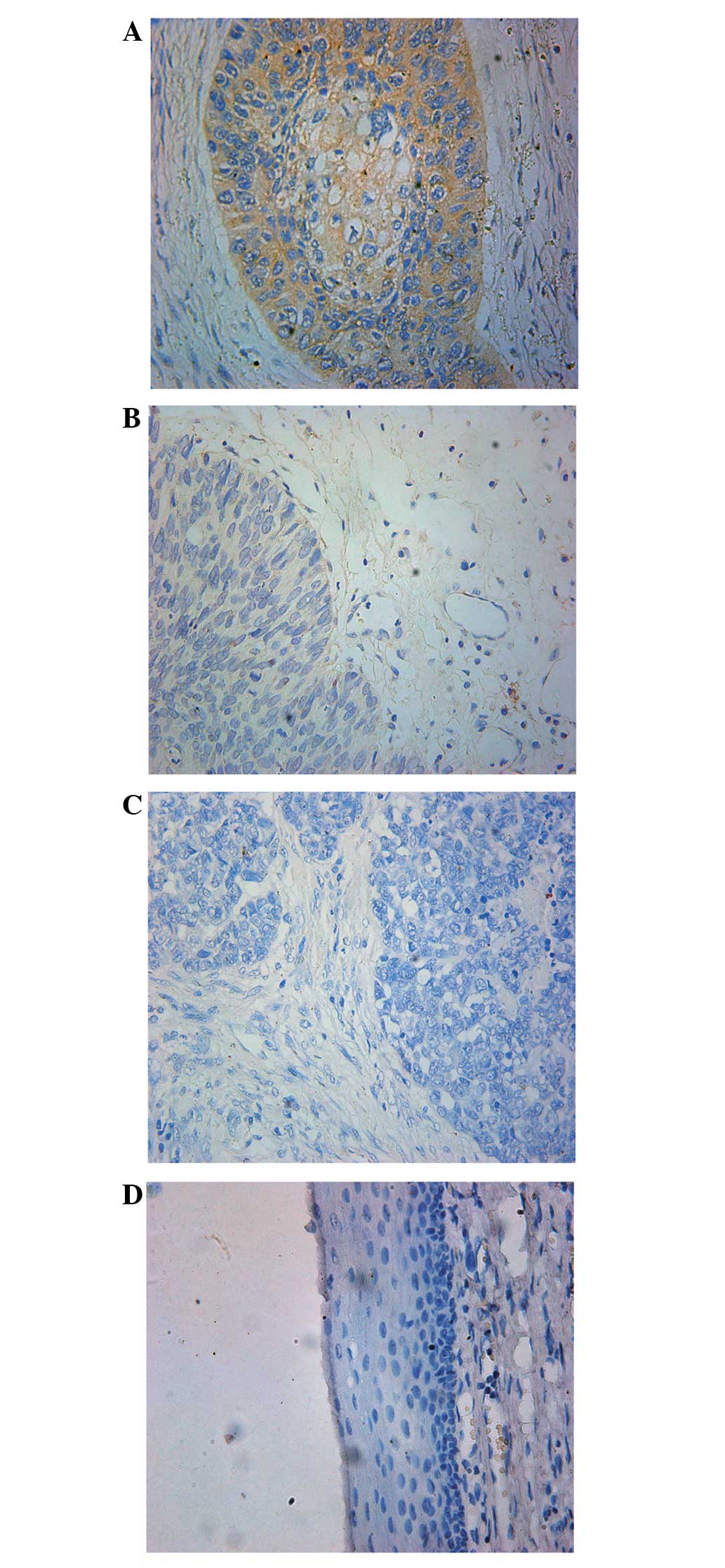
To investigate whether EpCAM is expressed in
hypopharyngeal carcinoma, immunohistochemistry was used to analyze
the 40 hypopharyngeal carcinoma tissues and 10 normal
hypopharyngeal epithelia samples. It was identified that EpCAM was
primarily located at the membrane of the hypopharyngeal carcinoma
cells, occasionally diffusing into the cytoplasm of cells, however,
EpCAM was absent in non-neoplastic tissues (Fig. 1).
