Introduction
Human papillomaviruses (HPVs) are small
double-stranded DNA viruses with a genome of ~8 kB. Over 90% of
human cervical carcinoma has been shown to be associated with high
risk HPVs, mainly the serotypes 16 and 18 (1). The mechanisms underlying the
carcinogenesis of high risk HPVs have been studied extensively,
showing that the E6 and E7 proteins are the oncoproteins, which
interact respectively with essential components of the cellular
regulatory machinery, leading to the dysregulated proliferation and
transformation of the infected cells (2). The viral E6 protein’s principal target
is cellular tumor suppressor p53, as a consequence of this
interaction, p53 is labeled with ubiquitin, leading to p53 entering
ubiquitin-mediated degradation system, therefore, the p53 growth
regulatory function is abolished (3). Thus, it has been well accepted that
HPV-E6 targeted p53 degradation resulting in p53 pathway failure,
together with E7 protein interacted with pRb, is responsible for
carcinogenesis (4,5).
p53 is a very important tumor suppressor protein, it
remains at low levels under normal conditions, only in response to
stress, such as UV radiation, DNA damage, hypoxia or virus
infection, p53 gene starts to be activated and the protein
expressed (6–8). Activation of p53 can be modulated at
different levels: increased p53 expression, transformation of the
protein from a latent to an active conformation through different
mechanisms, such as post-translational modification, and
translocation of p53 to the nucleus, where it acts as a
transcriptional factor (9,10). Little is known about whether the
overexpression of high risk HPV-E6 proteins alters the expression
and location of endogenous wild-type (wt) p53 protein, and what
happens next.
The traffic and distribution of E6 protein inside
the infected cells remains elusive. Some authors have shown that
the full-length high risk HPV-E6 was located in the nuclei in
transiently transfected COS cells by immunofluorescence staining
and considered it a nuclear protein (11). Some other studies found it to have
both nuclear and cytoplasmic distribution (12,13).
These confusing results were probably due to the lack of reliable
anti-HPV-16E6 antibodies, and the risk of introducing artifacts
into protein distribution from the fixation procedures (14). We used green fluorescent protein
(GFP) as a tag labeling HPV-16E6 (GFP-16E6) to track its
subcellular location in living cells to get ride of any artificial
interference. In the present experiment, an expression plasmid of
GFP with HPV-16E6 inserted was applied to transfect wt p53 cell
lines, such as MCF-7 and 293T cells, the experiment system would
provide a platform for tracing the E6 protein. Simultaneously, we
observed the expression, localization, and traffic of p53 protein
with immunofluorescence technique, to determine whether the
expression of E6 protein would affect the behavior of p53. By
immunoblotting, we studied the expression level of p53 in the
context of E6. Strikingly, the p53 was not degraded in 24 h in
pGFP-16E6 transfected cells.
We observed the stabilization and increased
expression of p53 in the presence of overexpressed E6 proteins
clearly in the short-term. To avoid the possible effect of
GFP-fusion protein on E6-p53 binding and the degradation of p53, we
used His tagged HPV-16E6 protein (His-16E6) at the same time. We
observed His-16E6 was mainly located in nuclei together with p53,
and the p53 was not degraded in 24 h in His-16E6 expressing cells.
Furthermore, at the later times of transfection, p53 was degraded
gradually whereas the other apoptosis associated proteins such as
bax, Bak, c-myc and cdc2 were increased and bcl-2 was decreased
compared with control. We further observed obvious apoptosis
induced by E6, which was proved to be dependent on p53 expression.
Taken together, in the transient expression system, the high risk
HPV-16E6 was located in nuclei together with endogenous wt p53,
which in turn induced apoptosis.
Materials and methods
Plasmid construction
Full length HPV-16E6 was amplified by PCR from HPV
type 16 complete genome, and then cloned in frame within the C
terminus of the mammalian expression vector pEGFP-C1 (Clontech, CA,
USA) and pcDNA4/To/myc-HisC (Invitrogen, CA, USA) respectively,
producing plasmids pGFP-16E6 and pcDNA4/To/myc-HisC-16E6.
Cell culture and transfection
The human breast adenocarcinoma MCF-7 cells and
human embryonic 293T kidney cells were maintained in RPMI1640
medium (Gibco) supplemented with 10% fetal bovine serum (FBS).
Human colon carcinoma HCT116 cells and HCT116 p53−/−
were maintained in DMEM (Gibco) supplemented with 10% FBS, at 37°C
in a humidified atmosphere of 5% CO2. The MCF-7 and 293T
cells were seeded at approximately 30% confluency on glass
coverslips in 12-well cell culture plates. The cells were
transiently transfected with plasmid pGFP-16E6 and pGFP overnight
using Lipofectamine 2000 transfection reagent (Invitrogen)
following the recommendations of the manufacturer. The reagent:DNA
ratio was 2:1.
