Introduction
Breast cancer is one of the most common malignancies
worldwide. It is desirable to explore new molecular targets and
develop novel targeted drugs for breast cancer patients.
Hypermethylated in cancer 1 (HIC-1), a tumor suppressor gene for
breast cancer, located on 17p13.3, encodes a transcriptional
suppressor protein, with five Kruppel-like
C2H2 zinc finger motifs and the N-terminal
protein-protein interaction domain, BTB/POZ (1). Epigenetic silencing of HIC-1 is
significant in the pathogenesis of epithelial cancers. The loss of
HIC-1 may be closely associated with the promotion of tumorigenesis
in a wide variety of cell types. Decreased expression of the HIC-1
gene is observed in non-small cell lung cancer (2), hepatocellular carcinoma (3), gastric cancer (4) and human medulloblastomas (5), and the loss of HIC-1 expression is a
common event in primary breast cancer (6). Inactivation of HIC-1 in breast
carcinomas is associated with tumor metastasis (7), and a previous study demonstrated that
restoring the HIC-1 expression by demethylation treatment impaired
the aggressiveness of head and neck squamous cell carcinoma
(8).
In 2006, Li et al (9) reported a novel function of
double-stranded RNA (dsRNA) molecules. The results indicated that
dsRNA induced sequence-specific transcriptional activation by
targeting gene promoter regions. This phenomenon was termed
RNA-induced gene activation (RNAa) and the dsRNA molecules were
denominated as small activating RNAs (saRNAs). Janowski et
al (10) reported similar
findings, in which multiple duplex RNAs, complementary to the
progesterone receptor (PR) promoter, activated PR protein
expression in T47D and MCF-7 human breast cancer cells (10). Subsequently, dsRNAs have been used
for the activation of various target genes in multiple
laboratories. Chen et al (11) and Yang et al (12) used dsRNAs to upregulate p21WAF1/CIP1
(p21) in human bladder cancer cells. In addition, Ren et al
(13) induced NKX3-1 in prostate
tumor cells by saRNA. More recently, our research group
successfully reactivated the HIC-1 tumor suppressor in gastric
cancer and in breast cancer (4,14). Our
previous studies disclosed that dsHIC1-2998, an saRNA, effectively
activated HIC-1 with evident suppression of cell growth and
induction of apoptosis in breast cancer. These findings indicate
the possibility that this saRNA may become a practical and highly
cost-effective approach for gene therapy.
In the present study, the efficacy of saRNA on
suppression of clonogenicity and invasiveness of breast cancer cell
lines was investigated by the reactivation of HIC-1. dsHIC1-2998,
which targets the HIC-1 promoter region, was used as an effective
saRNA. This study aims to increase the supporting evidence for
saRNA as a promising molecule for restoring the gene expression and
biological activity of tumor suppressors in breast cancer.
Materials and methods
saRNA design
The dsRNA targeting the region (2998 bp) above from
the transcription start site of human HIC-1 was designed based on
the rational design rules described (9,15) and
our previous reports (4,14). The sequences of saRNA-HIC-1
(dsHIC-1-2998) used in this experiment were as follows: Sense,
5′-CGGUUUCCMGGAGAAGUUATT-3′ and antisense,
5′-UAACUUCUCCAGGAAACCGTT-3′. A further RNA strand, unrelated to
that of the human dsRNA sequence, was used as a control (sense,
5′-ACGMGACACGUUCGGAGAATT-3′ and antisense,
5′-UUCUCCGAACGMGUCACGUTT-3′). All dsRNA sequences were synthesized
by Genepharma Inc. (Shanghai, China).
Cell culture and saRNA transfection
MCF-7 and MDA-MB-231 breast cancer cell lines were
originally obtained from the Institute of Biochemistry and Cell
Biology, Shanghai Chinese Academy of Science (Shanghai, China).
MCF-7 and MDA-MB-231 cells were cultured in Dulbecco’s modified
Eagle’s medium with 10% fetal bovine serum (Gibco-BRL, Invitrogen
Life Technologies, Carlsbad, CA, USA). Immediately prior to
transfection, the cells were trypsinized, diluted with growth
medium without antibiotics or serum, and seeded into six-well
plates at a density of 3.0×105 cells per well for MCF-7
cells and 4.0×105 cells per well for MDA-MB-231 cells.
