Introduction
Breast cancer is the most common malignancy in women
and is a significant worldwide health problem, accounting for
approximately 1.3 million new cases and more than 450,000
cancer-related deaths annually in the world (1). Although recent advancements in early
detection, prevention and treatments have effectively reduced
breast cancer incidence and improved patient survival, a number of
patients are still diagnosed at advanced stages of the disease and
chemotherapy is the mainstream method of treatment against these
advanced diseases. However, nearly 50% of such patients develop
multidrug resistance (MDR) to chemotherapeutic agents during the
course of treatment (2). Previous
studies have demonstrated different mechanisms responsible for drug
resistance, such as drug inactivation, extrusion of the drug by
enhanced drug efflux pumps, or changes in drug target during the
course of treatment (3–5). Thus, novel approaches to discover
molecular targeting therapeutic agents and to re-sensitize the
existing drugs to therapy may aid in effectively controlling
advanced breast cancer and may improve the prognosis of these
patients.
Therefore, our research focused on microRNAs
(miRNAs), which are a novel class of endogenous non-coding small
RNAs, and function in post-transcriptional regulation of the target
gene expressions by binding to the target mRNA to inhibit its
translation and/or to degrade the target mRNA molecules (6). Altered expression of different miRNAs
has been demonstrated to play an important role in cancer
development and progression, as well as in drug resistance. Thus,
detection of miRNA was used as a biomarker for early detection of
tumorigenesis, drug treatment outcome, prediction of prognosis and
disease progression (7).
Nevertheless, our research focused mainly on aberrant expression
and functions of miRNAs in drug responses. Indeed, previous studies
have shown that dysregulation of miRNA expression and function
affected the sensitivity of various types of cancer to
chemotherapies (8,9). For example, miR-34a expression was
able to modulate expression of Bcl-2 and cyclin D1 proteins, and
responded to the chemosensitivity of breast cancer MCF-7 cells to
docetaxel treatment (10). miR-451
and miR-27 were shown to be involved in the resistance of MCF-7
cells to the chemotherapeutic drug doxorubicin by mediated MDR-1
expression (11,12). Thus, targeting of miRNA expression
could be used as a novel therapeutic approach for the treatment of
breast cancer (13,14).
In the present study, we investigated a particular
miRNA, miR-195, which is localized at chromosome 17p13.1 and
clustered with other miRNAs, such as miR-497, to form the miR-15
family (15). Previous studies
suggested that miR-195 plays a tumor-suppressor role in various
types of cancer. Following overexpression in different cancer
cells, miR-195 was able to suppress cell cycle progression and
tumor cell invasion, but sensitized the tumor cells to treatment
with a different anticancer drug (16,17).
Thus, we first determined the role of miR-195 in the development of
anticancer drug resistance in breast cancer cells and then the
underlying molecular mechanism. Subsequently, we searched the
GenBank database for identification of the target gene of miR-195
and found that Raf-1 could be a potential target of miR-195.
Proto-oncogene serine/threonine-protein kinase
(Raf-1) functions as a part of the MAPK/ERK signal transduction
pathway. Once activated, Raf-1 phosphorylates and activates MEK1
and MEK2 protein kinases and then, in turn, phosphorylates and
activates the serine/threonine-specific protein kinases ERK1 and
ERK2 to control expression of various genes (such as Bcl-2 and
P-glycoprotein) in the regulation of cell cycle, cell migration,
apoptosis and differentiation (18–21).
To date, there has been no report showing Raf-1 mutations in the
clinic, but Raf-1 overexpression has frequently been reported in
different human tumors (22).
Several studies have shown that inhibition of key kinases of the
Ras/Raf/MEK/Erk signaling pathway were able to regulate tumor cell
proliferation and apoptosis (18,23).
Thus, in this study, we investigated whether and how miR-195
targets Raf-1 expression to sensitize breast cancer cells to
Adriamycin treatment.
Materials and methods
Breast cancer tissue samples
We obtained tissue specimens from 17 breast cancer
patients at the Second People's Hospital in Neijiang, Sichuan,
China. The tissue specimens were available from previously
untreated patients who received an Adriamycin-containing regimen
for at least six months as adjuvant therapy. The chemotherapy
regimen consisted of four 21-day cycles of AC (doxorubicin, 60
mg/m2 on day 1; cyclophosphamide, 600 mg/m2
on day 1) followed by four 21-day cycles of docetaxel (100
mg/m2 on day 1) or six 21-day cycles of FAC (5
fluorouracil, 500 mg/m2 on day 1; doxorubicin, 50
mg/m2 on day 1; cyclophosphamide, 500 mg/m2
on day 1), under the treatment of which 10 patients had a
recurrence at least 6 months after their primary occurrence and
were classed as multidrug-resistant, while the other seven patients
did not. The drug-resistant or -sensitive tumor tissues were
resected and used for this study. In addition, the 17 noncancerous
breast tissues were also collected from the Second People's
Hospital. The tissues were surgically resected and snap-frozen and
stored in liquid nitrogen until use. All cases of breast cancer
were pathologically confirmed. All patients provided written
informed consent for the use of their tissues and this study was
approved by the Hospital's Committee of Human Subject Protection.