Viable-cell imaging by confocal
microscopy
The MCF-7 and 293T cells were grown on glass
coverslips, transfected, and at 21 h post-transfection, coverslips
were mounted on modified glass slides with 10% fetal calf
serum-containing cell culture medium, and used immediately for
imaging. Images of live cells were collected with a Leica confocal
microscope (Leica Microsystems, Wetzler, Germany) at a
magnification of ×400. Fluorescent images were analyzed using Leica
Confocal Software (Leica Microsystems).
Immunocytochemistry
The cells were seeded on glass coverslips at a
density of 100,000 or 200,000 cells/well. Following standard
procedures, they were transfected with plasmid pGFP-16E6, pGFP,
pcDNA4/To/myc-HisC-16E6 and pcDNA4/To/myc-HisC, respectively. After
transfection, the cells were washed with PBS and fixed with 4%
paraformaldehyde for 10 min at room temperature. They were then
rehydrated three times with cold PBS, permeabilized with 1% Triton
X-100 for 5 min on ice, and rinsed with PBS and blocked. The pGFP
and pGFP-16E6 transfected cells were incubated with a primary
antibody against p53 (cell signaling: dilution, 1:500) overnight at
4°C. Subsequently, signal detection was performed using
Cy3-conjugated goat anti-rabbit IgG (Sigma; dilution, 1:200) in
blocking solution for 30 min at room temperature in the dark. Then,
the cells were washed three times with PBS and examined by confocal
microscopy.
The pcDNA4/To/myc-HisC and pcDNA4/To/myc-HisC-16E6
transfected cells were incubated with a primary antibody against
His (Clontech, dilution 1:500) overnight at 4°C, the secondary
antibody of HRP-conjugated goat anti-mouse IgG (Pierce, Rockford,
IL; dilution, 1:500) in blocking solution for 30 min at room
temperature. Reaction products were visualized with
3,3′-diaminobenzidine tetrahydrochloride (DAB) and the slides were
counterstained with hematoxylin.
The pcDNA4/To/myc-HisC and pcDNA4/To/myc-HisC-16E6
transfected cells were also incubated with primary antibodies
against p53 (cell signaling; dilution, 1:500) and against His
(Clontech, dilution 1:500) overnight at 4°C, the secondary
antibodies of FITC-conjugated goat anti-mouse IgG (Sigma; dilution,
1:200) and Cy3-conjugated goat anti-rabbit IgG (Sigma; dilution,
1:200) in blocking solution for 30 min at room temperature in the
dark. Then, the cells were washed three times with PBS and examined
by fluorescence microscopy.
Co-immunoprecipitation
Cells (107) were lysed in RIPA buffer on
ice for 20 min followed by 20 min of centrifugation at 16,000 × g.
Cell lysates 0.5 ml (1 mg of protein) were clarified with 20 μl of
Protein G PLUS-Agarose (Santa Cruz Biotechnology) for 1 h at room
temperature. After centrifugation supernatants were incubated with
2 μg of anti-p53 rabbit antibody overnight at 4°C. Protein G-PLUS
Agarose (20 μl) was then added to cell lysates and incubated for 2
h at room temperature. Beads were washed three times with PBS and
proteins were eluted by the addition of 40 μl 1X electrophoresis
sample buffer. Western blots were performed using anti-GFP mouse
antibody (Clontech, JL, CA).
Immunoblotting analysis
For each sample, 106 cells were collected
by centrifugation (1000 rpm for 5 min), washed once with ice cold
PBS, and lysed in 100 μl RIPA buffer containing 50 mM Tris-HCl (pH
7.4), 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA,
2.5 mM glycerophosphate, 1 mM PMSF, 10 mM NaF, and protease
inhibitors (Complete Mini, Roche Diagnostics, Mannheim, Germany).
Protein concentration was determined using the BCA reagents
(Pierce). Samples (30 μg) were analyzed on 12% SDS polyacrylamide
gels, transferred to PVDF membranes (Invitrogen), and blocked for 1
h at room temperature with 5% non-fat milk in TBS buffer [20 mM
Tris-HCl (pH 7.5), 0.5 M NaCl]. The membranes were then incubated
with the primary antibody overnight at 4°C. After three washes with
TBS, the membranes were incubated with the secondary antibody for
30 min at room temperature. After three additional washes, the
proteins were visualized by enhanced chemiluminescence (ECL)
(Amersham Pharmacia, Piscataway, NJ, USA). The amounts of proteins
were determined by densitometric scanning (Dinco and Rhenium
Biological Imaging System BIS 202).