The transfection of saRNA and control RNA was conducted at a
concentration of 50 nmol/l using Lipofectamine 2000 (Invitrogen
Life Technologies) according to the manufacturer’s reverse
transfection instructions. The cells were harvested for further
analysis. In this study, the mock group was transfected with
lipofectamine 2000 alone, while the control group was transfected
with non-specific dsRNA.
mRNA analysis by real-time polymerase
chain reaction (PCR)
Total RNA was extracted using TRIzol solution
(Invitrogen Life Technologies). Reverse transcription PCR was
performed in a 20-μl reaction system according to the
manufacturer’s instructions (Promega Corporation, Madison, WI,
USA). The cDNA was amplified using gene-specific primer sets in
conjunction with the SYBR Green PCR master mix (Applied Biosystems,
Foster City, CA, USA). Real-time PCR was performed in a reaction
mixture with a final volume of 20 μl containing 10 μl SYBR Green
PCR Master Mix, 1 μl of 5 mmol/l paired primer specific to the
target gene and 1 μl cDNA. The primers used for real-time PCR were
as follows: Forward, 5′-GACGGCGACGACTACAAGAG-3′ and reverse,
5′-GAATGCACACGTACAGGTTGTC-3′ for HIC-1; and forward,
5′-GGACCTGACCTGCCGTCTAG-3′ and reverse, 5′-GTAGCCCAGGATGCCCTTGA-3′
for GAPDH.
Protein analysis by western blotting
The cells were harvested and washed twice with PBS,
pH 7.4, and resuspended in lysis buffer [1 mM dithiothreitol, 0.125
mM EDTA, 5% glycerol, 1 mM phenylmethyl 5 μl fonylfluoride, 1 μg/ml
leupeptin, 1 μg/ml pepstatin, 1 μg/ml aprotinin, 1% Triton X-100
(Shanghai Chemical Co., Shanghai, China) in 12.5 mM Tris-HCl
buffer, pH 7.0] on ice. The cell extracts were centrifuged and the
protein concentration was determined using the bicinchoninic acid
protein assay kit (Pierce Biotechnology, Inc., Rockford, IL, USA)
according to the manufacturer’s instructions. Each protein extract
(50 μg) was electrophoresed on a 12.5% SDS-polyacrylamide gel,
transferred to polyvinylidene difluoride membranes in a buffer
containing 25 mM Tris-HCl, pH 8.3, 192 mM glycine and 20% (v/v)
methanol, and blocked in 5% (w/v) skimmed milk in Tris-buffered
saline-Tween 20 (TBST; 0.1% v/v) for 2 h at room temperature. This
was subsequently probed with specific primary antibodies (mouse
monoclonal anti-HIC-1, 1:800, ab55120, Abcam, Cambridge, England;
and mouse monoclonal anti-GAPDH, 1:5000; GW22763, Sigma-Aldrich,
St. Louis, MO, USA) overnight at 4°C. The primary antibodies were
removed and the blots were extensively washed with TBST three
times. The blots were incubated for 1 h at room temperature with
horseradish peroxidase-conjugated rabbit anit-mouse polyclonal
secondary antibody (1:5000; Sigma-Aldrich) in TBST. Following this,
the blots were washed for 30 min and developed using an Enhanced
Chemiluminescence kit (NENTM Life Science Products Inc., Boston,
MA, USA).
Clonogenicity assay
The cancer cells were transfected with saRNA or
control RNA for 12 h, and then transferred to six-well plates and
seeded at a density of 1.0×103 cells per well. The
plates were incubated at 37°C in a humidified atmosphere of 5%
CO2 for 12 days. The culture medium was changed every
three days. Clonogenicity was analyzed at 12 days following the
saRNA transfection. The plates were stained with 0.05% crystal
violet solution for 15 min, and the colonies were counted under the
inverted microscope and photographed. The experiments were
performed in triplicate, at minimum. Data are presented as the mean
± standard deviation (SD).
Scratch-healing assay
MDA-MB-231 and MCF-7 cells were seeded into six-well
plates at a density of 0.8×105 cells per well. Following
overnight incubation, the cells were transfected with 50 nmol/l
saRNA-HIC-1 or control RNA for 72 h until the cells reached full
confluence. A monolayer of cells was scratched by a 1-mm
micropipette tip, rinsed with PBS to remove cell debris and
cultured continuously in growth medium containing 1% fetal bovine
serum (Gibco-BRL, Invitrogen Life Technologies). The wound-closing
procedure was observed for 72 h. The wound width of each well was
calculated at every 24 h interval.
Invasion and migration assays
The cells were harvested following the 72-h
transfection of saRNA-HIC-1 or the control RNA, and were
resuspended in medium. For the MCF-7 cells, the cell concentrations
for the migration and invasion assay were 1.0×106 and
2.5×106 cells/ml, respectively. For the MDA-MB-231
cells, the cell concentrations for the migration and invasion
assays were 1.5×105 and 5×105 cells/ml,
respectively. In total, 0.2 ml cells was added to the top Transwell
chamber (24-well insert, 8-μm pore size; Millipore, Bedford, MA,
USA) and 0.6 ml medium with 10% fetal bovine serum was added to the
lower chamber as a chemoattractive factor. Subsequently, the cells
were incubated for 20–48 h. The cells that did not migrate through
the pores were removed by scraping the upper surface of the
membrane with a cotton swab. The cells that migrated to the lower
surface of the membrane were fixed with 100% methanol for 15 min
and stained with 0.1% crystal violet for a further 15 min. The
cells that migrated through the insert were counted at five random
fields and expressed as the mean number of cells per field. These
experiments were performed in triplicate.
