Introduction
Altered glycosylation has been reported in various
types of cancer and may have a role in cancer metastasis and
progression (1–3). Lectins are carbohydrate-binding
proteins that contain at least one non-catalytic domain that binds
reversibly with mono- or oligosaccharides with high specificity
(4). Therefore, lectins may be
useful tools for analyzing glycofiles and may be used as biomarkers
for a variety of types of cancer, including aggressive breast
(5,6), ovarian (7), pancreatic (8), prostate (9) and liver (10) cancer. The monocot mannose-binding
lectin, Pinellia pedatisecta agglutinin (PPA), accumulates
in the tuber of P. pedatisecta, a species of the Araceae
family. In our previous studies, recombinant PPA was used in
labeling fractions of myeloid leukemia cells (11) and was found to preferentially
recognize drug-resistant cancer cells (12). The binding of drug-resistant
K562/ADR leukemia cells with PPA enhanced the macrophage-induced
phagocytosis of the K562/ADR cells, and the target of PPA on the
K562/ADR cells was determined to be sarcolemmal membrane-associated
protein (12). Furthermore, the
exogenous expression of PPA using gene delivery has been found to
induce cancer cell death through interacting with the methylosome,
which contains methylosome protein 50 and protein arginine
methyltransferase 5 (13). These
studies indicate that PPA may be further developed to analyze
glycosylation profiles in different types of cancer.
Lectin blot analysis is a biochemical technique that
is similar to western blot analysis, in which tagged lectins are
used as probes to detect glycosylation profiles in biological
samples (14), including cancer
plasma (15), as well as sera and
tissue samples (16). In the
present study, a novel PPA-based lectin blot analysis technique was
developed using soluble Coxsackie-adenovirus receptor-PPA domain b
fusion protein (sCAR-PPAb) as a probe. PPA-based lectin blot
analysis detected typical glycofiles for various cancer cell lines,
including leukemia and solid tumor cell lines.
Materials and methods
Cells
All cell lines were obtained from the American Type
Culture Collection (Rockville, MD, USA). The HL-60 and Kasumi-1
human acute myeloid leukemia cell lines were maintained in
RPMI-1640 medium (Hyclone Laboratories, Logan, UT, USA)
supplemented with 10% fetal bovine serum (Life Technologies, Inc.,
Grand Island, NY, USA) and 1% L-glutamine (Life Technologies,
Inc.). The PLC, BEL-7404 and Huh7 human liver cancer cell lines and
the H1299 human lung cancer cell line were maintained in Dulbecco’s
modified Eagle’s medium (Hyclone Laboratories) supplemented with
10% fetal bovine serum and 1% L-glutamine.
Production and purification of sCAR-PPAb
protein
The construction of the pQE30-sCAR-PPA plasmid has
been reported previously (11). In
the present study, a PPA domain b fragment
(D146-S250) was used to replace the PPA
full-length fragment. The pQE30-sCAR-PPAb plasmid was transformed
into Escherichia coli strain M15, and the expression of
sCAR-PPAb was induced using isopropyl β-D-1-thiogalactopyranoside.
Inclusion bodies were subjected to protein purification using a
wash method. In brief, inclusion bodies were washed three times
with 4 ml wash buffer containing 1.46 mg/ml EDTA, 0.01 M Tris-HCl
(pH 8.0), 8 M urea and 0.7% β-mercaptoethanol (all Sigma-Aldrich,
St. Louis, MO, USA) for 12 h each. Following each wash,
supernatants were collected using centrifugation at 15,984 rpm for
15 min and analyzed using SDS-PAGE followed by Coomassie Brilliant
Blue (Sigma-Aldrich) staining. The purified proteins were then
renatured through dialyzing against phosphate-buffered saline at
4°C overnight. The production of sCAR-PPAb was then assessed using
western blot analysis with a mouse anti-6 histidine (6his)
monoclonal antibody (Santa Cruz Biotechnology, Inc., Santa Cruz,
CA, USA) and the IRDye® 800 donkey anti-mouse
immunoglobulin (Ig) G secondary antibody (Li-Cor, Inc., Lincoln,
NA, USA). Bands were analyzed using an Odyssey® Infrared
Imaging System (Li-Cor, Inc.).
