Introduction
Acute promyelocytic leukemia (APL) is characterized
by particular clinical features that are important in
differentiating it from other acute myeloid leukemias (AMLs) and
determining an accurate diagnosis. These clinical features include
unique hemorrhagic syndrome, disseminated intravascular coagulation
(DIC) and association with the translocation between chromosomes 15
and 17, resulting in the formation of two reciprocal fusion genes;
promyelocytic leukemia (PML)/retinoic acid α-receptor
(RARA) on chromosome 15 and RARA/PML on chromosome
17. These fusion genes are sensitive to retinoid differentiating
agents, such as all-trans retinoic acid (ATRA), and novel
antiapoptotic agents, including arsenic trioxide (1,2).
Although t(15;17) has been found in ~90% of APL
patients, variant translocations have been reported in a few APL
patients, which are described as simple translocations involving
chromosome 15 or 17 with any other chromosomes [t(15;v) or t(17;v)]
or complex translocations characterized by the involvement of
additional chromosome(s) in addition to chromosomes 15 and 17
[ct(15;17;v)] (3–5). In the last few years, a number of
studies have focused on the alternate translocation in APL, for
example t(5;17)(q35;q21) forming NPM/RARA, t(11;17)(q23;q21)
producing PLF/RARA fusion and t(11;17)(q13;q21) generating
NUMA/RARA, providing advanced insights into the pathogenesis
of APL (3,6,7).
However, little is known concerning the complex variant
translocations in APL. The current study reports a patient who
presented with the classical t(15;17) and complex variant
t(15;16;17)(q22;q24;q21), which were demonstrated by traditional
cytogenetic analysis, including G-banding karyotype and
fluorescence in situ hybridization (FISH). In addition,
cryptic losses on 3p25.3, 8q23.1 and 12p13.2-p13.1, and a gain of
chromosome 8q11.2, were revealed in the level of array comparative
genomic hybridization (CGH). This study was approved by the
Institutional Review Board (IRB) at the University of Oklahoma
Health Sciences Center (IRB no. 13100; Oklahoma City, OK, USA). The
patient provided consent.
Case report
Clinical presentation
The current study presents a 54-year-old female with
APL who was admitted to the University of Oklahoma Health Sciences
Center due to fever and bleeding gums. The peripheral blood
examination showed a hemoglobin count of 7.1 g/dl (normal range,
12–15.5 g/dl) and platelet count of 35×103/μl (normal
range, 150–450×103/μl), as well as a white blood cell
count of 11.95×103/μl (normal range,
3.5–10.5×103/μl) with 86% blasts, characterized by small
to large cells with irregular, lobulated and bilobed nuclei with
prominent nucleoli. The blast cells in the peripheral blood
exhibited scant to moderate rare blue granulated cytoplasm and the
bone marrow aspirate revealed no cellular particles. The touch
preparations showed suboptimal cellular morphology, but revealed
numerous blasts that were more frequently granulated than those
present in the peripheral smear. The core biopsy of the bone marrow
revealed a cellularity of >95% consisting of sheets of immature
cells. A diagnosis of APL, M3 variant was determined according to
the French-American-British Cooperative Group criteria (8).
Cytogenetics, FISH and array CGH
analyses
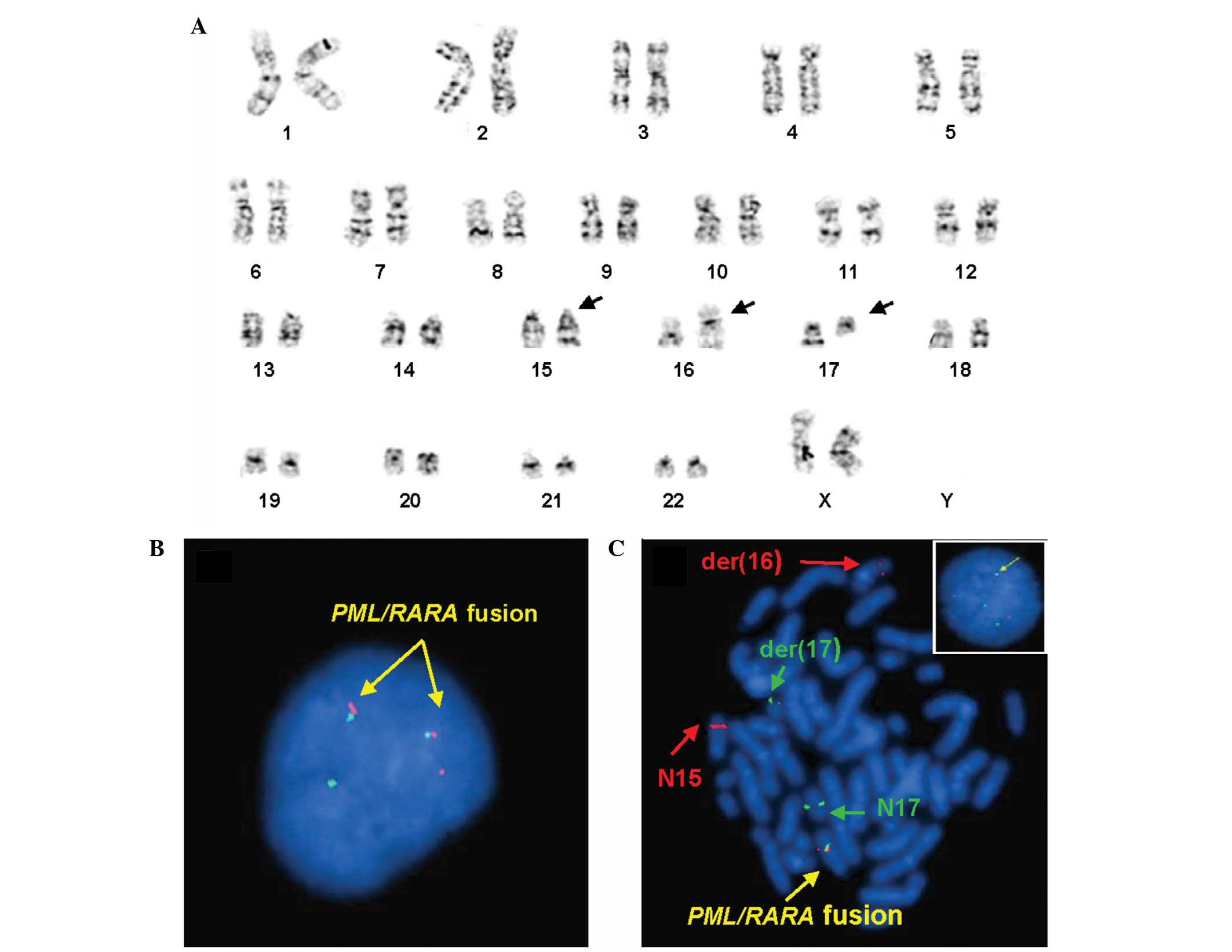
The karyotype analysis at diagnosis revealed that
the 20 metaphases analyzed exhibited a variant translocation among