Cell cycle analysis by flow
cytometry
The cells (1×106 cells/ml) were
transfected with saRNA or control RNA. At 96 h following
transfection, the cells were harvested and fixed in 70% ethanol at
−20°C overnight, and then stained with 250 μg/ml propidium iodide
(Sigma-Aldrich), 5 μg/ml RNase A (Sigma-Aldrich) and 5 mmol/l EDTA
in PBS (pH 7.4) for 30 min. The cell cycle analysis was performed
using the FACScan (Beckman Instruments, Fullerton, CA, USA). The
data was evaluated using the FlowJo software (Tree Star, Inc.
Ashland, OR, USA).
Statistical analysis
The results are presented as the mean ± SD.
Statistical analyses were performed using SPSS, version 15.0 (SPSS
Inc., Chicago, IL, USA). Student’s t-test and one-way analysis of
variance, followed by Dunnett’s multiple comparison tests, were
conducted. P<0.05 was considered to indicate a statistically
significant difference, indicated by asterisks in the figures.
Results
Reactivation of HIC-1 inhibits colony
formation of breast cancer cells
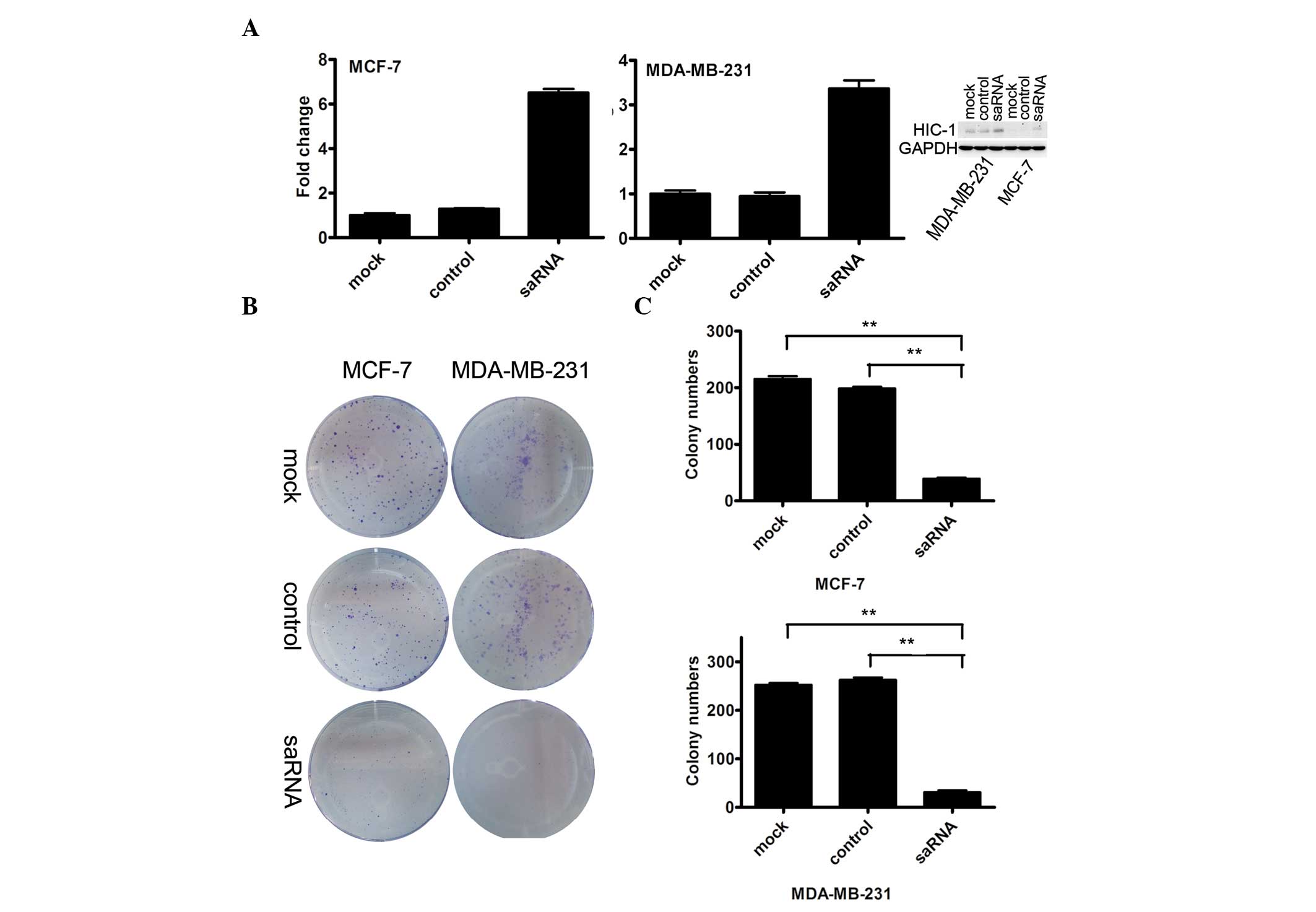
Initially, whether or not dsHIC1-2998 was an
effective saRNA was investigated. In total, 50 nmol/l saRNA was
transfected into MDA-MB-231 and MCF-7 cancer cell lines. The
restoration of HIC-1 mRNA was evaluated by real-time RT-PCR 96 h
following saRNA transfection. In MCF-7 cells transfected with HIC-1
mRNA, the HIC-1 mRNA level was upregulated 6.52-fold compared with
the mock-transfected cells. In MDA-MB-231 cells transfected with
HIC-1 mRNA, the HIC-1 mRNA level was upregulated 3.37-fold,
compared with the mock-transfected cells. The protein analysis
revealed that HIC-1 protein levels were also elevated based on the
saRNA transfection for the two cancer cell lines (Fig. 1A), compared with that of the control