PPA-based lectin blot analysis
Cell lysates were subjected to SDS-PAGE and
electroblotted onto nitrocellulose membranes. The membranes were
then blocked using Tris-buffered saline-Tween 20 (TBS-T) containing
5% bovine serum albumin at room temperature for 2 h, followed by
incubation with 1.5 μg/ml sCAR-PPAb at 4°C overnight. Membranes
incubated without sCAR-PPAb were used as the controls. The
membranes were then washed with TBS-T three times and incubated
with a mouse anti-6his monoclonal antibody (Santa Cruz
Biotechnology, Inc.) at 4°C overnight. The membranes were
subsequently washed and incubated with IRDye 800 donkey anti-mouse
IgG (Li-Cor, Inc.) secondary antibody for 1 h at room temperature
followed by analysis using an Odyssey Infrared Imaging System
(Li-Cor, Inc.).
Results
Production and purification of the
sCAR-PPAb fusion protein
The sCAR-PPAb protein contains a 6his-tag, a human
sCAR, a short flexible linker and a PPA domain b. In the present
study, a bacterial expression system was used to produce sCAR-PPAb.
The recombinant fusion protein was expressed as inclusion bodies in
E. coli M15 and purified using a three-step wash method.
Purification was verified using SDS-PAGE followed by Coomassie
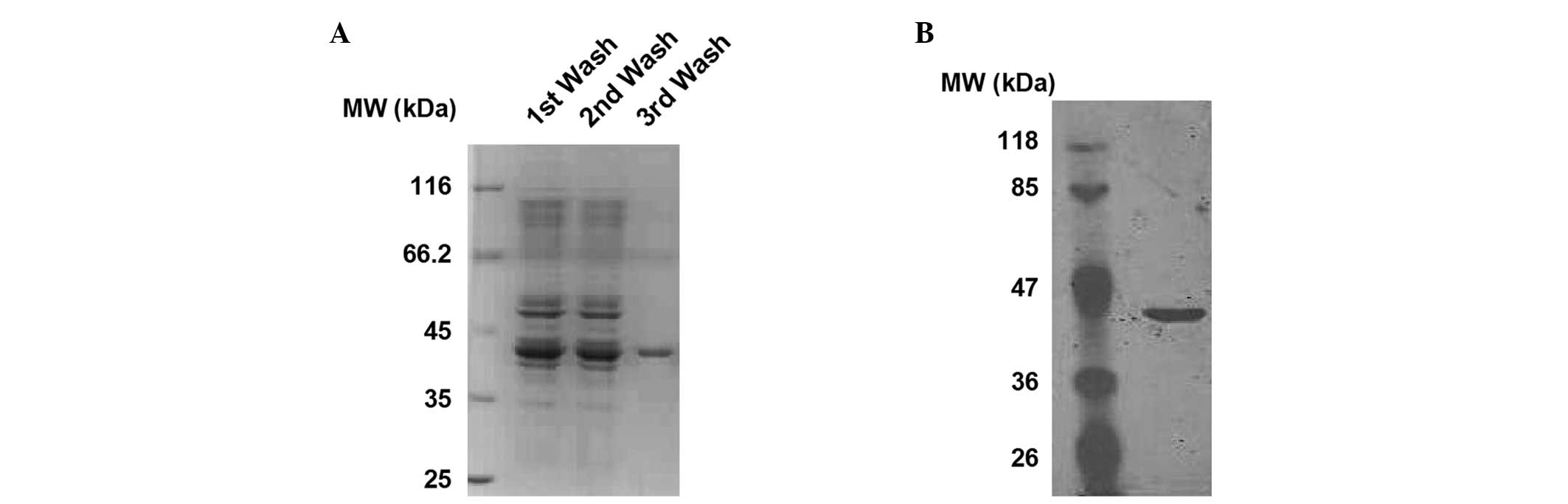
Brilliant Blue staining. As shown in Fig. 1A, a relatively pure protein, with a
molecular weight of ~42 kDa, was obtained subsequent to the third
wash. To further confirm the identity of the protein, western blot
analysis was performed using an anti-6his antibody. As shown in
Fig. 1B, the presence of the
6his-tag was verified. These findings show that the sCAR-PPAb
fusion protein was successfully expressed and purified.