three chromosome; chromosomes 15, 16 and 17. The karyotype was
designated as 46,XX,t(15;16;17)(q24;q24;q21)[20] (Fig. 1A). FISH analyses using the LSI
PML/RARA dual color and dual fusion probe (Abbott Molecular, Inc.,
Des Plaines, IL, USA) was applied to the uncultured and cultured
cells. On uncultured interphase cells, 12 out of 200 cells (6%)
exhibited two classical PML/RARA fusion signals, one
SpectrumOrange and one SpectrumGreen signal, which were the result
of the classical t(15;17) (Fig. 1B)
(http://www.abbottmolecular.com/us/siteMap.html#sthash.483IvlrV.dpuf).
While 168 out of 200 cells (84%) exhibited one fusion signal, two
SpectrumOrange signals and two SpectrumGreen signals, which were
the result of the complex variant translocation, t(15;16;17)
(Fig. 1C) (9). By combining the karyotyping and FISH
results, it was demonstrated on metaphase cells that a part of the
PML gene labeled with SpectrumOrange was present on the derivative
chromosome 16 and a part of the RARA gene labeled with
SpectrumGreen signal was present on the derivative chromosome 17
(Fig. 1C). Subsequent
cohybridization of whole chromosome painting probes (WCP) 15 and 16
(Cytocell, Ltd., Cambridge, UK) and cohybridization of WCP17
(Abbott Molecular, Inc.), as well as the subtelomeric probes for
chromosome 16p and q (Abbott Molecular, Inc.) on metaphase cells,
confirmed the complex translocations among chromosomes 15, 16 and
17 (Fig. 2A and B) (http://www.abbottmolecular.com/us/siteMap.html#sthash.483IvlrV.dpuf)
(10).
Array CGH analysis showed the acquired genomic
aberrations. No loss or gain was identified on chromosomes 15, 16,
or 17, but cryptic losses of genomic material on 3p25.3, 8q23.1 and
12p13.1-p13.2, and a cryptic gain of 8q11.23 were detected
(Fig. 3A–D). Several cancer-related
genes were located in these genomic imbalanced regions. A loss of
3p25.3 (10,364,050–10,670,236 bp hg18; ~0.3 Mb) encompasses the
plasma membrane Ca2+-ATPase 2 (ATP2B2) gene, a
loss of 8q23.1 (107,811,331–108,868,110 bp hg18; ~1.1 Mb) contains
the angiopoietin 1 (ANGPT1) gene, a loss of 12p13.2-p13.1
(11,619,439–14,472,130 bp hg18; ~2.9 Mb) involves ~23 genes,
including the ETS variant 6 (ETV6) gene which is highly
associated with t(12;21) in acute lymphoblastic leukemia, and a
gain of 8q11.23 (53,762,025–53,876,752 bp hg18; ~0.1 Mb) contains
the tumor suppressor gene retinoblastoma 1 inducible coiled-coil 1
(RB1CC1). Subsequent FISH confirmed the array CGH
observations. The cultured cells were hybridized with home-brew
probes using bacterial artificial chromosome clones, RP11-533J10
labeled with SpectrumOrange on 3p25.3 and RP11-79F7 labeled with
SpectrumGreen on 8q23.1 (Invitrogen Life Technologies, Carlsbad,
CA, USA), and 194 out of 200 cells (97%) presented with one
SpectrumOrange signal and 104 out of 200 cells (52%) showed one
SpectrumGreen signal, demonstrating the deletions of 3p25.3 and
8q23.1, respectively (Fig. 3E). The
percentage of the cells with deletions was close to the percentage
of cells with t(15;17) combined with t(15;16;17). These deletions
are likely to be somatically acquired and not constitutional
changes. In addition, hybridization with the LSI ETV6/AML1 dual
color ES probe (Abbott Molecular, Inc.) confirmed the deletion of
12p13.1-p13.2 involving the ETV6 genes (Fig. 3F) and 116 out of 200 (58%) cells
were observed with deletion of the ETV6 gene. However, gain
of 8q11.23 was not be confirmed by FISH due to the small size (only
100 bp) of the duplication.