The primary antibodies used were: anti-p53 (Cell
Signaling; dilution, 1:1,000), (the following were all from Santa
Cruz Biotechnology) anti-bax (dilution 1:500), anti-bcl-2 (dilution
1:500), anti-Bak (dilution 1:500), anti-c myc (dilution 1:500),
anti-cdc2 (dilution 1:500) and anti-β actin (dilution, 1:10,000).
The blots were counterstained with goat anti-mouse or goat
anti-rabbit IgG conjugated with HRP (Pierce).
Analysis of apoptosis by flow cytometry
using Annexin V and PI double staining
The transfected cells were harvested after 24, 48,
and 72 h by trypsinization, and apoptotic cells were assayed with
the Annexin V-APC Apoptosis Detection kit (Bender Medsystems,
Burlingame, CA). Briefly, 1×106 cells in 100 μl binding
buffer were stained with 5 μl Annexin V-APC and 10 μl PI (final
concentration, 1 μg/ml) by mixing and incubating on ice for 10 min
in the dark. The cells were analyzed by flow cytometry. The data
were processed using the Cell Quest software.
Statistics
All data were recorded as means ± standard
deviation, and analyzed by the SPSS 11.0 software. Analysis of data
was performed using one-way ANOVA for multiple comparisons.
P-values <0.05 were considered statistically significant.
Results
GFP-16E6 is a nuclear protein
Viral E6 coding regions were inserted within the C
terminus of the pGFP vector, producing plasmid pGFP-16E6. We
transiently transfected pGFP-16E6 in MCF-7 and 293T cells, which
allow E6 proteins to be expressed as GFP-16E6 fusion proteins. By
confocal microscopy, we observed the subcellular localization of
GFP-16E6 and GFP in viable cell images. The E6 fusion proteins may
have low or high expression levels at different times, and this
could affect the distribution of E6. Therefore, we observed the
localization and expression of proteins from 6 to 72 h
post-transfection. The results indicated that GFP-16E6 protein was
expressed essentially in the nucleus from 6 h post-transfection.
Its expression increased gradually, and reached its maximum
expression level at 21 h (P<0.001). Then, it decreased gradually
and disappeared after one week. During this whole period, no change
was observed in protein localization. As control, we observed the
expression of GFP alone. It exhibited a diffused signal, and was
present in both the nucleus and cytoplasm from 6 h to one week
post-transfection. In addition, its location did not change at any
time (Fig. 1A).
The 293T cells were also used to study E6
localization. The cellular distributions of GFP and GFP-16E6
proteins were similar to those in MCF-7 cells (Fig. 1A). The analysis of relative
fluorescence signal intensity of GFP-16E6 in different cell lines
is shown in Fig. 1B.
Co-localization of GFP-16E6 and p53
protein
Because high risk E6 can bind to p53 (15), we suspected that the GFP-16E6 and
p53 might locate together. Using MCF-7 and 293T cells, we
investigated endogenous wt p53 localization by
immunocytofluorescence technique. The results showed that p53
protein was mainly located in the nuclei of pGFP transfected cells.
In pGFP-16E6 transfected cells, the distribution of p53 protein was
not changed, and it was located in the nuclei together with
GFP-16E6. Fig. 2A shows
representative images of the co-localization of GFP-16E6 and p53
proteins.
GFP-16E6 interacts with p53 in vivo
In the present study, we observed p53 and E6 protein
located together, it was necessary to determine whether GFP-16E6
could interact with p53 in vivo. We investigated the
potential role of the interaction between endogenous wt p53 and E6
protein by performing immunoprecipitation assay with anti-p53
antibodies. Then, by western blot analyses with anti-GFP antibodies
it was shown that p53 interacted with GFP-16E6 protein. Moreover,
as a control, in GFP expressing cells, only the GFP protein that
lacked the E6 protein was unable to interact with p53 (Fig. 2B). Therefore, in present study, we
observed that p53 could interact with HPV-16E6 protein in
vivo. This is supported by a report that p53 has binding sites
for high risk HPV-E6 (15).
p53 is increased in 24 h transfection
with pGFP-16E6
It has been extensively shown that p53 was degraded
by 26S proteasome via the ubiquitin pathway (16). Since E6 interaction with p53, we
next to determine whether the overexpressed E6 affected this
process of p53 degradation. At 24 h post-transfection, the GFP and
GFP-16E6 expressing cells were treated with MG132, a potent
inhibitor of 26S, and then examined p53 level by immunoblotting.