Clinicopathological characteristics of these patients are presented
in Table I.
 | Table IClinical characteristics of the 30
patients studied. |
Table I
Clinical characteristics of the 30
patients studied.
|
Characteristics | All patients
(N=17) | % |
|---|
| Age at
diagnosis | 49 years
(28–82) | |
| Gender | Female | |
| Tumor stage | 6 | |
| T0 | 2 | 35 |
| T1 | 9 | 12 |
| T2 | 8 | 53 |
| N0 | 2 | 47 |
| N1 | 7 | 12 |
| N2 | 11 | 41 |
| M0 | 6 | 65 |
| M1 | 35 | |
| ER |
| Negative | 9 | 53 |
| Positive | 8 | 47 |
| PR |
| Negative | 10 | 59 |
| Positive | 7 | 41 |
| Recurrence |
| Yes | 10 | 59 |
| No | 7 | 41 |
Cell lines and culture
A human breast cancer cell line MCF-7 and a human
mammary gland epithelial cell line HBL-100 were obtained from
KeyGEN Biotech Co. (Nanjing, China). To generate the
multidrug-resistant subline MCF-7/ADR, MCF-7 cells were established
by serial passages and incubations with increasing Adriamycin
concentrations and were maintained in the presence of 500 ng/ml
Adriamycin, as previously described (24), and reached a multidrug-resistant
phenotype. These cell lines were maintained in Dulbecco's modified
Eagle's medium (DMEM; Invitrogen) with 10% fetal bovine serum (FBS)
(both from Invitrogen, Carlsbad, CA, USA) and
penicillin/streptomycin at 37°C in a humidified atmosphere with 5%
CO2.
RNA isolation and quantitative reverse
transcriptase-polymerase chain reaction (qRT-PCR)
Total cellular RNA from tissues and cultured cells
were isolated using a TRIzol Reagent (Invitrogen) according to the
manufacturer's instructions. Prior to RNA isolation, freezing
tissues were cut into 5 mm3 pieces of each sample and
then grinded into finely ground particles. The concentration of
these RNA samples was quantitated by measuring the absorbance at
260 vs. 280 nm. Next, the RNA samples were further isolated with a
mirPremier miRNA isolation kit (mirVana miRNA isolation kit from
Ambion, Austin, TX, USA). The stem-loop reverse transcription was
performed to amplify the mature miR-195 using the RT primer,
5′-TGTCAGGCAACCGTATTCACCGGAGTGGT GGGAAG-3′ and U6 small RNA with a
U6 RT primer, 5′-CGCTTCACGAATTTGCGTGTCAT-3′. Subsequently, reverse
transcription was carried out at 25°C for 10 min, 42°C for 1 h, and
85°C for 5 min using a kit from Takara Biotechnology Co. (Dalian,
China) for qPCR amplification. SYBR-Green (SYBR-Green I from
Invitrogen) was utilized according to the manufacturer's
instructions. hsa-miR-195 and U6 snRNA primers were obtained from
Ambion; hsa-miR-195 primers, 5′-CCTAGCAGCACAGAAA-3′ and
5′-GAGCAGGCTGGA GAA-3′; U6, 5′-CTCGCTTCGGCAGCACATA-3′ and 5′-CGC
TTCACGAATTTGCGTG-3′. Each sample was analyzed in triplicate and the
cycle number (CT) at which the amplicon concentration crossed a
defined threshold was determined for each individual miRNA. The
relative miR-195 levels were normalized to U6 levels and calculated
using the equation 2−ΔΔCt ± standard deviation (SD).
Lentivirus carrying Raf-1 siRNA, cell
infection and miR-195 mimics and inhibitor
We utilized an online siRNA design software
(http://design.RNAi.jp/) to determine Raf-1 target
oligonucleotide sequence (5′-GAGACATGAAATCCAA CAA-3′; GenBank
#NM_002880) and synthesized the sense and antisense Raf-1
oligonucleotides according to Shanghai Sangon Biological Corp.
(Shanghai, China); siRAF1 sense,
5′-GATCCGAGACATGAAATCCAACAATACTTCCTGTC
AGATATTGTTGGATTTCATGTCTCTTTTTG-3′ and antisense,
5′-AATTCAAAAAGAGACATGAAATCCAACAAT
ATCTGACAGGAAGTATTGTTGGATTTCATGTCTCG-3′. The lentiviral-shRNA vector
(pLenOR-GPH Raf-1 vector) was constructed by cloning a
PCR-amplified fragment of Raf-1 siRNA into the
BamHI/EcoRI site of pcDNA-GFP and then amplifying the
GFP-tagged Raf-1 siRNA with primers flanked by the restriction
enzyme sites and cloned into the pLenOR-GPH. Empty pLenOR-GPH
vector, without Raf-1 siRNA, was used as the negative control.
Lentivirus was produced with the Lenti-Pac™
Lentivirus expression system (Invabio), together with the transfer
plasmid pLenOR-GPH-Raf-1, packaging plasmid pRsv-REV, pMDlg-pRRE,
and pMD2G, all of which are lentivirus shuttle carriers.
Recombinant lentiviruses were produced by cotransfection of
siRNA-transferring plasmids and plasmids were packaged into 293FT
cells with the calcium phosphate method. Subsequently, the cell
culture supernatant was harvested after 48-h cultures; debris was
dumped by 4,000 × g centrifugation for 10 min at 4°C. The virus was
then purified by a Plus-20 kit (Millipore, USA) and stored at
−80°C. The production of control virus enveloping pLenOR-GPH
followed the same protocol.
For titer determination, 293T cells were seeded into
6-well plates at a density of 1×105 cells/well. The next
day, lentivirus was diluted 10 times in serum-free culture
consistently to prepare five concentrations of solution and
lentivirus was added in 6-well plates. Cells were harvested after 4
days and the titer of the virus was estimated by the flow
cytometric test. Titer [transduction unit (TU)/ml] = Cell quantity
×105 × GFP cell proportion × 1,000/M (M, 1 μl-contained
amount of virus).
To infect tumor cells with lentivirus, cells were
seeded into 6-well plates (5×105/well with 2 ml of DMEM
containing 10% FCS) overnight, washed with serum-free DMEM,
lenti-Raf-1 storage solution was diluted to appropriate
concentrations and then infected with a negative control lentiviral
vector or the lentiviral vector carrying Raf-1 siRNA at a
multiplicity of infection (MOI) of 20 and added to cells in the
presence of 8 μg/ml Polybrene (Sigma, St. Louis, MO, USA). After 12
h at 37°C, the cells were washed and 2 ml of fresh growth medium
was added. After 72 h, the cells were harvested for analyses.