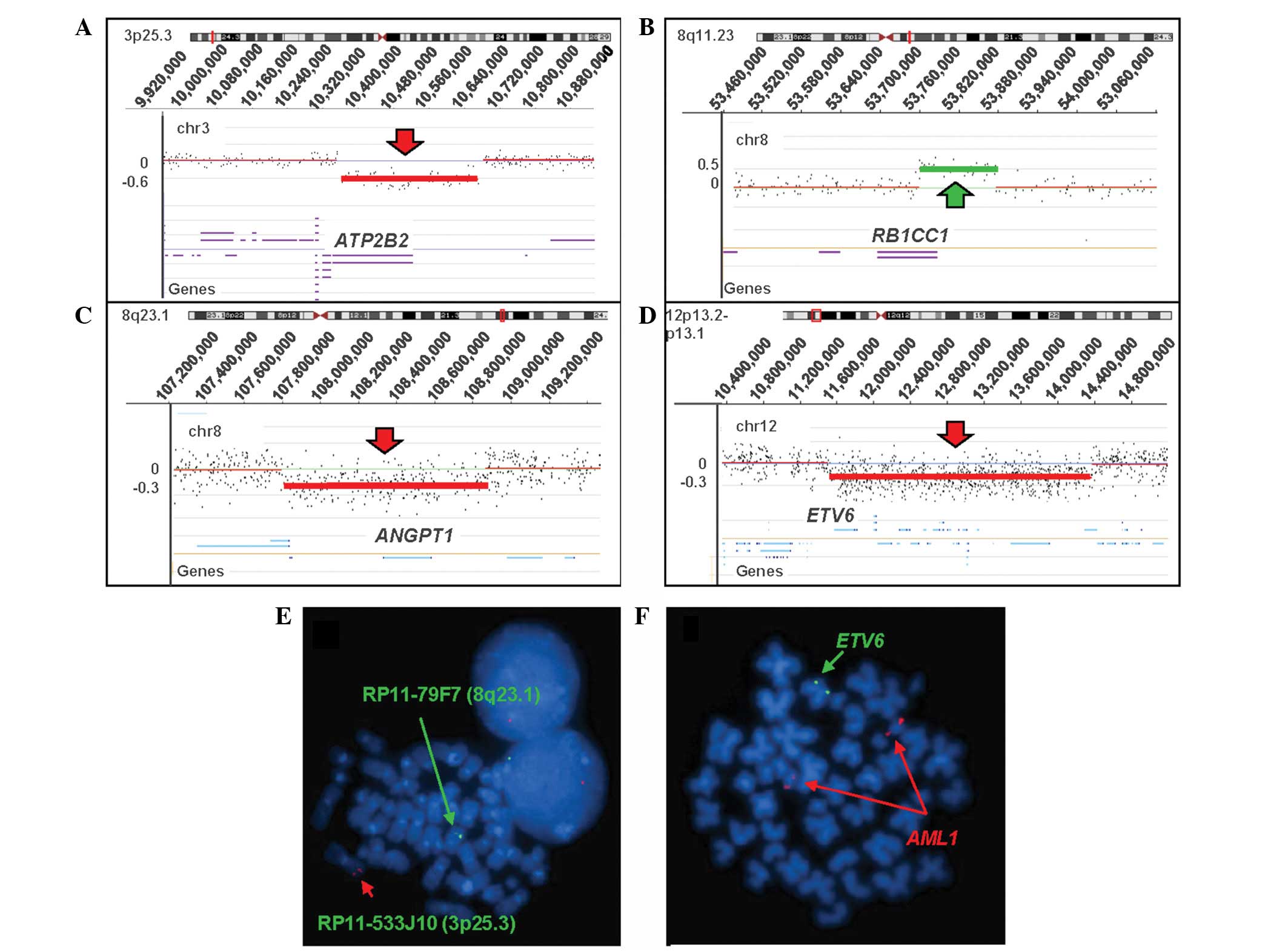 | Figure 3Results of the oligoarray CGH using
NimbleGen SegMNT and the RefSeq genes in the abnormal region
(University of California, Santa Cruz genome browser hg18). The
Y-axis indicates a gain or loss of genetic material, while the
X-axis indicates the genomic position of each feature on the
chromosome. The red arrow indicates loss and the green arrow
indicates gain and the significant gene in the lost or gained
region is listed. (A) Loss of 3p25.3 (10,364,050–10,670,236 bp,
hg18; ~0.3 Mb). (B) Gain of 8q11.23 (53,762,025–53,876,752bp, hg18;
~0.1 Mb), which is potentially constitutional rather than a
disease-related duplication. (C) Loss of 8q23.1
(107,811,331–108,868,110 bp, hg 18; ~1.1 Mb). (D) Loss of
12p13.2-p13.1 (11,619,439–14,472,130 bp, hg18; ~2.9 Mb).
Fluorescence in situ hybridization analyses using bacterial
artificial chromosome RP11-533J10 (3p25.3; SpectrumOrange),
RP11-79F7 (8p23.1; SpectrumGreen) and LSI ETV6 (SpectrumGreen)
confirmed the array CGH results of the losses of (E) 3p25.3 and
8q23.1, and (F) ETV6 gene. CGH, comparative genomic hybridization;
ETV6, ETS variant 6; AML1, acute myeloid leukemia-1. |
Discussion
It is rare that complex variant translocations
between chromosomes 15 and 17 [ct(15;17;v)] involving more than
three chromosomes in APL result in the PML/RARA fusion gene. To the
best of our knowledge, 45 cases have been reported (5,6,9,11–40),
which account for 10% of APLs lacking classical t(15;17) (6). The clinical characterizations of the
reported cases of APL with ct(15;17;v), including the present
study, are summarized in Table I.