The ubiquitination of p53 was readily detected in both GFP and
GFP-16E6 expressing cells upon MG132 treatment. The results showed
that the ubiquitination of p53 was significantly lower in GFP-16E6
cells than GFP control cells (Fig.
3). Thus, p53 was increased in 24 h in GFP-16E6 expressing
cells, which was partly due to the decreased ubiquitin-mediated
degradation of p53.
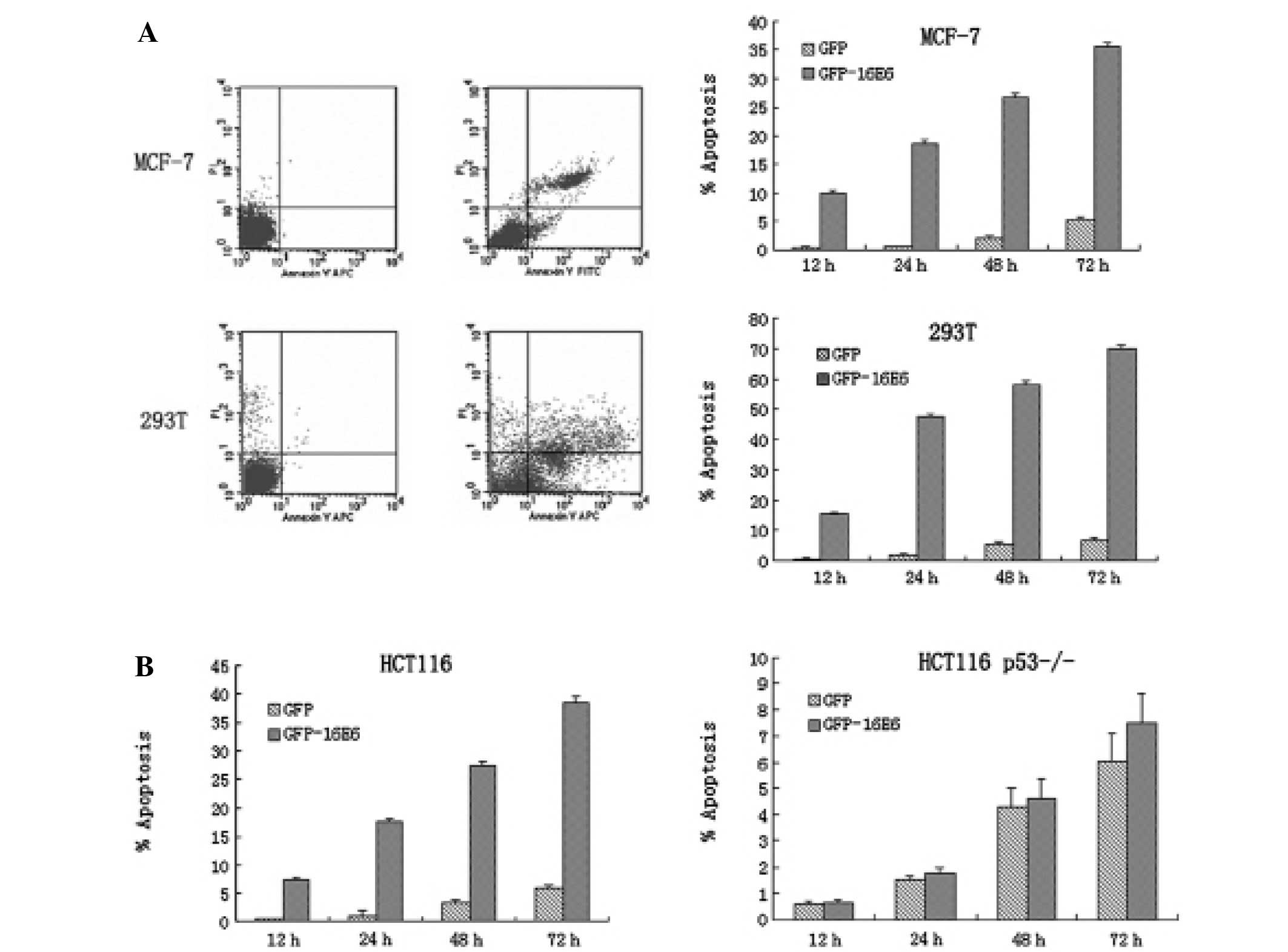
GFP-16E6 induces apoptosis in transfected
cells
With the overexpression of oncoprotein E6, we next
asked whether it can promote cell apoptosis along with the
expression of p53. By Annexin V and PI double-staining combined
with flow cytometry, we observed obvious apoptosis in GFP-16E6
expressing MCF-7 cells. Apoptosis occurred at 12 h
post-transfection, and it increased gradually. From 12 to 72 h
post-transfection, apoptosis was prominent compared with GFP alone
expressing cells (P<0.001). For 293T cells, we obtained similar
result to that in MCF-7 cells (Fig.