Plasmids carrying miR-195 mimic or inhibitor were
purchased from RiboBio Co., Ltd. (Ghuangzhou, China) and used to
transfect breast cancer cells. In brief, cells were plated at a
density of 5×105/ml in 6-well cell culture plates (BD
Biosciences, Mountain View, CA, USA) overnight and then transfected
with 50 nM of miR-195 mimic or inhibitor with Lipofectamine™ 2000
and a nonspecific miRNA mimic or inhibitor was used as negative
control plasmid. Twenty-four hours later, the cells were collected
for further analysis.
MTT cell viability assay
To perform
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT;
Sigma) cell viability assay, MCF-7, MCF-7/ADR and HBL-100 cells
infected with lentivirus or transfected with miR-195 plasmid were
seeded in 96-well plates (5×103 cells/well) and were
treated with various concentrations of Adriamycin (10, 50, 100, 200
and 500 ng/ml) at the indicated time-points. At the end of the
experiments, the cell cultures were supplemented with 150 μl of 0.5
mg/ml MTT assay and incubated for an additional 4 h. Then, dimethyl
sulfoxide (0.1% DMSO) was added to the cell culture to dissolve the
formazan crystals and incubated for 10 min at room temperature. The
absorbance rate of the cell cultures was read at 570 nm by using a
Vmax Microplate Reader (Bio-Rad, Hercules, CA, USA). Each
experiment was performed in triplicate and repeated at least once.
Cell viability (%) = 100 × (A1/A0), where A1 and A0 were the
absorbance rate of treated and untreated cells, respectively.
Flow cytometric assay
In vitro cell apoptosis and cell death assays
were assessed by flow cytometry with an Annexin V-fluorescein
isothiocyanate (FITC)/propidium iodide assay kit (BD Biosciences)
according to the manufacturer's protocol. Briefly, cells
(5×105 cells) were seeded in a 6-well plate and
transfected with or without miR-195 mimics or inhibitors, or
infected with Raf-1 siRNA lentivirus, or the control lentivirus,
and then incubated for 48 h. The medium was then replaced with
either fresh medium (control medium) or with medium supplemented
with various concentrations of Adriamycin. At the end of
experiments, cells were washed twice with phosphate-buffered saline
(PBS) and resuspended in Annexin V-binding buffer. Cell suspension
was then incubated with 5 μl of Annexin V-FITC and incubated for 10
min at 4°C in the dark. After adding 10 μl of propidium iodide and
incubating for another 10 min at 4°C in the dark, the cells were
read by a FACScan flow cytometer (BD Biosciences, San Jose, CA,
USA). The fraction of cell population in different quadrants was
analyzed using the quadrant statistics, i.e., cells in the lower
right quadrant represented early apoptosis and cells in the upper
right quadrant represented late apoptotic cells.
Prediction of miRNA targeting genes
The computer-based miRNA target detection programs,
i.e., TargetScan (http://www.targetscan.org/), PITA (http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html),
and miRanda (http://www.microrna.org/microrna/home.do), were used
to predict miR-195 binding sites of targeting gene mRNA according
to the website instructions.
Immunofluorescence staining analysis
MCF-7 and MCF-7/ADR cells infected with lentivirus
or transfected with miR-195 mimic plasmid were seeded in 6-well
plates (5×103 cells/well). The cells were fixed in 4%
paraformaldehyde for 10 min and then incubated in 1% BSA/10% normal
goat serum/0.3 M glycine in 0.1% PBS-Tween for 1 h to permeabilize
the cells and block non-specific protein-protein interactions. The
cells were incubated with the total Raf-1 (Abcam plc, Cambridge,
UK) at a 1/200 dilution for 12 h at 37°C and washed 3 times with 1X
PBS for 5 min each wash. Secondary antibody against total Raf-1
which is conjugated to a green fluorescence probe (Abcam plc) was
added and it was incubated for 2 h at 37°C.
Protein extraction and western
blotting
To extract total cellular protein from breast cancer
cell lines, cells were trypsinized, washed with DMEM and
centrifuged at 500 × g for 10 min at 4°C and lysed in a lysis
buffer containing 20 mmol/l of Tris-HCl (pH 7.5), 1% CHAPS, 150
mmol/l of NaCl, 10% glycerol, 1 mmol/l of
Na3VO4, and the complete protease inhibitor
mixture (Roche Diagnostics GmbH, Mannheim, Germany). Samples were
placed on ice for 20 min and then centrifuged at 12,000 × g for 20
min at 4°C. For western blotting, concentration of these protein
samples was measured by the BCA (Sigma) method, and equal amounts
of samples (30–50 μg/lane) were then loaded onto 8–12% of
SDS-polyacrylamide gel for electrophoresis and subsequently
transferred to nitrocellulose membranes (0.45 μm; Millipore). Next,
the membranes were incubated with 5% non-fat dry milk for 1 h at
room temperature, followed by incubation with a mouse monoclonal
anti-human Raf-1, Bcl-2 or P-glycoprotein antibody at a dilution
factor 1:500 (Abcam Inc., Cambridge MA, USA) or mouse anti-human
GAPDH monoclonal antibody at a dilution factor 1:500 (Abcam) at 4°C
overnight. The following day, the membranes were washed with 0.1%
Tween-20 in PBS (PBS-T) and then incubated with a secondary
antibody conjugated with goat anti-mouse IgG (1:3,000; Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA) in blocking buffer for 1
h, followed by exposure to the Enhanced Chemiluminescence Luminal
reagent (Santa Cruz Biotechnology, Inc.) briefly and then analyzed
with software Image-Pro Plus 5.1 (Media Cybernetics, Inc.,
Bethesda, MD, USA).