No distinct clinical features have been observed in APL with
ct(15;17;v) compared with APL with classical t(15;17). Generally,
the ratio between males and females is 1:1, the median age at
diagnosis is 45 years and 77% of ct(15;17;v) cases are accompanied
with DIC. This is similar to the DIC ratio of cases with classical
t(15;17). All observed ct(15;17;v) cases exhibited good responses
to ATRA. Among these ct(15;17;v), nine recurrent breakpoints on the
third or fourth chromosome consisted of the ct(15;17;v), including
1p36, 2q21, 3p21, 4q21, 11q13, 18q12, 20p13, 22q11.2 and Xq13. The
current study presents the third case of APL with ct(15;17;v)
involving chromosome 16. The first was described in a human
immunodeficiency virus patient with secondary APL and the second
was described in an APL patient with four chromosome translocations
involving chromosomes 5, 15, 16 and 17 (23,29).
However, the breakpoint of chromosome 16 in the current case
(16q24) was different from those identified in the two previously
reported cases. The breakpoint 16q24 has been described in AML with
t(16;21)(q24;q22), which leads to the fusion gene
RUNX1/MTG16 and is predominantly associated with
therapy-related AML (41). Whether
the MTG16 gene is located at the breakpoint and forms a
fusion gene in the present case requires clarification.
 | Table IClinical characterization of reported
acute promyelocytic leukemia cases with variant t(15;17). |
Table I
Clinical characterization of reported
acute promyelocytic leukemia cases with variant t(15;17).
| Author (ref.) | Cytogenetic
abnormality | Gender | Age, years | FAB | DIC | Cell clones | Treatment | Relapse | Survival, months |
|---|
| Berger et al
(11) |
t(15;17;4)(q22;q12;q21) | M | NA | M3 | NA | 46,XY,
t(15;17;4)(q22;q12;q21) | CNA | NA | |
| Bernstein et
al (12) |
t(2;15;17)(q21?;q25?6?;q21?2?) | F | 23 | M3v | ± | 46,XX
46,XX,t(2;15;17)( q21?;q25?6?;q21?2?) 9p+ | C + | 6.5 | |
| Bernstein et
al (13) |
t(3;15;17)(p21;q25?6?;q21?2?) | M | 28 | M3 | + | 46,XY
46,XY,t(3;15;17)( p21;q25?6?;q21?2?) 4p+ | CNR | 0.2 | |
| Ohyashiki et
al (14) |
t(1;5;15;17)(p36;q31;q22;q12) | F | 34 | M3 | + | 46,XX
47,XX,+X, t(1;5;15;17)(p36;q31;q22;q12) | CNR | 1 | |
| Callen et al
(15) |
t(X;15;17)(q13;q21;q21) | F | 48 | M3v? | − | 46,XX/46,XX,
t(X;15;17)(q13;q21;q21)
Relapse: 46,XX, t(X;15;17)(q13;q21;q21) | C + | 13 | |
| Bjerrum et al
(16) |
t(2;17;15)(q21;q21;q22or23or24) | M | 10 | M3 | ± | 46,XY,
t(2;17;15)(q21;q21;q22or23or24)
46,XY,t(15;17)(q22or23or24;q21) | C + | 34 | |
| | | | | | 46,XY,
t(2;17;15)(q21;q21;q22or23or24),8p+ | A − | 10+ | |
| Osella et al
(17) |
t(1;15;17)(p36;q22;q21.1) | M | 46 | M3 | + | 46,XY
46,XY, t(1;15;17)(p36;q22;q21.1)
46,XY, t(1;15;17)(p36;q22;q21.1),+8 | | | |
| Saitoh et al
(18) |
t(15;19;17)(q22;p13;q12) | F | 16 | M3 | NA | 46,XX
46,XX, t(15;19;17)(q22;p13;q12) | A − | 7+ | |
| Wan et al
(19) |
t(X;17;15)(q13;q12;q21) | F | 30 | M3 | + | 46,XX,
t(X;17;15)(q13;q12;q21) | C − | 2+ | |
| Fujishima et
al (20) |
t(2;15;17)(q21;q22;q21) | M | 77 | M3 | + | 46,XY,
t(2;15;17)(q21;q22;q21) | C+A | − | 12+ |
| Liu et al
(21) |
t(4;15;17)(q21;q22;q21) | M | 37 | M3 | + | 46,XY
46,XY, t(4;15;17)(q21;q22;q21) |
As2O3 | − | 10+ |
| Misawa et al
(22) |
t(7;17;15)(p22;q21.1orq12;q22) | M | 35 | M3 | + | 46,XY,
t(7;17;15)(p22;q21.1orq12;q22) | C − | 10+ | |
| Zaccaria et
al (23) |
t(13;15;17;20)(q22;q22;q12;p13) | F | 74 | M3 | + | 46,XX
47,XX,+8, t(1;5;15;17)(p36;q31;q22;q12) | ANR | 0.2 | |
|
t(5;15;16;17)(q22;q22;p13;q12) | M | 45 | M3 | + | 46,XY