4A).
By western blotting, we examined p53 level in
GFP-16E6 cells at 12, 24, 48 and 72 h post-transfection. The result
indicated, for GFP-16E6 expressed cells, p53 was increased in 24 h
post-transfection, and then it degraded gradually at later times.
Accordingly, the other apoptosis associated proteins, such as bax,
Bak, c-myc and cdc2 were increased and the bcl-2 was decreased
compared with GFP control cells (Fig.
5). On the other hand, p53 in GFP-16E6 expressing MCF-7 cells
was degraded more than 293T cells. The various activity of E6 to
target and degrade p53 was partly due to p53 conformation in
different cells (17). Thus, our
data indicated the GFP-16E6 could induce apoptosis in wt p53 cell
lines.
p53 is necessary for GFP-16E6 induced
apoptosis
Our result showed there was obvious apoptosis
induced by E6 along with the expression of p53. To further
investigate whether p53 was necessary for apoptosis, we transfected
pGFP-16E6 in both HCT116 and HCT116 p53−/− cells. We
observed, in the context of HPV-16E6, there was obvious apoptosis
in HCT116 cells, whereas there was no obvious apoptosis in HCT116
p53−/− cells (Fig. 4B).
Therefore, we concluded p53 was necessary for GFP-16E6 induced
apoptosis.
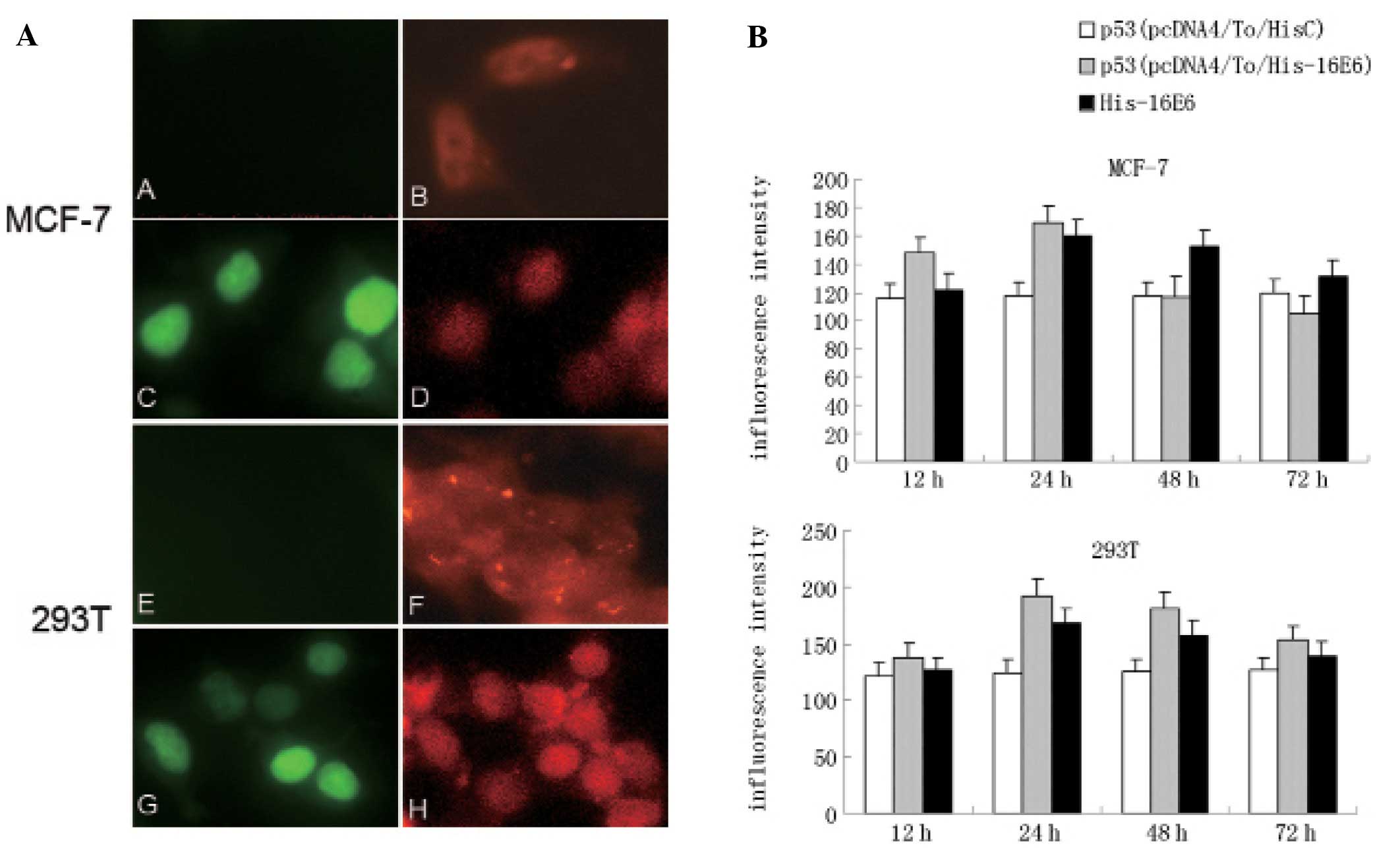
p53 is increased in 24 h by His tagged
HPV-16E6 protein expression
To avoid the possible effect of GFP-fusion protein
on E6-p53 binding and the degradation of p53, we used His with
HPV-16E6 fusion protein for further research. By
immunocytochemistry stain, we observed the His-16E6 expression in
both MCF-7 and 293T cells (Fig. 6).