cells. Therefore, dsHIC1-2998 was confirmed as effective
saRNA-HIC-1.
Subsequently, 50 nmol/l saRNA-HIC-1 or control RNA
was transfected into MCF-7 and MDA-MB-231 cells for 12 h and the
clonogenicity was analyzed at 12 days following the saRNA
transfection. The size of the colonies formed in saRNA-HIC-1 group
was smaller than that in the control groups (Fig. 1B). The colonies containing at least
50 cells in five fields were randomly counted. The number of
colonies was significantly reduced in the saRNA-HIC-1 transfection
group in MCF-7 cells (39.0 vs. 198.7 and 215.2; the mock and
control groups, respectively; P<0.001) and MDA-MB-231 cells
(31.0 vs. 262.7 and 252.3; the mock and control groups,
respectively, P<0.001), compared with the control groups
(Fig. 1C).
Reactivation of HIC-1 inhibits cell
migration and cell invasion of breast cancer cells
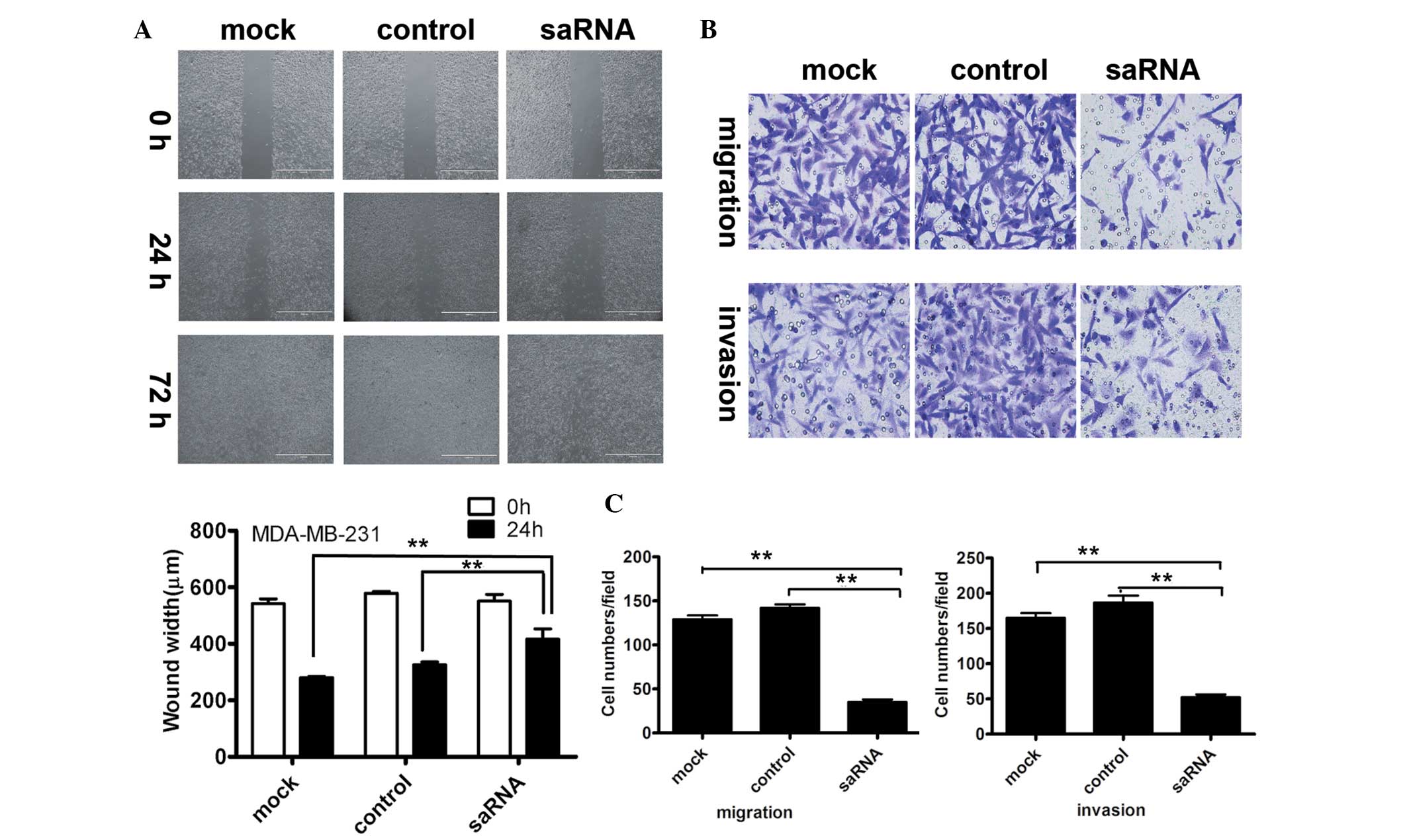
Initially, the cell migration ability was analyzed
using a wound closure experiment in MDA-MB-231 cells and MCF-7
cells following saRNA-HIC-1 transfection. The wound-closing
procedure was serially observed for 72 h following the introduction
of the wound on the plate. As shown in Fig. 2A, the speed of wound-closing was
slower in saRNA-HIC-1-transfected cells, compared with that in the
control groups (untransfected and mock-transfected HIC-1 cells). At
72 h, the wounds of the control groups were completely closed. This
indicated that the upregulation of HIC-1 expression inhibited cell
migration in vitro. The results for the MCF-7 cell line
could not be obtained due to its low migration capacity.
Following this, the cell migration and invasion
ability were analyzed for the saRNA-HIC-1 transfectant on