47,XY, t(5;15;16;17)(q22;q22;p13;q12) | C+A | + | 24 |
| Ogawa et al
(24) |
t(15;17;18)(q21q22;q12;q12) | M | 53 | M3 | + |
46,XY,del(15)(q21q22),der(17)t(17;18)(q12;q12),
der(18)t(17;18)(q12;q12)ins(18;15)(q12;q21q22) | C − | 3+ | |
| Chen et al
(25) |
t(11;15;17)(q13;q22;q12) | F | 15 | M3 | NA | 46,XX,
t(11;15;17)(q13;q22;q12) | NA | NA | NA |
|
t(11;15;17)(q13;q22;q12) | M | 41 | M3 | NA | 46,XY
46,XY, t(11;15;17)(q13;q22;q12) | NA | NA | NA |
|
t(5;15;17)(q13;q22;q12) | M | 50 | M3 | NA | 46,XY
46,XY, t(5;15;17)(q13;q22;q12) | NA | NA | NA |
| Calabrese et
al (26) |
t(15;17;21)(q22;q12;q22) | M | 38 | M3 | NA | 46,XY,
t(15;17;21)(q22;q12;q22) | C+BMT | + | 3 |
|
der(4)t(4;17)(q11;p11),der(15)t(15;17)(q22;q21),der(17)t(15;17)(q22;q21)t(4;17)(q11;p11) | F | 22 | M3 | NA |
46,XX,der(4)t(4;17)(q11;p11),der(15)t(15;17)(q22;q21),der(17)t(15;17)(q22;q21)t(4;17)(q11;p11) | C+A | − | 44+ |
| Park et al
(27) |
t(1;15;17)(p31;q22;q21) | M | 46 | M3 | + | 46,XY | | | |
| | | | | |
46,XY,del(1)(p22),del(3)(p25),der(17)t(1;15;17)(17pter→17q21::15q21→15q22::1p36→1p31::1
5q21→15q22::17q21→17pter) | C+A | + | 9 |
| Yamamoto et
al (28) |
t(15;20;17)(q22;p13;q21) | M | 39 | M3 | + | 46,XY
46,XY, t(15;20;17)(q22;p13;q21) | C+A | − | 12+ |
| Kudva et al
(29) |
t(15;16;17)(q22;q13;q21) | M | 27 | M3v | − | 46,XY
46,XY, t(15;16;17)(q22;q13;q21) | C+A | − | 40+ |
| Eclache et
al (30) |
t(6;15;17)(q25;q22;q21) | F | 56 | M3 | NA | 46,XX
46,XX, t(6;15;17)(q25;q22;q21) | C+A | − | 5+ |
| Xu et al
(5) | t(15;17) relapsed
with t(4;15;17)(q27-28;q22;q21) | F | 49 | M3
M5a | NA |
46,XX,dup(8)(q?),t(4;15;17)(q27-28;q22;q21) | C+A | + | 10 |
| García-Casado et
al (31) |
t(15;17(q22;q21),t(17;20)(q21;q12) | F | 31 | M3 | − | 46,XX | | | |
| | | | | | 46,XX,der(15)
t(15;17(q22;q21),t(17;20)(q21;q12) | C+A | − | 60+ |
| Yoo et al
(32) |
t(10;17;15;22)(q22;q21;q22;q11.2) | F | 47 | M3 | NA |
46,XX,t(10;17;15;22)(q22;q21;q22;q11.2) | C+A | − | 1+ |
| Miyazaki et
al (33) | t(15;17) involved
8q21 and 14p? | M | 78 | M3v | − | 46,XY
46,XY,t(15;17)(q22;q21)
46,XY,der(15)t(15;17)(q22;q21),der(17)(17pter→1
7q21::15q22→15qter::8q21→8qter)
46,XY,der(15)t(15;17)(q22;q21),der(17)(17pter→17q21::15q22→15qter::8q21→8qter::14p?→14qter) | C+A | − | 14+ |
| Abe et al
(34) |
t(5;17;15)(q11;q21;q22) | F | 22 | M3v | + | 46,XX
46,XX, t(5;17;15)(q11;q21;q22) | C+A | − | 12+ |
| Stavropoulou et
al (35) | t(15;17)
,t(17;18) | F | 46 | M3 | NA |
46,XX,der(15)t(15;17)(q22;q21),der(17)t(15;17)(q
22;q21)t(17;18)(q21;q12),der(18)t(15;17)(q22;q21)t(17;18)(q21;q12) | C+A | − | 2 |
| Kato et al
(36) |
t(15;22;17)(q22;q11.2;q21) | M | 44 | M3 | + | 46,XY
46,XY,t(15;22;17)(q22;q11.2;q21) | C+A | − | 1+ |
| Grimwade et
al (6) |
t(1;17;15)(p32;q21;q22) | M | NA | M3 | NA | 46,XY
46,XY, t(1;17;15)(p32;q21;q22)
46,idem,add(21)(p13) | NA | NA | NA |
| Grimwade et
al (6) |
t(7;17;15)(q22;q21;q22) | F | NA | M3v | NA | 46,XX,
t(7;17;15)(q22;q21;q22) | NA | NA | NA |
|
t(6;17;15)(p21;q21;q22) | M | NA | M3v | NA | 46,XY
46,XY, t(6;17;15)(p21;q21;q22) | NA | NA | NA |
|
t(8;17;15)(q22;q21;q22) | F | NA | M3 and M3r | NA | 46,XX
46,XX, t(8;17;15)(q22;q21;q22)47,idem,+8 | NA | NA | NA |
|
t(13;17;15)(p13;q21;q22) | F | NA | M3v | NA | 46,XX
46,XX, t(13;17;15)(p13;q21;q22) | NA | NA | NA |
|
t(5;17;15)(q14;q21;q22) | F | NA | M3 | NA |
46,XX,t(5;17;15)(q14;q21;q22
48,idem,+8,+21 | NA | NA | NA |
| Tirado et al
(36) |
t(15;17;17)(q22;q23;q21) | F | 52 | M3 | − | 46,XX,
t(15;17;17)(q22;q23;q21) | NA | − | 12+ |
| Freeman et
al (37) |
t(3;17;15)(q27;q21;q22) | M | 78 | M3 | − | 46,XY