EpCAM overexpression correlates with
tumor size and lymph node metastasis in hypopharyngeal
carcinoma
EpCAM overexpression was markedly increased in the
cancer tissues (26 of the 40) when compared with the normal
epithelia (0 of the 10) (P=0.001; Table
I). In addition, EpCAM overexpression was found to correlate
with tumor size stage (P=0.007) and lymph node metastasis
(P=0.029).
 | Table ICorrelation between EpCAM
overexpression and clinicopathological features. |
Table I
Correlation between EpCAM
overexpression and clinicopathological features.
| | EpCAM overexpression,
n (%) | |
|---|
| |
| |
|---|
| Characteristics | n | − | + | P-value |
|---|
| Normal
epithelium | 10 | 10 (100.0) | 0 (0.0) | 0.001 |
| Hypopharyngeal
carcinoma | 40 | 14 (35.0) | 26 (65.0) | |
| Gender | | | | 0.652 |
| Male | 38 | 13 (34.2) | 25 (65.8) | |
| Female | 2 | 1 (50.0) | 1 (50.0) | |
| Age | | | | 0.591 |
| ≥60 | 28 | 10 (35.7) | 18 (64.3) | |
| <60 | 12 | 4 (33.3) | 8 (66.7) | |
| T classification | | | | 0.007 |
| T1+T2 | 14 | 9 (64.3) | 5 (35.7) | |
| T3+T4 | 26 | 5 (19.2) | 21 (80.8) | |
| N classification | | | | 0.029 |