Statistical analysis
The data are expressed as the mean ± SEM.
Statistical comparison of the data was performed using the t-test
between two groups or one-way ANOVA or using a post hoc Tukey's
test for multiple comparisons between more than two groups. Data
were analyzed using an SPSS statistical package for Windows (SPSS
Inc., Chicago, IL, USA). A value of P<0.05 was considered to
indicate a statistically significant difference.
Results
Reduced miR-195 expression in breast
cancer tissues is associated with chemotherapy response
In this study, we first determined expression levels
of miR-195 in HBL-100, MCF-7 and MCF-7/ADR cell lines and our data
showed that miR-195 expression was significantly lower in MCF-7 and
MCF-7/ADR cells than in HBL-100 (Fig.
1A). Then, we assessed miR-195 expression in clinical breast
tissue samples, which were obtained from chemotherapy-sensitive and
-resistant breast cancer tissues using qRT-PCR. Our data showed
that levels of miR-195 expression were significantly reduced in
breast cancer and drug-resistant tissue specimens compared to
distant non-cancerous tissues in all 17 cases (Fig. 1B). These data suggest that reduced
miR-195 expression may be associated with breast cancer development
and drug resistance, particularly Adriamycin resistance.
Association of miR-195 with sensitivity
of breast cancer cells to Adriamycin treatment
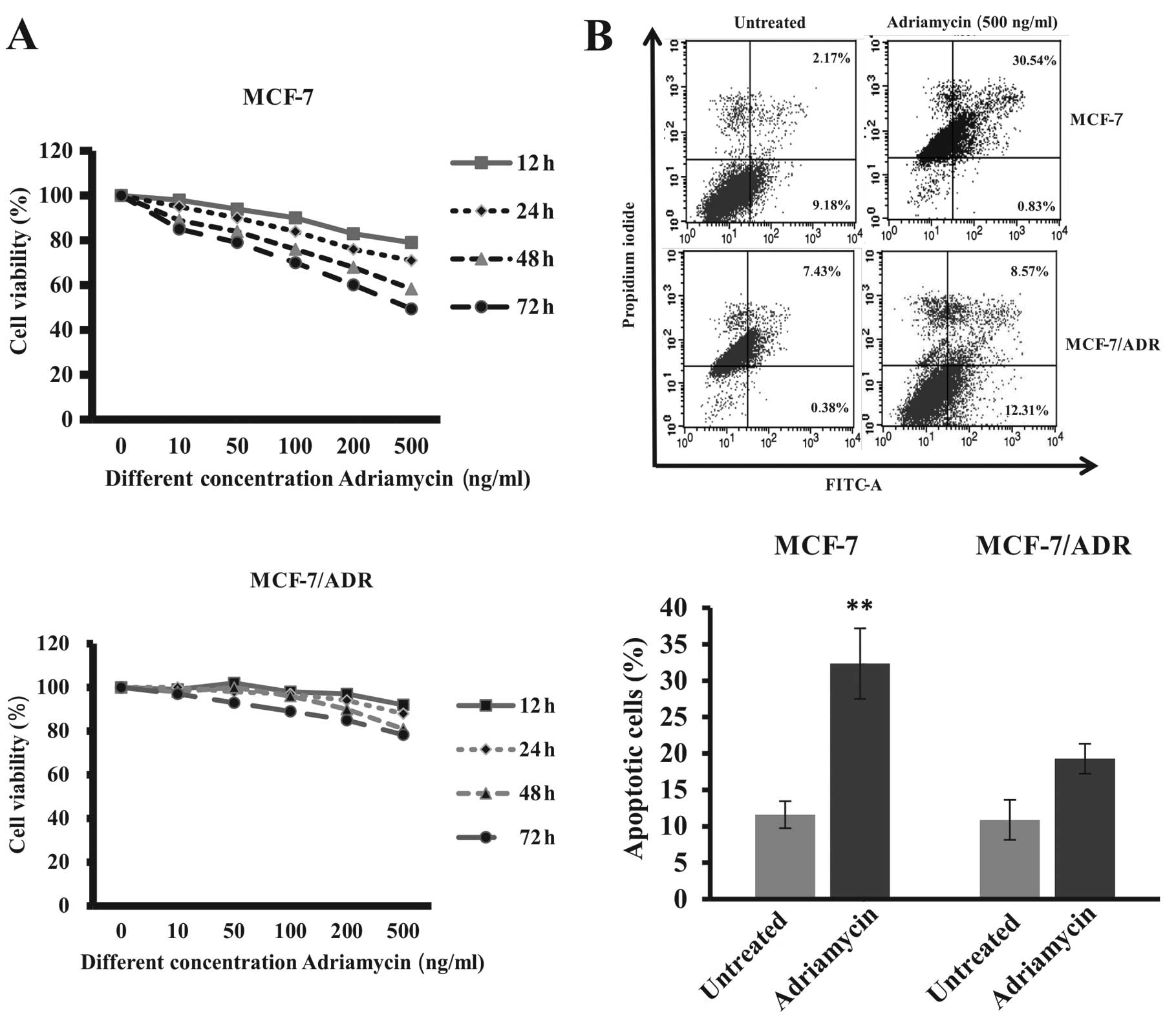
Fig. 2 shows the
sensitivity of breast cancer cells to Adriamycin treatment.
Parental breast cancer MCF-7 cells were sensitive to Adriamycin
treatment in a dose- and time-dependent manner, whereas the
drug-resistant MCF-7 subline MCF-7/ADR cells were resistant to
Adriamycin treatment detected by MTT cell viability and flow
cytometric apoptosis assays. Moreover, Adriamycin treatment
decreased the fraction of MCF-7 cells in G0/G1 and S phases, while
they increased the fraction of cells in G2/M phases, suggesting
G2/M arrest of Adriamycin-treated cancer cells (25).