46,XY, t(3;17;15)(q27;q21;q22) | C+A | − | NA |
|
t(8;17;15)(q24.3;q12;q22) | F | 49 | M3 | + | 46,XX
46,XX, t(8;17;15)(q24.3;q12;q22) | C+A | − | NA |
| Huret et al
(38) |
t(4;15;17)(q21;q26;q22) relapsed | M | 67 | M3 | + | 46,XY, 46,XY,
t(4;15;17)(q21;q26;q22)
46,XY, add(1)(p?),t(1;18)(p36;p10),add(10)(p?),
t(15;17)(q26;q22) | C | + | 25+ |
| McKinney et
al (39) |
t(3;17;15)(p21;q21;q22) | F | 51 | M3 | + |
46,XX,t(3;17;15)(p21;q21;q22) | C | + | 33 |
|
t(3;17;15)(p12;q21;q22) | M | 63 | M3 | + | 46,XY
46,XY, t(3;17;15)(q12;q21;q22) | C | + | 14 |
| Galieni et
al (40) |
t(1;17;15)(q23;q23;q22) | M | 41 | M3 | NA | 46,XY | | | |
| | | | | | 46,XY,
t(1;17;15)(q23;q23;q22)47,idem, +10 | C+A | − | 8+ |
In addition, the association between classical
t(15;17) and ct(15;17;v) clones remains unclear. Several previous
studies have assumed that the complicated rearrangements originate
from the single standard translocation and then quickly outgrow
this stem line (16,26,33).
The current study is the first to report the presentation of two
abnormal clones, classical t(15;17) and ct(15;17;v), simultaneously
in an adult APL patient, providing indirect evidence that the
variant translocation possibly evolves from the classical
t(15;17).
It is well known that chromosomal imbalances,
including the deletion(s) or amplification(s) of key driver
gene(s), may promote the malignant transformation of leukemia
(42–44). In the reported 45 APL cases of
ct(15;17;v), eight cases presented with trisomy 8 or partial
trisomy, including 8p+, 4p+, 9p+, +10 and +X. However, deletions,
particularly small or cryptic deletions, have not yet been
reported. In the present study, array CGH revealed the cryptic
chromosome aberrations, including the deletions of ATP2B2,
ANGPT1 and ETV6, and a gain of RB1CC1 genes, but
no imbalances of the breakpoints of chromosomes 15, 16 or 17. FISH
analysis not only confirmed the deletions of ATP2B2, ANGPT1
and ETV6, but also demonstrated that these deletions were
somatically acquired events since normal cells coexist with
abnormal cells, which may potentially be involved in the
leukemogenesis. However, the gain of 8q11.23 has not been confirmed
due to the one size of duplication, which is too small to confirm
by FISH. Furthermore, the possibility of a constitutional gain
rather than disease-related gain cannot be ruled out. A divergence
of ratio has also been identified between the deletions of
ATP2B2 (97%), ANGPT1 (52%), ETV6 (90%) and the
PML/RARA fusion (classical and variant combined; 90%) by FISH,
suggesting that these unbalanced genetic events may occur
asynchronously in leukemogenesis.
In conclusion, the current study was the first to
identify the classical t(15;17) and complex variant t(15;16;17) in
an adult patient with APL using FISH. Furthermore, cryptic genomic
alterations involving leukemia-related genes, such as ATP2B2,
ANGPT1, ETV6 and RB1CC1, were inferred in the level of
array CGH and confirmed by FISH. It may be proposed that the
malignant transformation of APL with complex variant translocation
presents the following multi-step progression: i) Classical
translocation; ii) formation of complex variant translocation by
third chromosome involvement; and iii) further genomic changes. The
further genomic changes, with the exception of the fusion of
PML/RARA in APL, may be implicated in the heterogenicity of therapy
outcome. Advanced study on these genes is likely to aid the
elucidation of the oncogene or tumor suppressor gene candidates
that potentially affect the prognosis of APL.