Furthermore, using immonofluorescent technique, we clearly observed
His-16E6 was mainly located in the nuclei together with p53
(Fig. 7A). Comparing with
pcDNA4/To/myc-HisC control cells, the p53 protein expression level
was increased in 24 h along with the His-16E6 expression (Fig. 7B).
Discussion
The present study provides a novel observation that
the transiently expressed high risk HPV-16E6 with GFP fusion
protein induced apoptosis in wild-type (wt) p53 cells. We showed
HPV-16E6 was a nuclear protein, and the endogenous wt p53 was
located in nuclei together with HPV-16E6. Furthermore, there was
obvious apoptosis induced by HPV-16E6 which was dependent on p53
expression.
Many studies on the localization of E6 protein have
led to contradictory results, most probably due to the low level of
endogenous E6 protein and the poor reactivity of the available
anti-E6 antibodies. In the present study, we used GFP with HPV-16E6
fusion proteins to dynamically trace the traffic and localization
of E6 proteins in different cell lines. GFP is a convenient,
genetically encoded intrinsic fluorescent molecular label that has
been widely and successfully used to study protein distribution in
cells (18). Our results suggested
that GFP-16E6 was mainly expressed in the nucleus of transfected
cells. This was consistent with the study by Tao et al who
showed that the high risk full-length E6 protein was distributed
predominantly in the nucleus of transfected COS-1 cells (14). Thus, it seems likely that the
localization of HPV-16E6 in the nucleus is consistent with E6
having some transcription factors (19–21)
including p300/CBP, IRF-3, c-Myc or transcriptional co-activators
(22) as cellular binding partners
which were mainly located in the nuclei.
The tumor suppressor p53 causes cell cycle arrest or
apoptosis in response to DNA damage and other forms of stress
(23). The ability to localize into
the nucleus is essential for p53 to act as a transcription factor.
Previous studies have shown the p53 interacting with HPV-E6 playing
a very important role in carcinogenesis. We next asked whether the
presence of E6 altered the subcellular localization of p53. About
50% of tumor cells contain mutated p53 but only wt p53 is detected
in the HPV sequence positive tumors (24). Therefore, we chose MCF-7 and 293T
cells, which are wt p53 cell lines, they can partly stimulate
HPV-infected cells. We observed the endogenous wt p53 was mainly
located in the nuclei together with HPV-16E6, furthermore, we
proved E6 interaction with p53 in vivo. These data were
consistent with authors who claimed that E6 did not alter the
cellular localization of p53, and it was co-localized with p53
(12).
Next, we examined the level of endogenous wt p53 in
the context of HPV-16E6. Of note, the result indicated, the
endogenous wt p53 was not degraded but increased in 24 h in
GFP-16E6 expressing cells. Also, the same result was obtained
several times in both MCF-7 and 293T cells. To avoid the possible
effect of GFP-fusion protein on E6-p53 binding and the degradation
of p53, we expressed the His with HPV-16E6 fusion protein further
for further investigation. We obtained similar result to GFP tagged
HPV-16E6. The His-16E6 was mainly located in the nuclei together
with p53, and the p53 was increased in 24 h in His-16E6 expressing
cells. This agreed with the GFP tag available for HPV-E6 protein
research, and it did not effect the interaction of p53 and E6
(14,25,26).