Association of Raf-1 upregulation with
breast cancer cell resistance to Adriamycin
Since miRNA plays a role in the regulation of cell
growth, differentiation and apoptosis through inhibition of
targeting gene expressions, we performed a GenBank search for
miR-195 targeting gene and found that Raf-1 could be a target of
miR-195. Thus, we performed western blot analyses to analyze the
expression of Raf-1 and Raf-1-related genes in these two breast
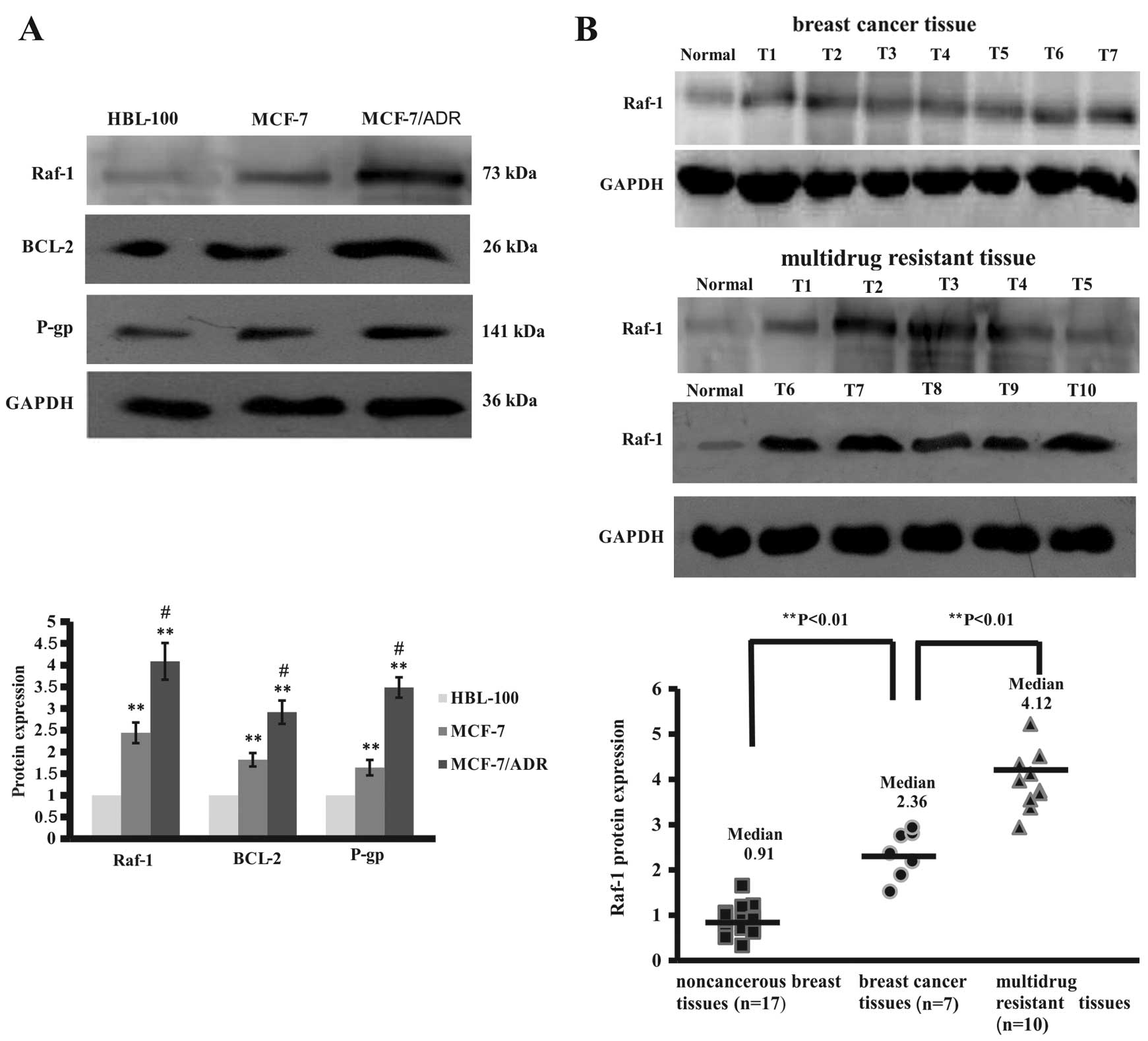
cancer cell lines and a normal breast cell line. Our data showed
that expression of Raf-1 protein levels was significantly higher in
breast cancer cells than in normal breast cells (Fig. 3A). However, drug resistant MCF-7/ADR
cells expressed higher levels of Raf-1 protein than HBL-100 and
MCF-7 cells (Fig. 3A). Similarly,
expression of Raf-1-related proteins, such as Bcl-2 and
P-glycoprotein, was significantly higher in MCF-7/ADR cells than in
HBL-100 and MCF-7 cells (Fig. 3A).
Moreover, we assayed Raf-1 expression in breast cancer tissue
specimens and found that these 10 chemotherapy-resistant breast
cancer tissues expressed high levels of Raf-1 protein compared to
the 7 chemotherapy-sensitive breast cancer tissues and the distant
normal breast tissues (Fig. 3B).
These data suggest that increased expression of Bcl-2 and
P-glycoprotein may be due to upregulated and constitutively active
Raf-1 protein in breast cancer cells, associated with sensitivity
of tumor cells to drug treatment.