PPA-based lectin blot analysis
distinguishes typical glycofiles in various types of cancer
cell
A lectin blot was generated using sCAR-PPAb as the
primary probe to detect the glycosylation pattern for a variety of
cancer cells, including leukemia and solid tumor cell lines. As
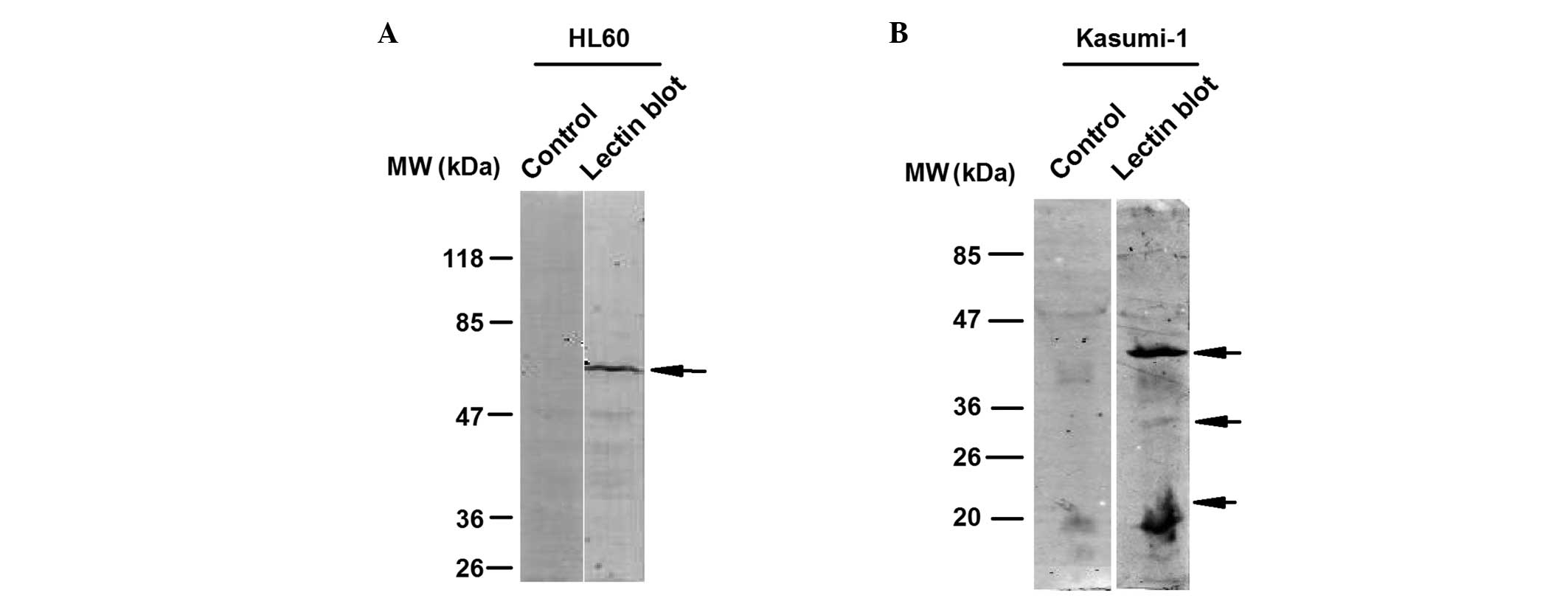
shown in Fig. 2A, the HL60 leukemia
cell line was subjected to PPA-based lectin blot analysis and a
typical band with a molecular weight of between 47 and 85 kDa was
detected. Kasumi-1 leukemia cells were also subjected to PPA-based
lectin blot analysis, in order to analyze whether other leukemia
cells exhibited different detection patterns. Lectin blot analysis
revealed three typical bands with various molecular weights in the
Kasumi-1 cells (Fig. 2B), which
were different to those of the HL60 cells, indicating that
PPA-based lectin blot analysis is capable of distinguishing between
glycosylation patterns in different leukemia cell lines.