| N0 | 11 | 7 (63.6) | 4 (36.4) | |
| N+ | 29 | 7 (24.1) | 22 (75.9) | |
Downregulation of EpCAM expression
significantly decreases the invasion and migration potential of
FaDu cells in vitro
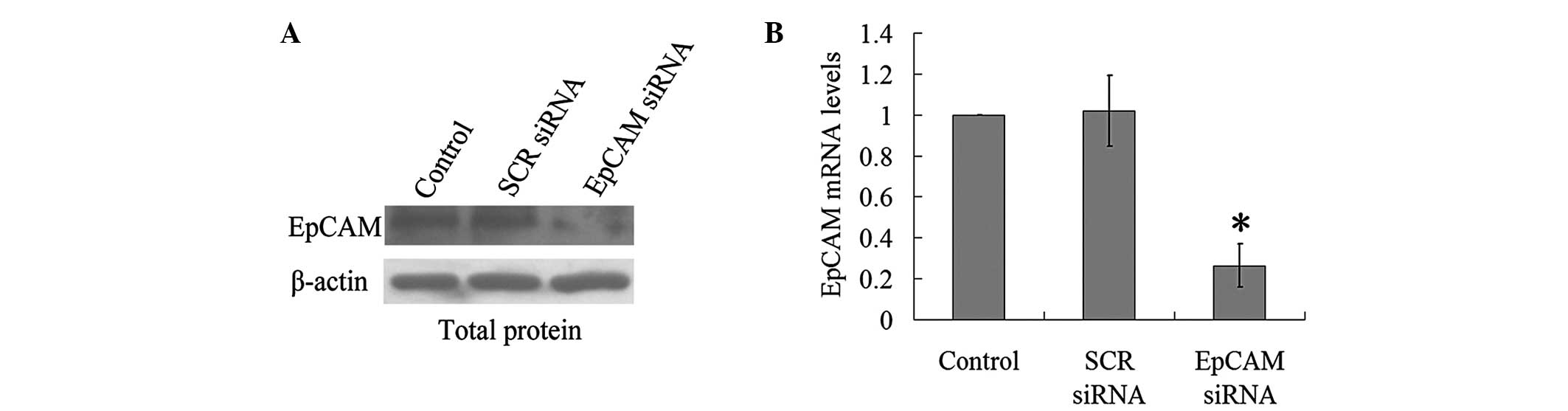
Firstly, an EpCAM siRNA was used to silence EpCAM,
and the results showed that EpCAM siRNA lead to a marked decrease
in EpCAM expression (Fig. 2). To
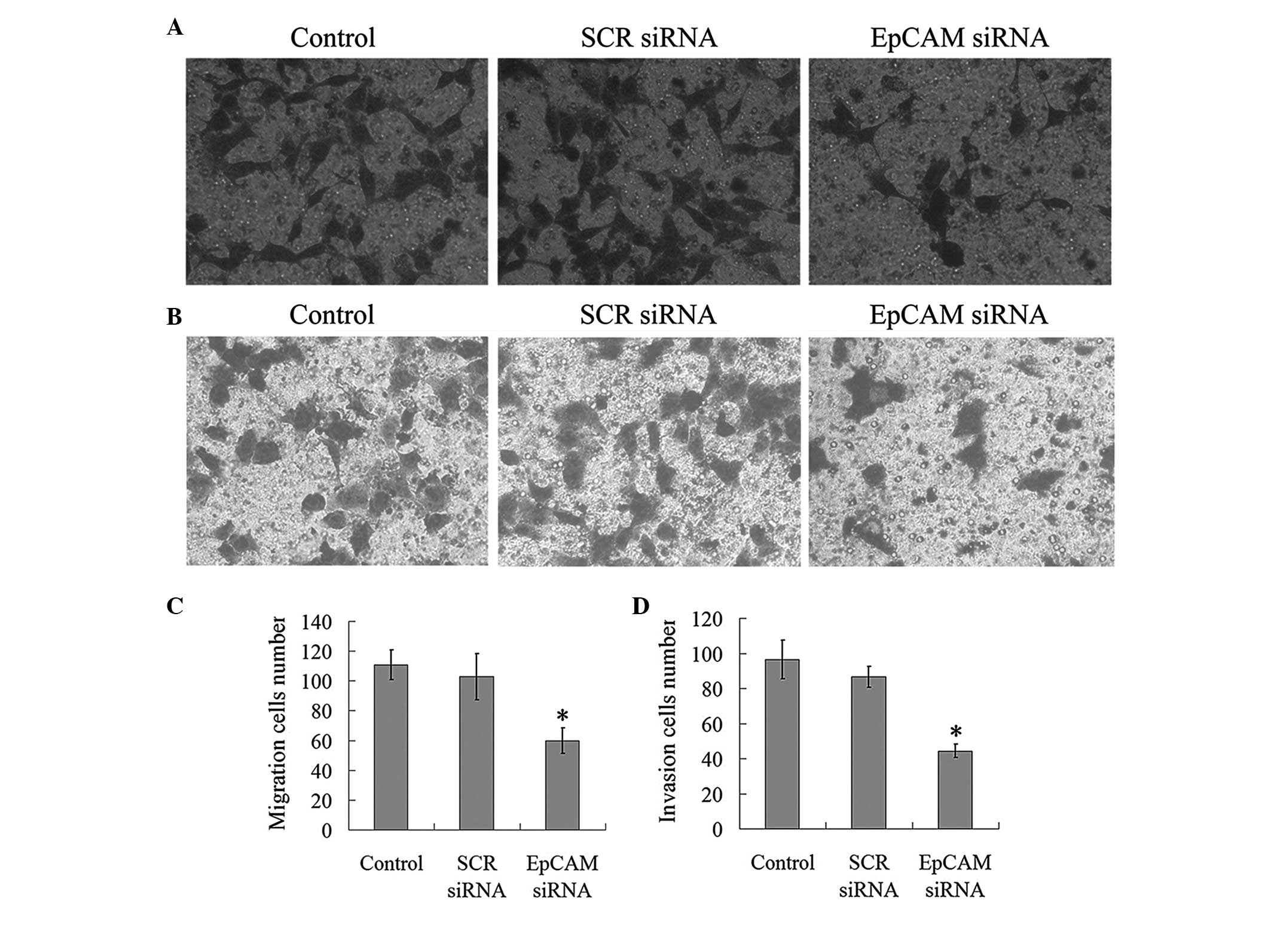
further investigate the effect of EpCAM downregulation on the
invasion and migration potential of the FaDu cells, Transwell
assays were performed. As indicated in Fig. 3, the number of migratory cells in
the EpCAM siRNA treatment groups (59.83±8.42) decreased
significantly when compared with the control (110.83±10.01) and SCR
siRNA treatment (102.89±15.55) groups (P<0.05). In the invasion
assays, the number of invasive cells in the EpCAM siRNA group
(44.40±3.75) was significantly less than that of the control
(96.61±10.98) and SCR siRNA (86.64±5.97) groups (P<0.05;
Fig. 3). These results revealed
that the downregulation of EpCAM expression inhibits the migration
and invasive ability of FaDu cells.