Effect of miR-195 on the regulation of
breast cancer cell viability and apoptosis
Next, we determined the effects of miR-195
expression on the regulation of breast cancer cell viability and
apoptosis by transient transfection of miR-195 mimics and inhibitor
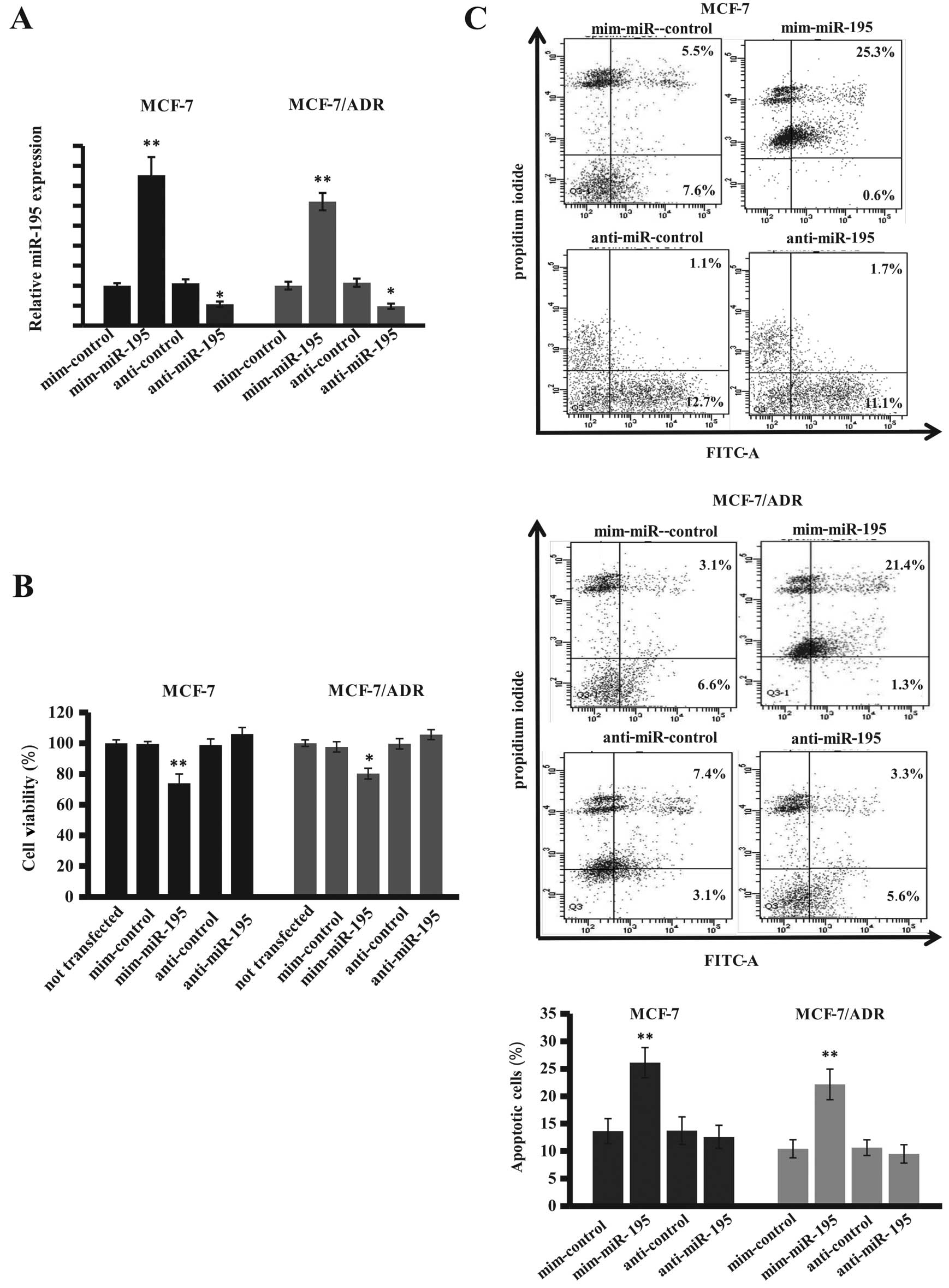
into these two breast cancer cell lines. Our data showed that the
expression of miR-195 was induced ~3-fold by the miR-195 mimic
compared with the negative control, whereas the miR-195 inhibitor
reduced miR-195 expression in these two breast cancer cell lines
(Fig. 4A). Cell viability MTT assay
showed expression of miR-195 reduced tumor cell viability, whereas
the specific miR-195 inhibitor failed to induce tumor cell
viability (Fig. 4B). The results
show that miR-195 mimics inhibited viability of these cells with an
average inhibition rate of 25.6% for MCF-7 cells and 18.2% for
MCF-7/ADR cells compared to the control cells. Furthermore,
transfection of miR-195 mimics clearly induced apoptosis of breast
cancer cells in these two breast cancer cell lines compared to the
negative control vector-transfected tumor cells (Fig. 4C). These data demonstrated that
miR-195 mimics were equally effective in these two breast cancer
cell lines, suggesting that miR-195 plays a role in breast cancer
resistance to drug treatment.