MDA-MB-231 and MCF-7 cell lines using a Transwell chamber. As shown
in Fig. 2B, MDA-MB-231 cells in the
saRNA-HIC-1 group exhibited a weaker migration ability with fewer
cells compared with the control groups. The cells in the
saRNA-HIC-1 group exhibited weaker invasive ability, with fewer
cells compared with the control groups. The cell counting revealed
that cell numbers for cell migration (35 vs. 142 and 129; the mock
and control groups, respectively; P<0.001) or cell invasion
(52.3 vs. 186.7 and 165; the mock and control groups,
respectively; P<0.001) in saRNA-HIC-1 group were significantly
lower than that of the control groups (Fig. 2C). The results for the MCF-7 cell
line could not be obtained due to its low migration and invasion
capacity.
Upregulation of HIC-1 expression via
saRNA induces cell cycle arrest in breast cancer cells
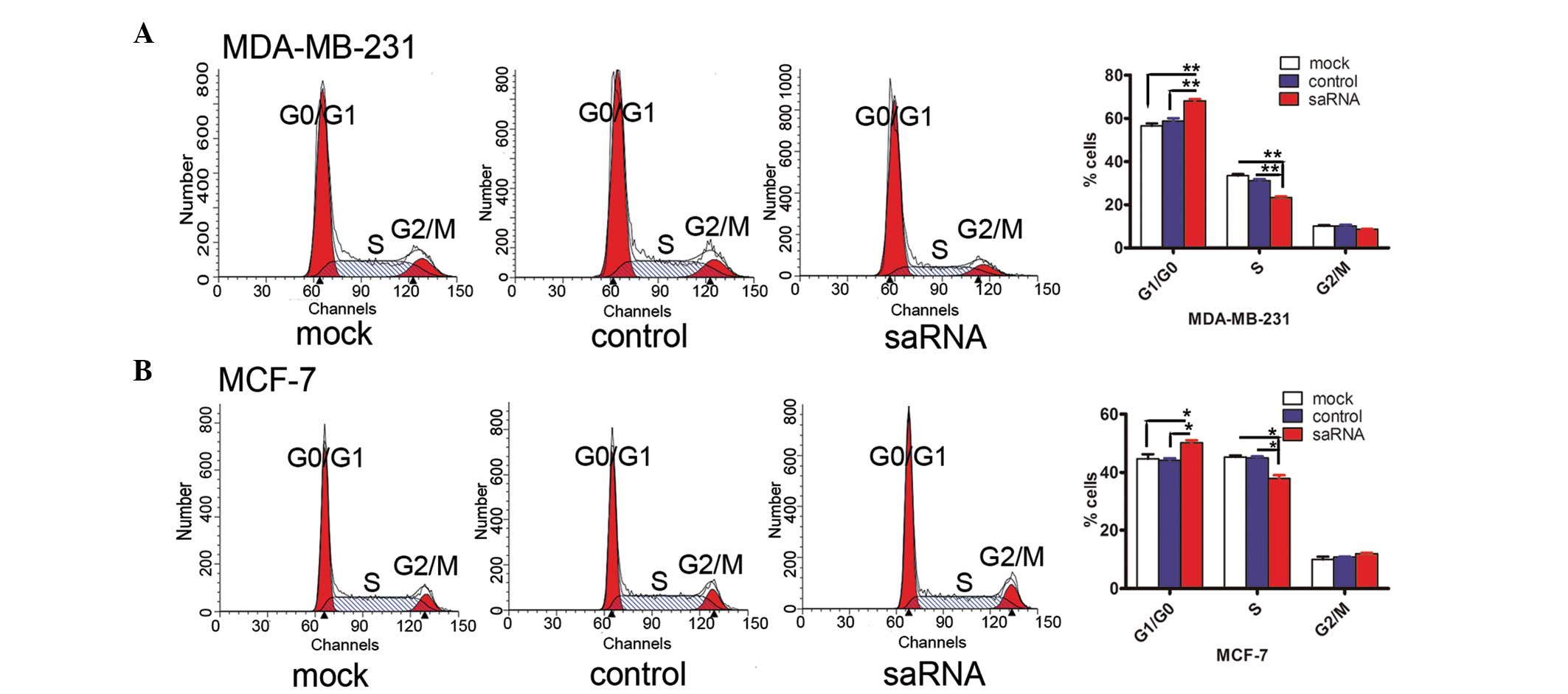
The cell cycle fraction was investigated using flow
cytometry based on saRNA-HIC-1 transfection for 96 h for the two
cancer cell lines. In the MDA-MB-231 cells, saRNA-HIC-1
transfection caused a significant increase in the G1/G0 fraction
(68.64 vs. 59.64 and 55.63%; the mock and control groups,
respectively) with concurrent decline in S (22.89 vs. 30.58 and
33.94%) and G2/M fractions (8.47 vs. 9.78 and 10.43%; the mock and
control groups, respectively), compared with the controls (Fig. 3). However, a significant increase in
the G1/G0 fraction (50.79 vs. 43.75 and 43.63%; the mock and
control groups, respectively) with a concurrent decline in the S
fraction (37.07 vs. 45.38 and 44.90%; the mock and control groups,
respectively) and slight increase in the G2/M fraction (12.14 vs.
10.87 and 9.27%; the mock and control groups, respectively) were