Downregulation of EpCAM expression
inhibits the proliferation ability and tumorigenicity of FaDu cells
in vitro
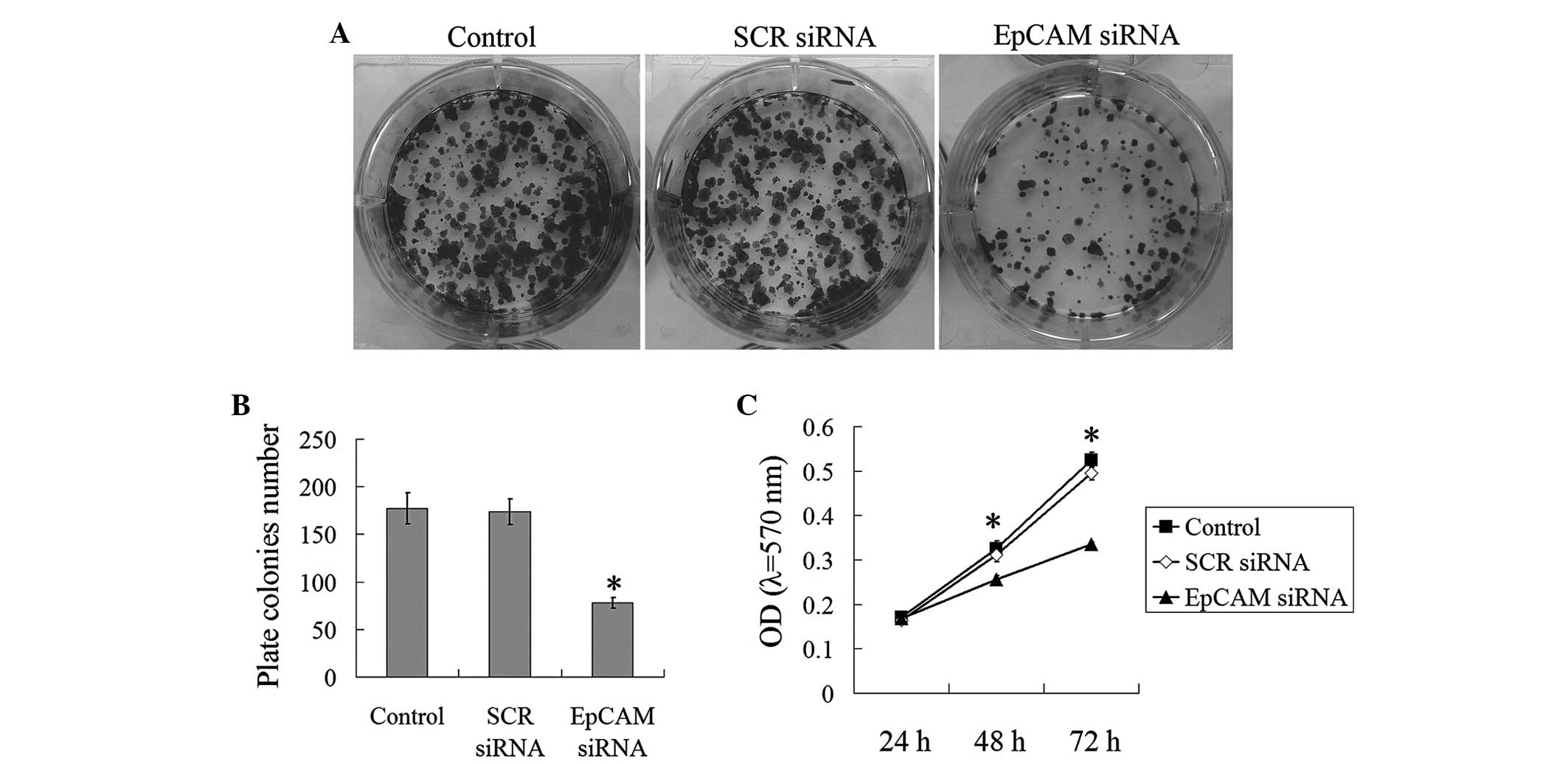
To measure the function of EpCAM downregulation on
the proliferation ability and tumorigenicity of the FaDu cells
in vitro, the MTT and plate colony formation assays were
performed. As shown in Fig. 4, in
the plate colony assay, the plate colony numbers of the control,
SCR siRNA and EpCAM siRNA groups were 177.33±16.50, 173.67±13.51
and 78.00±5.57, respectively (P<0.05). In the MTT assay at 48 h
and 72 h, the absorbance of the control (0.326±0.017 for 48 h and
0.525±0.017 for 72 h) and SCR siRNA groups (0.311±0.016 for 48 h
and 0.495±0.015 for 72 h) were significantly higher than that of
the EpCAM siRNA group (0.256±0.008 for 48 h and 0.335±0.005 for 72
h) (P<0.05; Fig. 4C). These
results clearly indicated that the downregulation of EpCAM
expression inhibits the proliferation ability and tumorigenicity of
FaDu cells.