References
|
1
|
Degos L, Dombret H, Chomienne C, et al:
All-trans-retinoic acid as a differentiating agent in the treatment
of acute promyelocytic leukemia. Blood. 85:2643–2653. 1995.
|
|
2
|
Shen ZX, Chen GQ, Ni JH, et al: Use of
arsenic trioxide (As2O3) in the treatment of acute promyelocytic
leukemia (APL): II. Clinical efficacy and pharmacokinetics in
relapsed patients. Blood. 89:3354–3360. 1997.
|
|
3
|
Redner RL: Variations on a theme: the
alternate translocations in APL. Leukemia. 16:1927–1932. 2002.
|
|
4
|
Brunel V, Lafage-Pochitaloff M, Alcalay M,
Pelicci PG and Birg F: Variant and masked translocations in acute
promyelocytic leukemia. Leuk Lymphoma. 22:221–228. 1996.
|
|
5
|
Xu L, Zhao WL, Xiong SM, et al: Molecular
cytogenetic characterization and clinical relevance of additional,
complex and/or variant chromosome abnormalities in acute
promyelocytic leukemia. Leukemia. 15:1359–1368. 2001.
|
|
6
|
Grimwade D, Biondi A, Mozziconacci MJ, et
al: Characterization of acute promyelocytic leukemia cases lacking
the classic t(15;17): results of the European Working Party. Groupe
Français de Cytogénétique Hématologique, Groupe de Français
d’Hematologie Cellulaire, UK Cancer Cytogenetics Group and BIOMED 1
European Community-Concerted Action ‘Molecular Cytogenetic
Diagnosis in Haematological Malignancies’. Blood. 96:1297–1308.
2000.
|
|
7
|
Rohr SS, Pelloso LA, Borgo A, et al: Acute
promyelocytic leukemia associated with the PLZF-RARA fusion gene:
two additional cases with clinical and laboratorial peculiar
presentations. Med Oncol. 29:2345–2347. 2012.
|
|
8
|
Bennett JM, Catovsky D, Daniel MT, et al:
Proposals for the classification of the acute leukaemias.
French-American-British (FAB) co-operative group. Br J Haematol.
33:451–458. 1976.
|
|
9
|
Kato T, Hangaishi A, Ichikawa M, Motokura
T, Takahashi T and Kurokawa M: A new three-way variant
t(15;22;17)(q22;q11.2;q21) in acute promyelocytic leukemia. Int J
Hematol. 89:204–208. 2009.
|
|
10
|
Ried T, Schröck E, Ning Y and Wienberg J:
Chromosome painting: a useful art. Hum Mol Genet. 7:1619–1626.
1998.
|
|
11
|
Berger R, Flandrin G, Bernheim A, et al:
Cytogenetic studies on 519 consecutive de novo acute nonlymphocytic
leukemias. Cancer Genet Cytogenet. 29:9–21. 1987.
|
|
12
|
Bernstein R, Mendelow B, Pinto MR, Morcom
G and Bezwoda W: Complex translocations involving chromosomes 15
and 17 in acute promyelocytic leukaemia. Br J Haematol. 46:311–314.
1980.
|
|
13
|
Bernstein R, Pinto MR, Morcom G, et al:
Karyotype analysis in acute nonlymphocytic leukemia (ANLL):
comparison with ethnic group, age, morphology, and survival. Cancer
Genet Cytogenet. 6:187–199. 1982.
|
|
14
|
Ohyashiki K, Oshimura M, Uchida H, et al:
Cytogenetic and ultrastructural studies on ten patients with acute
promyelocytic leukemia, including one case with a complex
translocation. Cancer Genet Cytogenet. 14:247–255. 1985.
|
|
15
|
Callen DF, Dale BM, Sage RE and Ford JH: A
complex translocation in acute promyelocytic leukemia. Cancer Genet
Cytogenet. 16:45–48. 1985.
|
|
16
|
Bjerrum OW, Philip P, Pressler T and
Tygstrup I: Acute promyelocytic leukemia with t(15;17) and
t(2;17;15). Cancer Genet Cytogenet. 28:107–111. 1987.
|
|
17
|
Osella P, Wyandt H, Vosburgh E and
Milunsky A: Report of a variant t(1;15;17)(p36;q22;q21.1) in a
patient with acute promyelocytic leukemia. Cancer Genet Cytogenet.
57:201–207. 1991.
|
|
18
|
Saitoh K, Miura I, Kobayashi Y, et al: A
new variant translocation of t(15;17) in a patient with acute
promyelocytic leukemia (M3): t(15;19;17)(q22;p13;q12). Cancer Genet
Cytogenet. 102:15–18. 1998.
|
|
19
|
Wan TS, Chim CS, So CK, Chan LC and Ma SK:
Complex variant 15;17 translocations in acute promyelocytic
leukemia. A case report and review of three-way translocations.
Cancer Genet Cytogenet. 111:139–143. 1999.
|
|
20
|
Fujishima M, Takahashi N, Miura I, et al:
A PML/RARA chimeric gene on chromosome 2 in a patient with acute
promyelocytic leukemia (M3) associated with a new variant
translocation: t(2;15;17)(q21;q22;q21). Cancer Genet Cytogenet.
120:80–82. 2000.
|
|
21
|
Liu S, Li Q, Pang W, et al: A new complex
variant t(4;15;17) in acute promyelocytic leukemia: fluorescence in
situ hybridization confirmation and literature review. Cancer Genet
Cytogenet. 130:33–37. 2001.
|
|
22
|
Misawa S, Lee E, Schiffer CA, Liu Z and
Testa JR: Association of the translocation (15;17) with malignant
proliferation of promyelocytes in acute leukemia and chronic
myelogenous leukemia at blastic crisis. Blood. 67:270–274.
1986.
|
|
23
|
Zaccaria A, Testoni M, Martinelli G, et
al: Four-chromosomes complex translocations in acute promyelocytic
leukemia: description of two cases. Eur J Haematol. 52:129–133.