observed in the MCF-7 cells. Overall, these results indicated that
the reactivation of the HIC-1 gene by saRNA induces G1/G0 phase
arrest.
Discussion
It is known that short 21-nucleotide dsRNA molecules
may silence endogenous human genes in a sequence-specific manner.
This method, termed RNA interference (RNAi), develops rapidly and
is extensively used in experimental medicine. RNAi exhibits
significant capacity in the silencing of oncogenes. The mechanism
behind RNAi involves the knockdown of endogenous human genes. RNAi
has exhibited potential in the field of tumor therapy; however, no
dependable method has been established for the restoration of
endogenous tumor suppressor genes, with the exception of
vector-mediated gene engineering. RNAa is mediated by small dsRNA
fragments. The RNAa technology exhibits the opposite efficacy of
RNAi by activating, as opposed to silencing, the target genes. As a
novel technique, RNAa has successfully activated several target
genes in various human diseases including those involved in
cancers, such as p21, E-cadherin, VEGF, WT1 and several others
(16–21). Although the exact mechanism of RNAa
remains unclear, as a new molecular tool, saRNA is currently under
use in the study of gene function and has exhibited promising
initial results. Mao et al (22) reported that the upregulation of
E-cadherin by saRNA inhibits cell invasion and migration of 5637
human bladder cancer cells. Restoring the E-cadherin gene in
MDA-MB-453 breast cancer cells induced apoptosis and inhibited cell
proliferation (21). Activation of
the p21 gene in a variety of cancer cells, including prostate,
bladder, liver, pancreas and lung cancer cells, inhibited cell
proliferation and clonogenicity (18,19,23,24).