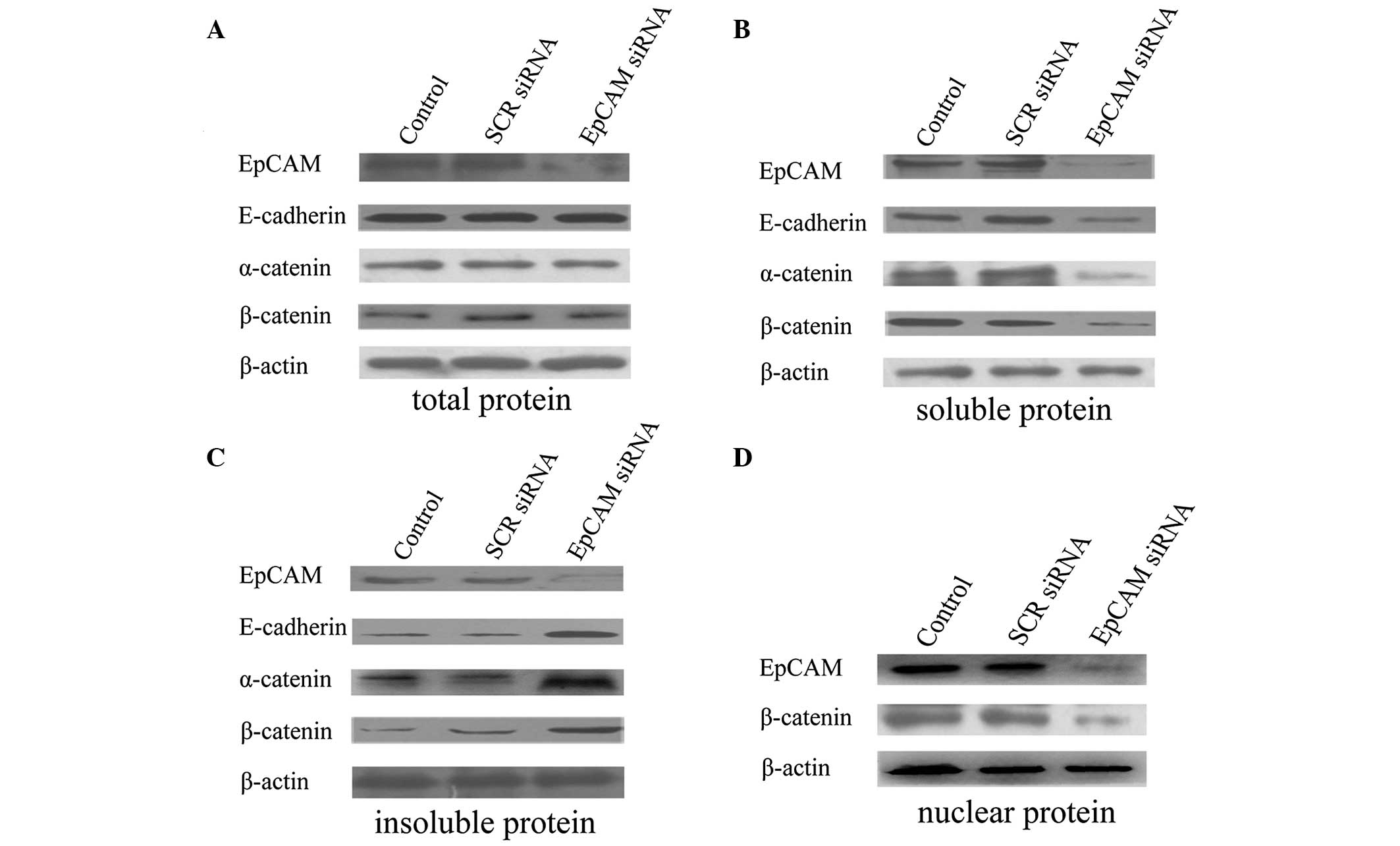
Downregulation of EpCAM expression
increases the E-cadherin, α-catenin and β-catenin expression of the
insoluble protein (cytoskeleton)
To further investigate the mechanism by which the
downregulation of EpCAM expression inhibits the migration and
invasion ability, the expression of EpCAM, E-cadherin, α-catenin,
β-catenin and β-actin at the protein level was analyzed by western
blot analysis. No evident alterations in E-cadherin, α-catenin, and
β-catenin total proteins were observed when compared with the
control and SCR siRNA treatment groups (Fig. 5A). However, the EpCAM siRNA
treatment gave rise to an apparent increase of E-cadherin,
α-catenin and β-catenin in insoluble protein (cytoskeleton;
Fig. 5C) and an apparent decrease
of E-cadherin, α-catenin and β-catenin in soluble protein (no
anchorage to cytoskeleton; Fig.
5B). These results indicated that EpCAM siRNA treatment
possibly enhances the anchorage of E-cadherin, α-catenin and
β-catenin to the cytoskeleton.
Downregulation of EpCAM expression
inhibits β-catenin expression in the nucleus
To further investigate the mechanism by which the
downregulation of EpCAM expression inhibits the proliferation and
tumorigenicity, the expression of β-catenin in the nucleus was
analyzed. The results revealed that the downregulation of EpCAM
decreases the expression of β-catenin in the nucleus (Fig. 5D).
Discussion
Although EpCAM has been well studied as a