miR-195 directly targets Raf-1
expression
We have shown the importance of both miR-195 and
Raf-1 in breast cancer cell viability and drug resistance. To
provide a direct link between these genes, we employed a multiple
bioinformatics analysis of miR-195 targeting genes using PITA,
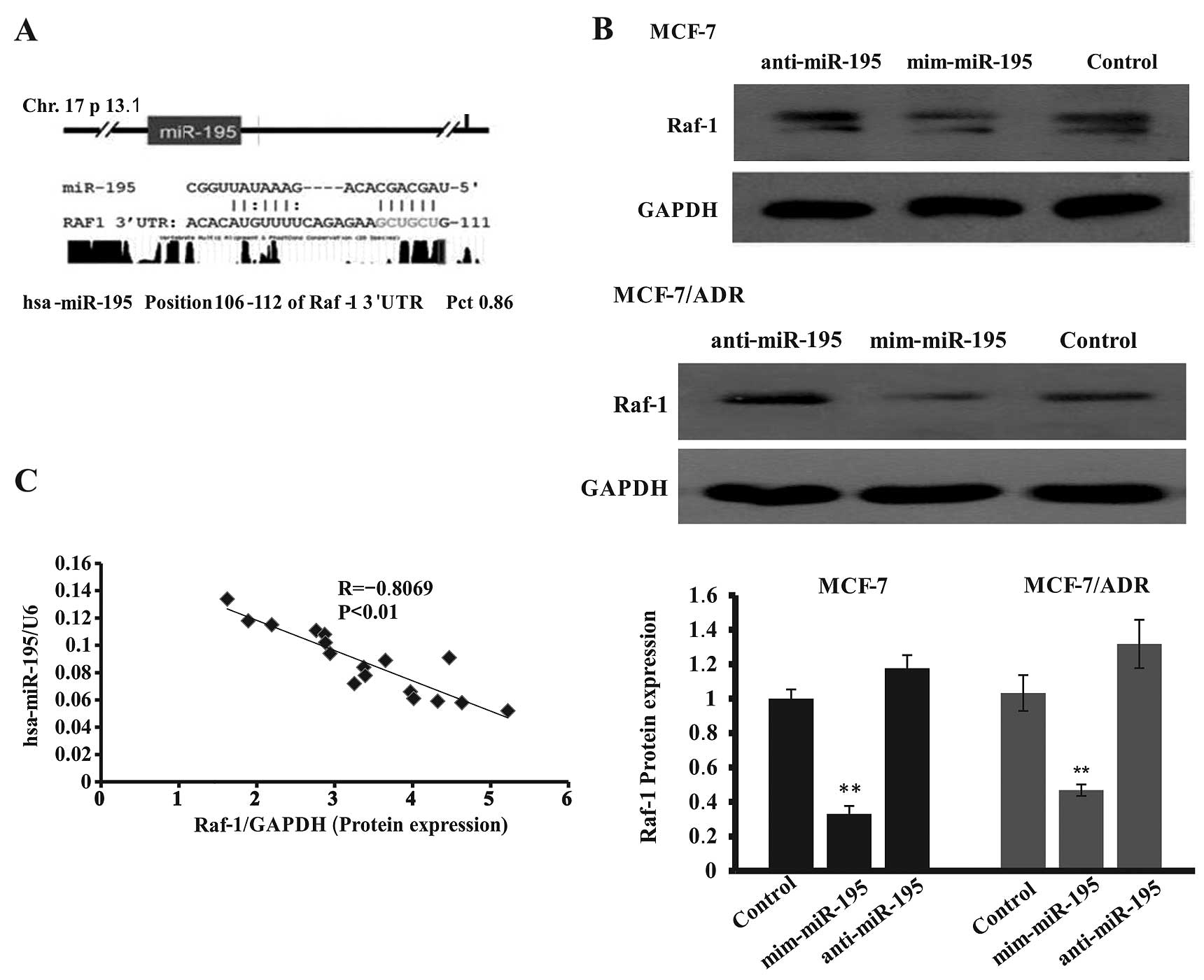
TargetScan, and miRanda. PITA analysis showed that miR-195 targeted
Raf-1 expression with a lower interaction-free energy (ddG, 2.07
kcal/mol; difference between free binding energy of a miRNA to the
target, dGduplex and free energy lost by opening the
target site, dGopen) and the lower free binding energy
(dGduplex, −12.1 kcal/mol) (Table II). The same miR-195 response
element (MRE) was identified by TargetScan and the probability of
conserved targeting of miRNA (PCT value) was 0.86, the context
score was −0.29, and the context score percentile was 89. miRanda
data showed that miR-195 has six nucleotides at the 5′ ‘seed’
region complementary to bases 106–112 of the Raf-1 3′-UTR (Fig. 5A). Thus, we detected Raf-1
expression after transfection of miR-195 mimics or inhibitor in
these breast cancer cell lines and found that expression of Raf-1
protein level significantly reduced transfection with mim-miR-195
(68.7±7.1% in MCF-7 cells and 61.8±6.2% in MCF-7/ADR compared to
the negative control cells). By contrast, miR-195 inhibitor
transfection also slightly increased Raf-1 expression (Fig. 5B). In addition, we also analyzed the
association of miR-195 with Raf-1 expression in 17 breast cancer
tissues and the data showed that miR-195 levels were inversely
associated with Raf-1 levels (Fig.
5C).
 | Table IIBioinformatics analysis. |
Table II
Bioinformatics analysis.
| Organism | RefSeq | Gene name | microRNA | Position |
dGduplex | dGopen
(kcal/mol) | ddG |
|---|
| Human | NM_002880 | Raf-1 | hsa-miR-195 | 111 | −12.1 | −12.01 | −0.082 |
Knockdown of Raf-1 expression alters
ability of breast cancer cell survival
To further investigate the significance of this
miR-195-regulated Raf-1 expression in breast cancer cells, we
designed and constructed Raf-1 siRNA to knock down Raf-1
expression. We found that Raf-1 siRNA significantly reduced Raf-1
protein levels (73.3±2.8% in MCF-7 cells and 68.5±3.4% in MCF-7/ADR
cells) (Fig. 6A), which was similar
to transfection with miR-195 mimic (Fig. 5C). We next evaluated the effect of
Raf-1 knockdown on the regulation of breast cell viability by
transient transfection with miR-195 mimic or siRNA-Raf-1. The data
showed that Raf-1 knockdown inhibited breast cancer cell viability
(average inhibition rate of 23.4% for MCF-7 cells and 16.2% for
MCF-7/ADR cells compared to control cells), similar to that of
miR-195 mimics transfection (Figs.