Restoration of the p21 gene enhanced apoptotic cell death and
caused G0/G1 arrest in T24 and J82 bladder cancer cells (11). Recently, lipid
nanoparticle-formulated dsp21-322-2′F revealed an inhibiting effect
on bladder tumors in vivo (25). saRNAs have exhibited similar
benefits to RNAi as a therapeutic molecule.
HIC-1 is a transcriptional repressor involved in the
regulation of growth control, cell survival and DNA damage response
(26). HIC-1 has been observed to
be epigenetically silenced in human cancers including breast cancer
(27). Hypermethylation is a
significant inactivation mechanism for a number of tumor
suppressors. Boulay et al revealed that the loss of HIC-1 is
involved in stress-induced migration and invasion in breast cancer
(7). HIC-1 promoter
hypermethylation is associated with tumor aggressiveness and poor
survival. The restoration of HIC-1 expression by a demethylation
reagent, caused the suppression of cancer progression in head and
neck squamous cell carcinoma (8).
Our research group has had an interest in this novel
molecular technique since it was established. As demonstrated in
our previous studies, the HIC-1 tumor suppressor was initially
reactivated in gastric cancer cells. The reactivation of HIC-1 was
observed to suppress cell migration and induce cell cycle arrest in
the G0/G1 phase as well as apoptosis (4). Subsequently, the HIC-1 tumor
suppressor was successfully reactivated in breast cancer cells.
dsHIC1-2998 was further confirmed as effective saRNA for gastric
cancer and breast cancer cells. The saRNA-HIC-1 effectively
activated the HIC-1 gene with evident suppression of cell growth
and induction of apoptosis in breast cancer (14). In the current study, further
evidence has been obtained, confirming that saRNA-HIC-1 effectively
inhibits clonogenicity in both MCF-7 and MDA-MB-231 cells. However,
the change of invasiveness in MDA-MB-231 cells is based on HIC-1
activation, while the change of invasiveness of MCF-7 cells is
unclear. The result reflects how different cell lines have varying
biological behavious.
In conclusion, cell models were created for the
restoration of the tumor suppressor gene, HIC-1, in breast cancer
cells. Using these cell models, the effects of upregulating the
HIC-1 gene were explored in multiple biological features, including
tumor growth, migration, invasion and the cell cycle. These
findings provide evidence that HIC-1 may potentially be a target
for gene therapy against breast cancer. The upregulation of HIC-1
by saRNA molecules may be a therapeutic strategy for the
suppression of breast cancer progression. The targeted activation
of tumor suppressor genes by saRNA may provide a new therapeutic
option that could significantly improve the treatment of breast
cancer.
Acknowledgements
The authors thank Professor Long-Cheng Li, of the
Department of Urology, Helen Diller Family Comprehensive Cancer
Center, University of California, USA, for the suggestive
instruction for saRNA design. This study was supported, in part, by
grants from the National Natural Science Foundation of China (grant
nos. 81172329 and 81372644), the Chinese National High Tech Program
(grant nos. 2012AA02A504, 2012AA02A203 and 2011ZX09307-001-05),
Shanghai Excellent Academic Leader Plan (grant no. 11XD1403600) and
Shanghai Science and Technology Commission (grant no.
14ZR1424400).
References
|
1
|
Deltour S, Guerardel C, Stehelin D and
Leprince D: The carboxy-terminal end of the candidate tumor
suppressor gene HIC-1 is phylogenetically conserved. Biochim
Biophys Acta. 1443:230–232. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hayashi M, Tokuchi Y, Hashimoto T, et al:
Reduced HIC-1 gene expression in non-small cell lung cancer and its
clinical significance. Anticancer Res. 21:535–540. 2001.PubMed/NCBI
|
|
3
|
Kanai Y, Hui AM, Sun L, et al: DNA
hypermethylation at the D17S5 locus and reduced HIC-1 mRNA
expression are associated with hepatocarcinogenesis. Hepatology.
29:703–709. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pan S, Wang Z, Chen Y, et al: Inactivation
of tumor suppressor gene HIC1 in gastric cancer is reversed via
small activating RNAs. Gene. 527:102–108. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rood BR, Zhang H, Weitman DM and Cogen PH:
Hypermethylation of HIC-1 and 17p allelic loss in medulloblastoma.
Cancer Res. 62:3794–3797. 2002.PubMed/NCBI
|
|
6
|
Fujii H, Biel MA, Zhou W, Weitzman SA,
Baylin SB and Gabrielson E: Methylation of the HIC-1 candidate
tumor suppressor gene in human breast cancer. Oncogene.
16:2159–2164. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Boulay G, Malaquin N, Loison I, et al:
Loss of Hypermethylated in Cancer 1 (HIC1) in breast cancer cells
contributes to stress-induced migration and invasion through β-2
adrenergic receptor (ADRB2) misregulation. J Biol Chem.