cancer-associated antigen, the clinicopathological significance of
EpCAM overexpression in cancer remains unclear. On the one hand,
certain studies have reported that the expression of EpCAM improves
the patient survival rate in specific cancer types, including renal
cell and thyroid cancers (18). On
the other hand, in gastric, breast and tongue cancer, high EpCAM
expression has been identified as an indicator of advanced stage
and poor prognosis (7,19,20).
Various biological explanations may be indicated for these mixed
results, including the possibility that EpCAM could exhibit varying
functions in different organs and affected tissues.
The present study investigated the expression
pattern of EpCAM in hypopharyngeal carcinoma. EpCAM overexpression
was observed in the majority of the hypopharyngeal carcinoma
tissues, whereas it was absent in the normal hypopharyngeal
epithelia. Furthermore, a significant correlation was identified
between high EpCAM expression and advanced tumor size stage or
lymph node metastasis. To further confirm the results obtained by
immunohistochemical staining, a series of in vitro assays
were performed in the hypopharyngeal carcinoma FaDu cell line. The
silencing of EpCAM expression using siRNA was found to suppress the
invasion, migration, proliferation and tumorigenicity of the FaDu
cells. Therefore, the aforementioned observations in the tissues
and cell lines provides compelling evidence that EpCAM expression
promotes hypopharyngeal carcinoma progression and metastasis.