This result was supported by Kawamata et al who reported
that p53 protein expression levels in normal cervical keratinocytes
were not degraded by the introduction of HPV-16E6, probably due to
a tight transcriptional regulation of p53 (27). This was also supported by increasing
evidence, which clearly suggesting that the expression of E6 does
not necessarily equate to a p53 null background (28,29).
In present study, we clearly observed p53 was
located in the nuclei together with HPV-16E6. Furthermore, we
observed in both GFP tag and His tag expressed system, p53 was
increased in 24 h transfected with HPV-16E6. This confirmed that
the infected cells recognize virus replication as a DNA damage
stress and elicit host surveillance mechanism which ultimately
induces activation of p53 (30). We
observed obvious apoptosis induced by HPV-16E6 along with the
expression of p53. This finding agreed with the possibility that
p53 can transactivate other genes to induce apoptosis in response
to the overexpression of E6 (31).
Accordingly, our result showed the apoptosis associated proteins,
such as bax, Bak, c-myc and cdc2 (18) were upregulated, whereas the bcl-2
was downregulated by HPV-16E6 expression. To investigate whether
apoptosis-induced by E6 was dependent on p53 expression, we took
advantage of HCT116 and HCT116 p53−/− cells, which are a
pair of cells: one contain wt p53, the other one is wt p53 null.
Also, we proved p53 was necessary for E6 induced apoptosis. It
seems likely the activity of p53 is a key event for anti-virus
response. There are other viruses, such as EB and Africa Swine
Fever virus, they both can induce apoptosis which was dependent on
p53 activation (32,33).
In conclusion, we observed that transiently
expressed GFP-16E6 was located in the nuclei together with the
endogenous wt p53 protein, which in turn induced apoptosis.
Therefore, our experiments provided new insight into the
interaction of high risk HPV-E6 and endogenous wt p53.
References
|
1
|
Antonsson A, Spurr TP, Chen AC, et al:
High prevalence of human papillomaviruses in fresh frozen breast
cancer samples. J Med Virol. 83:2157–2163. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jones DL, Thompson DA and Munger K:
Destabilization of the RB tumor suppressor protein and
stabilization of p53 contribute to HPV type 16 E7-induced
apoptosis. Virology. 239:97–107. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Honda R and Yasuda H: Activity of MDM2, a
ubiquitin Ligase, toward p53 or itself is dependent on the RING
finger domain of the ligase. Oncogene. 19:1473–1476. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
DiMaio D: The role of human
papillomaviruses in cancer. J Neurovirol. 14:50. 2005.
|
|
5
|
Liu YM, McKalip A and Herman B: Human
papillomavirus type 16 E6 and HPV-16 E6/E7 sensitize human
keratinocytes to apoptosis induced by chemotherapeutic agents:
Roles of p53 and caspase activation. J Cell Biochem. 78:334–349.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Meek DW: The p53 response to DNA damage.
DNA Repair. 3:1049–1056. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chang YC, Liao CB, Hsieh PY, Liou ML and
Liu YC: Expression of tumor suppressor p53 facilitates DNA repair
but not UV-induced G2/M arrest or apoptosis in Chinese hamster
ovary CHO-K1 cells. J Cell Biochem. 103:528–537. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wei J, O’Brien D, Vilgelm A, et al:
Interaction of Helicobacter pylori with gastric epithelial
cells is mediated by the p53 protein family. Gastroenterology.
134:1412–1423. 2008.
|
|
9
|
Mayer C and Grummt I: Cellular stress and
nucleolar function. Cell Cycle. 4:1036–1038. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Qian H, Wang T, Naumovski L, Lopez CD and
Brachmann RK: Groups of p53 target genes involved in specific p53
downstream effects cluster into different classes of DNA binding
sites. Oncogene. 21:7901–7911. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sherman L and Schlegel R: Serum- and
calcium-induced differentiation of human keratinocytes is inhibited
by the E6 oncoprotein of human papillomavirus type 16. J Virol.
70:3269–3279. 1996.PubMed/NCBI
|
|
12
|
Liang XH, Volkmann M, Klein R, Herman B
and Lockett SJ: Co-localization of the tumor-suppressor protein p53
and human papillomavirus E6 protein in human cervical carcinoma
cell lines. Oncogene. 8:2645–2652. 1993.PubMed/NCBI
|
|
13
|
Kelley ML, Keiger KE, Lee CJ and
Huibregtse JM: The global transcriptional effects of the human
papillomavirus E6 protein in cervical carcinoma cell lines are
mediated by the E6AP ubiquitin ligase. J Virol. 79:3737–3747. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tao M, Kruhlak M, Xia S, Androphy E and
Zheng ZM: Signals that dictate nuclear localization of human
papillomavirus type 16 oncoprotein E6 in living cells. J Virol.