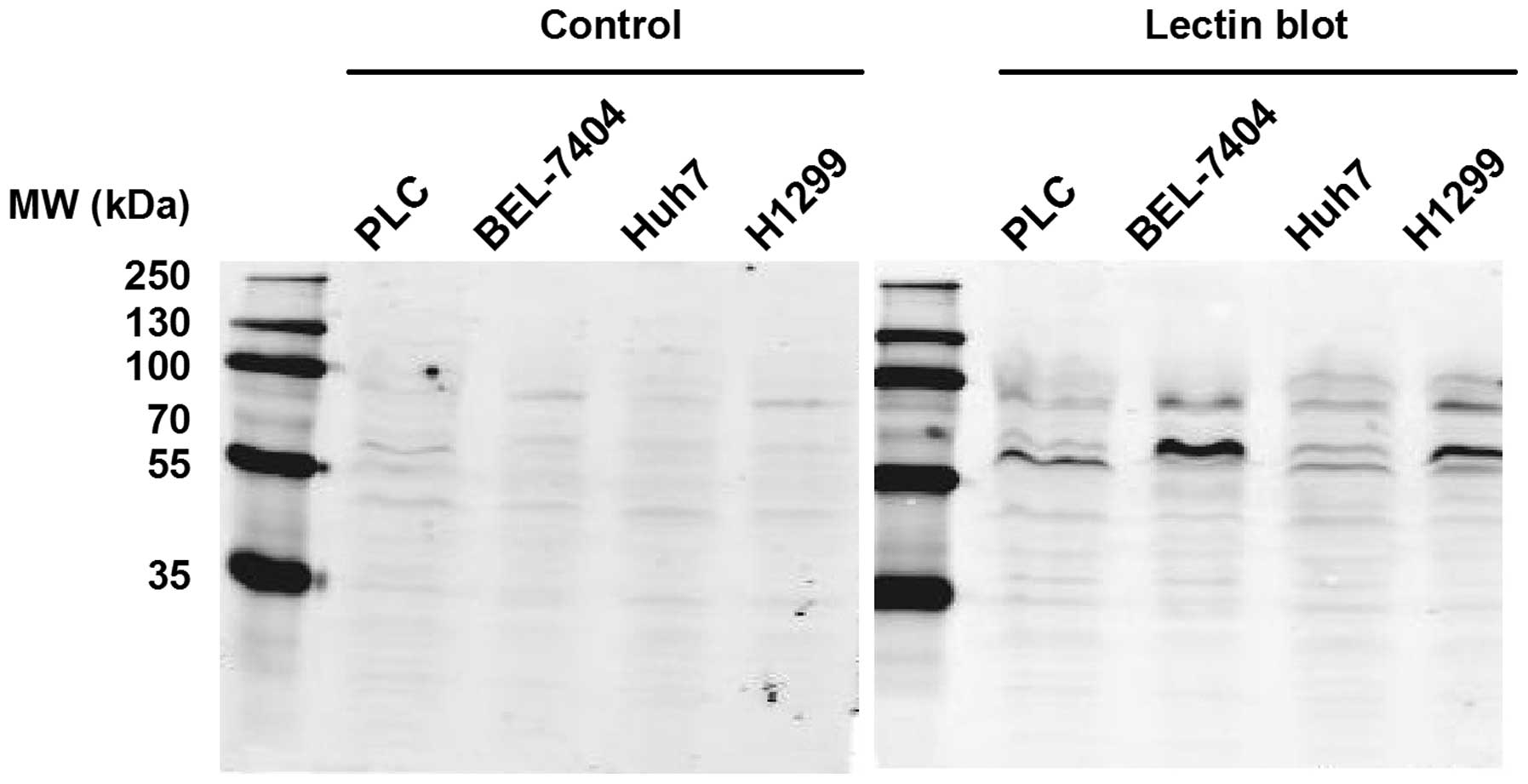
The solid tumor cell lines, including the PLC,
BEL-7404 and Huh7 liver cancer cell lines and the H1299 lung cancer
cell line, were further analyzed using PPA-based lectin blot
analysis. As shown in Fig. 3, these
four cell lines exhibited different glycosylation patterns and the
bands were primarily between 55 and 100 kDa. These findings
indicate that PPA-based lectin blot analysis could be used to
distinguish between glycosylation patterns in various cancer cell
types, including types of blood cancer and solid tumors.
Discussion
Due to their oligosaccharide specificity, lectins
have been used in a variety of biological techniques, including
lectin array, lectin blot analysis and lectin-based chromatography
(17). In previous studies, lectin
blot analysis has been used to detect the glycosylation of
proteins, including an Aspergillus oryzae lectin blot for
probing N-glycans containing core fucose (18), an Aleuria aurantia lectin
blot for detecting the fucosylation of β-haptoglobin and for
providing biomarkers for colon cancer (19), and a phytohemagglutinin lectin blot
for analyzing changes in N-glycan patterns for integrins, which is
involved in epithelial to mesenchymal transition of epithelial
cells (20). All these lectin blot
analysis techniques were mainly used for detecting the
glycosylation of specific molecules. However, unlike the lectin
blots used in these previous studies, the PPA-based lectin blot
analysis technique used in the present study successfully
recognized typical glycosylation patterns in various cancer cell
lines and the glycofiles were found to differ significantly among
these cell lines. The glycofiles detected using PPA-based lectin
blot analysis may provide a ‘glycosylation fingerprint’ for a
variety of cancer cells and may be valuable for cancer prognosis
and diagnosis.
Acknowledgements
The present study was supported by the College
Students Science and Technology Innovation project (Xin Miao Talent
Project) of Zhejiang Province.
References
|
1
|
Mattaini KR and Vander Heiden MG: Cancer.