The mechanism by which EpCAM enhances metastasis in
cancer remains unclear. EpCAM is a Ca2+-independent
homophilic CAM that early studies in cells deficient in
intercellular adhesion interactions, such as mouse fibroblast cell
lines, have found to be able to mediate cell aggregation, prevent
cell scattering and also direct cell segregation when introduced
into cells (21). Based on these
adhesive functions, EpCAM was considered to be an inhibitor of
tumor metastasis, which appears to present a paradox. Further
understanding of EpCAM regulation on E-cadherin mediated-adhesion
has clarified this subject. In the epithelium, E-cadherin molecules
function by connecting neighboring cells, thus forming a bridge
between their cortical actin cytoskeleton to maintain mechanical
coupling within the cells and establish intercellular adhesion
(22). Furthermore, the regulation
of the intercellular adhesion is dependent on the recruitment of
α-catenin and β-catenin to the adhesion sites of the adhesion
complexes, and their anchorage to the actin cytoskeleton. Winter
et al (14) revealed that
EpCAM expression inhibits E-cadherin-mediated cell-to-cell adhesion
by disrupting the link between α-catenin and F-actin. In this
manner, EpCAM relaxes the tight intercellular adhesions and
promotes metastasis, differentiation and tissue maintenance. In
addition, Osta et al (7)
reported a similar phenomenon in breast cancer. In the present
study, it was observed that the downregulation of EpCAM improves
the anchorage of E-cadherin, α-catenin and β-catenin to the actin
cytoskeleton (insoluble protein fraction) in FaDu cells, while the
expression of these proteins in the cytoplasm (soluble protein
fraction) was markedly decreased. Thus, we hypothesize that in
hypopharyngeal carcinoma, the downregulation of EpCAM expression
inhibiting tumor invasion and migration may transfer cytoplasmic
E-cadherin, α-catenin and β-catenin anchorage to the actin
cytoskeleton, tightening cell-to-cell adhesion.
However, the mechanism by which EpCAM expression
contributes to proliferation and colony formation is not entirely
understood. Chaves-Pérez et al (23) recently provided evidence that EpCAM
overexpression induces the rapid upregulation of the oncogenes
c-myc and cyclin D1, which induce cellular proliferation. These
transcriptional factors are activated by the nuclear translocation
of β-catenin. De novo expression of EpCAM in HKE293 cells
induces a redistribution of β-catenin from the cytoplasm to the
nucleus (15). Maetzel et al
(15) further investigated the
association between EpCAM and β-catenin, and the cleavage of EpCAM
led to EpCAM EpICD nuclear translocation in a complex with
β-catenin and T-cell factor. Within the nucleus, the EpICD complex
interacts with Lef-1 and contacts DNA to activate the target genes.
In the present study, the downregulation of EpCAM expression was
found to result in the decreased expression of β-catenin in the
nucleus of the FaDu cells, which is consistent with the findings in
the HKE293 cells. This may confirm that the downregulation of EpCAM
leads to a decrease of the complex and β-catenin in the nucleus,
and the subsequent inactivation of c-myc, cyclin D1 and other
target genes, in order to inhibit the cell proliferation and colony
formation ability. Therefore, the aforementioned results may
account for the mechanism of EpCAM expression promoting FaDu cell
proliferation and colony formation.
In conclusion, the results of the present study
demonstrated for the first time that EpCAM is overexpressed in
hypopharyngeal carcinoma and that EpCAM expression is tightly
associated with tumor size and lymph node metastasis. Furthermore,
silenced EpCAM may suppress the invasion, migration, proliferation
and colony abilities of hypopharyngeal carcinoma in vitro.
This study clearly indicates that EpCAM is a promising target for
hypopharyngeal carcinoma therapy.
Acknowledgements
The present study was supported by the Shandong
Provincial International Science and Technology Cooperation Project
of China (grant no. 2010GHZ20202).
References
|
1
|
Clark JI, Hofmeister C, Choudhury A, et
al: Phase II evaluation of paclitaxel in combination with
carboplatin in advanced head and neck carcinoma. Cancer.
92:2334–2340. 2001.
|
|
2
|
Hoffman HT, Porter K, Karnell LH, et al:
Laryngeal cancer in the United States: changes in demographics,
patterns of care, and survival. Laryngoscope. 116(Suppl 111):
S1–S13. 2006.
|
|
3
|
Ma J, Lu S, Yu L, et al: FaDu cell
characteristics induced by multidrug resistance. Oncol Rep.
26:1189–1195. 2011.
|
|
4
|
Baeuerle PA and Gires O: EpCAM (CD326)
finding its role in cancer. Br J Cancer. 96:417–423. 2007.
|
|
5
|
Schnell U, Cirulli V and Giepmans BN:
EpCAM: structure and function in health and disease. Biochim
Biophys Acta. 1828:1989–2001. 2013.
|
|
6
|
Went P, Vasei M, Bubendorf L, et al:
Frequent high-level expression of the immunotherapeutic target
Ep-CAM in colon, stomach, prostate and lung cancers. Br J Cancer.