77:13232–13247. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li X and Coffino P: High-risk human
papillomavirus E6 protein has two distinct binding sites within
p53, of which only one determines degradation. J Virol.
70:4509–4516. 1996.PubMed/NCBI
|
|
16
|
Momand J and Zambetti GP: Mdm-2: ‘big
brother’ of p53. J Cell Biochem. 64:343–352. 1997.
|
|
17
|
Butz K, Shahabeddin L, Geisen C,
Spitkovsky D, Ullmann A and Hoppe-Seyler F: Functional p53 protein
in human papillomavirus-positive cancer cells. Oncogene.
10:927–936. 1995.PubMed/NCBI
|
|
18
|
Chalfie M, Tu Y, Euskirchen G, Ward WW and
Prasher DC: Green fluorescent protein as a marker for gene
expression. Science. 263:802–805. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zheng Y, Zhang J and Rao Z: Ribozyme
targeting HPV16 E6E7 transcripts in cervical cancer cells
suppresses cell growth and sensitizes cells to chemotherapy and
radiotherapy. Cancer Biol Ther. 3:1129–1135. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gross-Mesilaty S, Reinstein E, Bercovich
B, et al: Basal and human papillomavirus E6 oncoprotein-induced
degradation of Myc proteins by the ubiquitin pathway. Proc Natl
Acad Sci USA. 95:8058–8063. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ronco LV, Karpova AY, Vidal M and Howley
PM: Human papillomavirus 16 E6 oncoprotein binds to interferon
regulatory factor-3 and inhibits its transcriptional activity.
Genes Dev. 12:2061–2072. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kumar A, Zhao Y, Meng G, et al: Human
papillomavirus oncoprotein E6 inactivates the transcriptional
coactivator human ADA3. Mol Cell Biol. 22:5801–5812. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ferrigno P and Silver PA: Regulated
nuclear localization of stress-responsive factors: how the nuclear
trafficking of protein kinases and transcription factors
contributes to cell survival. Oncogene. 18:6129–6134. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hollstein M, Rice K, Greenblatt MS, et al:
Database of p53 gene somatic mutations in human tumors and cell
lines. Nucleic Acids Res. 22:3551–3555. 1994.PubMed/NCBI
|
|
25
|
Diaz D, Santander MA and Chavez JA: HPV-16
E6 and E7 oncogene expression is downregulated as a result of Mdm2
knockdown. Int J Oncol. 41:141–146. 2012.PubMed/NCBI
|
|
26
|
Hwang SJ, Suh MJ, Yoon JH, et al:
Identification of characteristic molecular signature of Mullerian
inhibiting substance in human HPV-related cervical cancer cells.
Int J Oncol. 39:811–820. 2011.PubMed/NCBI
|
|
27
|
Kawamata Y, Mitsuhashi A, Unno Y, et al:
HPV 16-E6-mediated degradation of intrinsic p53 is compensated by
upregulation of p53 gene expression in normal cervical
keratinocytes. Int J Oncol. 21:561–567. 2002.PubMed/NCBI
|
|
28
|
Fouret P, Dabit D, Sibony M, et al:
Expression of p53 protein related to the presence of human
papillomavirus infection in precancer lesions of the larynx. Am J
Pathol. 146:599–604. 1995.PubMed/NCBI
|
|
29
|
Isaacs JS, Chen P, Garza A, Hansen MF,
Barrett JC and Weissman BE: Failure of HPV E6 to rapidly degrade
p53 in human HeLa x PNET cell hybrids. Oncogene. 14:1669–1678.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shin YC, Nakamura H, Liang X, et al:
Inhibition of the ATM/p53 signal transduction pathway by Kaposi’s
sarcoma-associated herpesvirus interferon regulatory factor 1. J
Virol. 80:2257–2266. 2006.PubMed/NCBI
|
|
31
|
Accardi R, Dong W, Smet A, et al: Skin
human papillomavirus type 38 alters p53 functions by accumulation
of deltaNp73. EMBO Rep. 7:334–340. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li L, Guo L, Tao Y, et al: Latent membrane
protein 1 of Epstein-Barr virus regulates p53 phosphorylation
through MAP kinases. Cancer Lett. 255:219–231. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Granja AG, Nogal ML, Hurtado C, et al:
Modulation of p53 cellular function and cell death by African swine
fever virus. J Virol. 78:7165–7174. 2004. View Article : Google Scholar : PubMed/NCBI
|