287:5379–5389. 2012. View Article : Google Scholar :
|
|
8
|
Brieger J, Pongsapich W, Mann SA, et al:
Demethylation treatment restores hic1 expression and impairs
aggressiveness of head and neck squamous cell carcinoma. Oral
Oncol. 46:678–683. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li LC, Okino ST, Zhao H, et al: Small
dsRNAs induce transcriptional activation in human cells. Proc Natl
Acad Sci USA. 103:17337–17342. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Janowski BA, Younger ST, Hardy DB, Ram R,
Huffman KE and Corey DR: Activating gene expression in mammalian
cells with promoter-targeted duplex RNAs. Nat Chem Biol. 3:166–173.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen Z, Place RF, Jia ZJ, Pookot D, Dahiya
R and Li LC: Antitumor effect of dsRNA-induced p21(WAF1/CIP1) gene
activation in human bladder cancer cells. Mol Cancer Ther.
7:698–703. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang K, Zheng XY, Qin J, et al:
Up-regulation of p21WAF1/Cip1 by saRNA induces G1-phase arrest and
apoptosis in T24 human bladder cancer cells. Cancer Lett.
265:206–214. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ren S, Kang MR, Wang J, et al: Targeted
induction of endogenous NKX3-1 by small activating RNA inhibits
prostate tumor growth. Prostat. 72:1591–1601. 2013. View Article : Google Scholar
|
|
14
|
Zhao F, Pan S, Gu Y, et al: Small
activating RNA restores the activity of the tumor suppressor HIC-1
on breast cancer. PLoS One. 9:e864862014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang V, Qin Y, Wang J, et al: RNAa is
conserved in mammalian cells. PLoS One. 5:e88482010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Portnoy V, Huang V, Place RF and Li LC:
Small RNA and transcriptional upregulation. Wiley Interdiscip Rev
RNA. 2:748–760. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hu J, Chen Z, Xia D, Wu J, Xu H and Ye ZQ:
Promoter-associated small double-stranded RNA interacts with
heterogeneous nuclear ribonucleoprotein A2/B1 to induce
transcriptional activation. Biochem J. 447:407–416. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kosaka M, Kang MR, Yang G and Li LC:
Targeted p21(WAF1/CIP1) activation by RNAa inhibits hepatocellular
carcinoma cells. Nucleic Acid Ther. 22:335–343. 2012.PubMed/NCBI
|
|
19
|
Qin Q, Lin YW, Zheng XY, et al:
RNAa-mediated overexpression of WT1 induces apoptosis in HepG2
cells. World J Surg Oncol. 10:112012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen R, Wang T, Rao K, et al:
Up-regulation of VEGF by small activator RNA in human corpus
cavernosum smooth muscle cells. J Sex Med. 8:2773–2780. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Junxia W, Ping G, Yuan H, et al: Double
strand RNA-guided endogeneous E-cadherin up-regulation induces the
apoptosis and inhibits proliferation of breast carcinoma cells in
vitro and in vivo. Cancer Sci. 101:1790–1796. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mao Q, Li Y, Zheng X, et al: Up-regulation
of E-cadherin by small activating RNA inhibits cell invasion and
migration in 5637 human bladder cancer cells. Biochem Biophys Res
Commun. 375:566–570. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wei J, Zhao J, Long M, et al: p21WAF1/CIP1
gene transcriptional activation exerts cell growth inhibition and
enhances chemosensitivity to cisplatin in lung carcinoma cell. BMC
Cancer. 10:6322010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu ZM, Dai C, Huang Y, et al: Anti-cancer
effects of p21WAF1/CIP1 transcriptional activation induced by
dsRNAs in human hepatocellular carcinoma cell lines. Acta Pharmacol
Sin. 32:939–946. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kang MR, Yang G, Place RF, et al:
Intravesical delivery of small activating RNA formulated into lipid
nanoparticles inhibits orthotopic bladder tumor growth. Cancer Res.
72:5069–5079. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dehennaut V, Loison I, Dubuissez M,
Nassour J, Abbadie C and Leprince D: DNA double-strand breaks lead
to activation of hypermethylated in cancer 1 (HIC1) by SUMOylation
to regulate DNA repair. J Biol Chem. 288:10254–10264. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cheng G, Sun X, Wang J, et al: HIC1
silencing in triple-negative breast cancer drives progression
through misregulation of LCN2. Cancer Res. 74:862–872. 2014.
View Article : Google Scholar
|
|
28
|
Parrella P, Scintu M, Prencipe M, et al:
HIC1 promoter methylation and 17p13.3 allelic loss in invasive
ductal carcinoma of the breast. Cancer Lett. 222:75–81. 2005.
View Article : Google Scholar : PubMed/NCBI
|