94:128–135. 2006.
|
|
7
|
Osta WA, Chen Y, Mikhitarian K, et al:
EpCAM is overexpressed in breast cancer and is a potential target
for breast cancer gene therapy. Cancer Res. 64:5818–5824. 2004.
|
|
8
|
Flatmark K, Borgen E, Nesland JM, et al:
Disseminated tumour cells as a prognostic biomarker in colorectal
cancer. Br J Cancer. 104:1434–1439. 2011.
|
|
9
|
Ni J, Cozzi PJ, Duan W, et al: Role of the
EpCAM (CD326) in prostate cancer metastasis and progression. Cancer
Metastasis Rev. 31:779–791. 2012.
|
|
10
|
Ji J, Yamashita T, Budhu A, et al:
Identification of microRNA-181 by genome-wide screening as a
critical player in EpCAM-positive hepatic cancer stem cells.
Hepatology. 50:472–480. 2009.
|
|
11
|
Pauli C, Münz M, Kieu C, et al:
Tumor-specific glycosylation of the carcinoma-associated epithelial
cell adhesion molecule EpCAM in head and neck carcinomas. Cancer
Lett. 193:25–32. 2003.
|
|
12
|
Litvinov SV, Velders MP, Bakker HA,
Fleuren GJ and Warnaar SO: Ep-CAM: a human epithelial antigen is a
homophilic cell-cell adhesion molecule. J Cell Biol. 125:437–446.
1994.
|
|
13
|
Balzar M, Prins FA, Bakker HA, Fleuren GJ,
Warnaar SO and Litvinov SV: The structural analysis of adhesions
mediated by Ep-CAM. Exp Cell Res. 246:108–121. 1999.
|
|
14
|
Winter MJ, Nagelkerken B, Mertens AE,
Rees-Bakker HA, Briaire-de Bruijn IH and Litvinov SV: Expression of
Ep-CAM shifts the state of cadherin-mediated adhesions from strong
to weak. Exp Cell Res. 285:50–58. 2003.
|
|
15
|
Maetzel D, Denzel S, Mack B, et al:
Nuclear signalling by tumour-associated antigen EpCAM. Nat Cell
Biol. 11:162–171. 2009.
|
|
16
|
Visvader JE and Lindeman GJ: Cancer stem
cells in solid tumours: accumulating evidence and unresolved
questions. Nat Rev Cancer. 8:755–768. 2008.
|
|
17
|
Gires O, Klein CA and Baeuerle PA: On the
abundance of EpCAM on cancer stem cells. Nat Rev Cancer.
9:1432009.
|
|
18
|
van der Gun BT, Melchers LJ, Ruiters MH,
de Leij LF, McLaughlin PM and Rots MG: EpCAM in carcinogenesis: the
good, the bad or the ugly. Carcinogenesis. 31:1913–1921. 2010.
|
|
19
|
Kroepil F, Dulian A, Vallböhmer D, et al:
High EpCAM expression is linked to proliferation and lauren
classification in gastric cancer. BMC Res Notes. 6:2532013.
|
|
20
|
Yanamoto S, Kawasaki G, Yoshitomi I,
Iwamoto T, Hirata K and Mizuno A: Clinicopathologic significance of
EpCAM expression in squamous cell carcinoma of the tongue and its
possibility as a potential target for tongue cancer gene therapy.
Oral Oncol. 43:869–877. 2007.
|
|
21
|
Litvinov SV, Bakker HA, Gourevitch MM,
Velders MP and Warnaar SO: Evidence for a role of the epithelial
glycoprotein 40 (Ep-CAM) in epithelial cell-cell adhesion. Cell
Adhes Commun. 2:417–428. 1994.
|
|
22
|
Dufour S, Mège RM and Thiery JP:
α-catenin, vinculin, and F-actin in strengthening E-cadherin
cell-cell adhesions and mechanosensing. Cell Adh Migr. 7:345–350.
2013.
|
|
23
|
Chaves-Pérez A, Mack B, Maetzel D, et al:
EpCAM regulates cell cycle progression via control of cyclin D1
expression. Oncogene. 32:641–650. 2013.
|