1994.
|
|
24
|
Ogawa S, Mitani K, Sato Y, et al:
Detection of the PML/RAR alpha fusion gene in acute promyelocytic
leukemia with a complex translocation involving chromosomes 15, 17,
and 18. Cancer Genet Cytogenet. 69:113–117. 1993.
|
|
25
|
Chen Z, Morgan R, Stone JF and Sandberg
AA: Identification of complex t(15;17) in APL by FISH. Cancer Genet
Cytogenet. 72:73–74. 1994.
|
|
26
|
Calabrese G, Min T, Stuppia L, et al:
Complex chromosome translocations of standard t(8;21) and t(15;17)
arise from a two-step mechanism as evidenced by fluorescence in
situ hybridization analysis. Cancer Genet Cytogenet. 91:40–45.
1996.
|
|
27
|
Park JP and Fairweather RB: Complex
t(1;15;17) in acute promyelocytic leukemia with duplication of RAR
alpha and PML sequences. Cancer Genet Cytogenet. 89:52–56.
1996.
|
|
28
|
Yamamoto K, Hamaguchi H, Nagata K,
Kobayashi M, Takashima T and Taniwaki M: A new complex
translocation (15;20;17)(q22;p13;q21) in acute promyelocytic
leukemia. Cancer Genet Cytogenet. 101:89–94. 1998.
|
|
29
|
Kudva GC, Maliekel K, Richart JM, et al:
Acute promyelocytic leukemia and HIV-1 infection: case report and
review of the literature. Am J Hematol. 77:287–290. 2004.
|
|
30
|
Eclache V, Viguie F, Frocrain C, et al: A
new variant t(6;15;17)(q25;q22;q21) in acute promyelocytic
leukemia: fluorescence in situ hybridization confirmation. Cancer
Genet Cytogenet. 159:69–73. 2005.
|
|
31
|
García-Casado Z, Cervera J, Valencia A, et
al: A t(17;20)(q21;q12) masking a variant t(15;17)(q22;q21) in a
patient with acute promyelocytic leukemia. Cancer Genet Cytogenet.
168:73–76. 2006.
|
|
32
|
Yoo SJ, Seo EJ, Lee JH, Seo YH, Park PW
and Ahn JY: A complex, four-way variant t(15;17) in acute
promyelocytic leukemia. Cancer Genet Cytogenet. 167:168–171.
2006.
|
|
33
|
Miyazaki K, Kikukawa M, Kiuchi A, Shin K,
Iwamoto T and Ohyashiki K: Complex translocations derived stepwise
from standard t(15;17) in a patient with variant acute
promyelocytic leukemia. Cancer Genet Cytogenet. 176:127–130.
2007.
|
|
34
|
Abe S, Ishikawa I, Harigae H and Sugawara
T: A new complex translocation t(5;17;15)(q11;q21;q22) in acute
promyelocytic leukemia. Cancer Genet Cytogenet. 184:44–47.
2008.
|
|
35
|
Stavropoulou C, Georgakakos VN, Manola KN,
et al: 5′RARA submicroscopic deletion from new variant
translocation involving chromosomes 15, 17, and 18, in a case of
acute promyelocytic leukemia. Cancer Genet Cytogenet. 182:50–55.
2008.
|
|
36
|
Tirado CA, Golembiewski-Ruiz V,
Horvatinovich J, et al: Cytogenetic and molecular analysis of an
unusual case of acute promyelocytic leukemia with a
t(15;17;17)(q22;q23;q21). Cancer Genet Cytogenet. 145:31–37.
2003.
|
|
37
|
Freeman CE, Mercer DD, Ye Y, Van Brunt J
3rd and Li MM: Cytogenetic and molecular characterization of
complex three-way translocations in acute promyelocytic leukemia.
Beijing Da Xue Xue Bao. 41:477–479. 2009.(In Chinese).
|
|
38
|
Huret JL, Couet D, Guilhot F, Brizard A
and Tanzer J: A two-step t(4;der(15)) t(15;17) complex
translocation in an acute promyelocytic leukaemia and review of the
literature. Leuk Res. 11:761–765. 1987.
|
|
39
|
McKinney CD, Golden WL, Gemma NW, Swerdlow
SH and Williams ME: RARA and PML gene rearrangements in acute
promyelocytic leukemia with complex translocations and atypical
features. Genes Chromosomes Cancer. 9:49–56. 1994.
|
|
40
|
Galieni P, Marotta G, Vessichelli F, et
al: Variant t(1;15;17)(q23;q22;q23) in a case of acute
promyelocytic leukemia. Leukemia. 10:1658–1661. 1996.
|
|
41
|
De Braekeleer E, Douet-Guilbert N, Le Bris
MJ, Morel F, Férec C and De Braekeleer M: RUNX1-MTG16 fusion gene
in acute myeloblastic leukemia with t(16;21)(q24;q22): case report
and review of the literature. Cancer Genet Cytogenet. 185:47–50.
2008.
|
|
42
|
Tybäkinoja A, Elonen E, Piippo K, Porkka K
and Knuutila S: Oligonucleotide array-CGH reveals cryptic gene copy
number alterations in karyotypically normal acute myeloid leukemia.
Leukemia. 21:571–574. 2007.
|
|
43
|
Zagaria A, Anelli L, Albano F, Vicari L,
Schiavone EM, et al: Molecular cytogenetic characterization of
deletions on der(9) in chronic myelocytic leukemia. Cancer Genet
Cytogenet. 167:97–102. 2006.
|
|
44
|
Paulsson K, Cazier JB, Macdougall F, et
al: Microdeletions are a general feature of adult and adolescent
acute lymphoblastic leukemia: Unexpected similarities with
pediatric disease. Proc Natl Acad Sci USA. 105:6708–6713. 2008.
|