Glycosylation to adapt to stress. Science. 337:925–926. 2012.
|
|
2
|
Kim YS, Ahn YH, Song KJ, et al:
Overexpression and β-1,6-N-acetylglucosaminylation-initiated
aberrant glycosylation of TIMP-1: a ‘double whammy’ strategy in
colon cancer progression. J Biol Chem. 287:32467–32478. 2012.
|
|
3
|
Saeland E, Belo AI, Mongera S, et al:
Differential glycosylation of MUC1 and CEACAM5 between normal
mucosa and tumour tissue of colon cancer patients. Int J Cancer.
131:117–128. 2012.
|
|
4
|
Sharon N and Lis H: Lectins as cell
recognition molecules. Science. 246:227–234. 1989.
|
|
5
|
Fry SA, Afrough B, Lomax-Browne HJ, et al:
Lectin microarray profiling of metastatic breast cancers.
Glycobiology. 21:1060–1070. 2011.
|
|
6
|
Drake PM, Schilling B, Niles RK, et al:
Lectin chromatography/mass spectrometry discovery workflow
identifies putative biomarkers of aggressive breast cancers. J
Proteome Res. 11:2508–2520. 2012.
|
|
7
|
Wu J, Xie X, Liu Y, et al: Identification
and confirmation of differentially expressed fucosylated
glycoproteins in the serum of ovarian cancer patients using a
lectin array and LC-MS/MS. J Proteome Res. 11:4541–4552. 2012.
|
|
8
|
Li C, Simeone DM, Brenner DE, et al:
Pancreatic cancer serum detection using a lectin/glyco-antibody
array method. J Proteome Res. 8:483–492. 2009.
|
|
9
|
Batabyal SK, Majhi R and Basu PS: Clinical
utility of the interaction between lectin and serum prostate
specific antigen in prostate cancer. Neoplasma. 56:68–71. 2009.
|
|
10
|
Ahn YH, Shin PM, Oh NR, et al: A
lectin-coupled, targeted proteomic mass spectrometry (MRM MS)
platform for identification of multiple liver cancer biomarkers in
human plasma. J Proteomics. 75:5507–5515. 2012.
|
|
11
|
Li GC, Li N, Zhang YH, et al:
Mannose-exposing myeloid leukemia cells detected by the sCAR-PPA
fusion protein. Int J Hematol. 89:611–617. 2009.
|
|
12
|
Chen K, Yang X, Wu L, et al: Pinellia
pedatisecta agglutinin targets drug resistant K562/ADR leukemia
cells through binding with sarcolemmal membrane associated protein
and enhancing macrophage phagocytosis. PLoS One. 8:e743632013.
|
|
13
|
Lu Q, Li N, Luo J, et al: Pinellia
pedatisecta agglutinin interacts with the methylosome and
induces cancer cell death. Oncogenesis. 1:e292012.
|
|
14
|
Cao J, Guo S, Arai K, Lo EH and Ning M:
Studying extracellular signaling utilizing a glycoproteomic
approach: lectin blot surveys, a first and important step. Methods
Mol Biol. 1013:227–233. 2013.
|
|
15
|
Qiu Y, Patwa TH, Xu L, et al: Plasma
glycoprotein profiling for colorectal cancer biomarker
identification by lectin glycoarray and lectin blot. J Proteome
Res. 7:1693–1703. 2008.
|
|
16
|
Ferguson RE, Jackson DH, Hutson R, et al:
Detection of glycosylation changes in serum and tissue proteins in
cancer by lectin blotting. Adv Exp Med Biol. 564:113–114. 2005.
|
|
17
|
Clark D and Mao L: Cancer biomarker
discovery: lectin-based strategies targeting glycoproteins. Dis
Markers. 33:1–10. 2012.
|
|
18
|
Mun JY, Lee KJ, Kim YJ, et al: Development
of fluorescent probes for the detection of fucosylated N-glycans
using an Aspergillus oryzae lectin. Appl Microbiol
Biotechnol. 93:251–260. 2012.
|
|
19
|
Park SY, Lee SH, Kawasaki N, et al: α1-3/4
fucosylation at Asn 241 of β-haptoglobin is a novel marker for
colon cancer: a combinatorial approach for development of glycan
biomarkers. Int J Cancer. 130:2366–2376. 2012.
|
|
20
|
Xu Q, Isaji T, Lu Y, et al: Roles of
N-acetylglucosaminyltransferase III in epithelial-to-mesenchymal
transition induced by transforming growth factor β1 (TGF-β1) in
epithelial cell lines. J Biol Chem. 287:16563–16574. 2012.
|