6B vs. 4B). We used
immunofluorescence staining to examine Raf-1 expression levels in
MCF-7 and MCF-7/ADR cells. We also found that the expression of
Raf-1 was markedly reduced with substantially reduced intensity of
fluorescence in the cells that transfected with Raf-1 siRNA or
miR-195 mimic, whereas non-transfection cells contained more Raf-1
than the parental cells transfected with Raf-1 siRNA or miR-195
mimic (Fig. 6C).
Expression of miR-195 alters sensitivity
of breast cancer cells to anticancer drugs by downregulation of
Raf-1 expression
To directly link the effects of miR-195 expression
on the regulation of breast cancer cell sensitivity to drug
treatment through downregulation of Raf-1 expression, we first
determined miR-195 transfection and Adriamycin treatment in these
breast cell lines. Our data showed that miR-195 mimic-transfected
tumor cells significantly reduced survival rate compared to control
cells, whereas anti-miR-195-transfected cell lines slightly
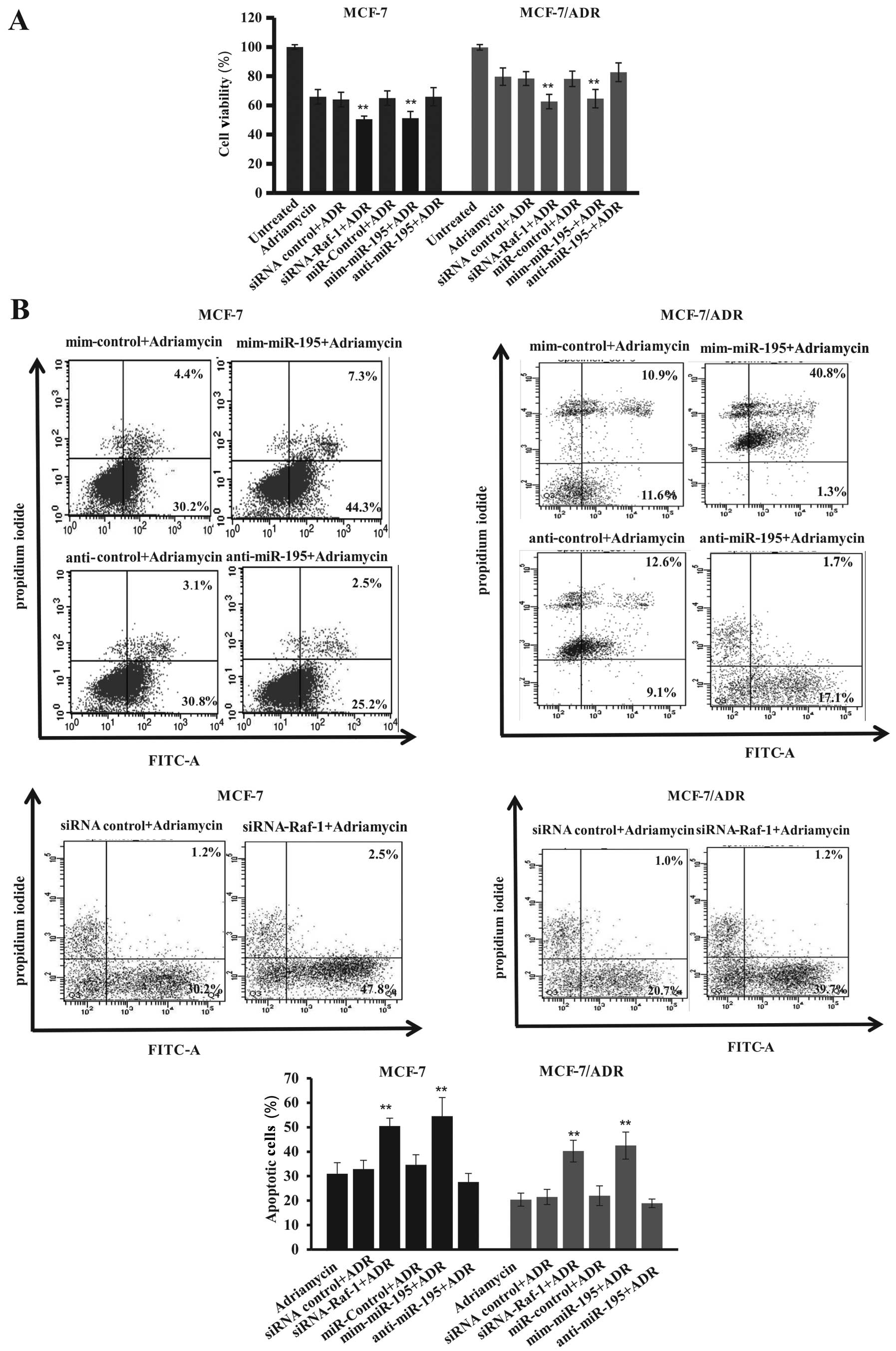
increased cell viability compared to control cells (Fig. 7A and B), which showed more additive
effects of Adriamycin plus miR-195 mimics on inhibition of tumor
cell survival but induction of apoptosis. Moreover, knockdown of
Raf-1 expression significantly suppressed the survivability of
MCF-7 and MCF-7/ADR cells compared with control vector-transfected
cells (Fig. 7A and B). We also
found that Raf-1 knockdown enhanced Adriamycin-induced tumor cell
apoptosis, i.e., apoptosis rates increased from 30±3% in
control-siRNA treated to 50±5% in MCF-7-Raf-1 siRNA transfected and
Adriamycin-treated tumor cells and from 21±3% in control-siRNA
treated to 40±4% in MCF-7/ADR-Raf-1 siRNA transfected and
Adriamycin-treated tumor cells (Fig.
7B). Similar findings were also extended to apoptosis of human
MCF-7 and MCF-7/ADR cells after transfection with miR-195 mimic,
i.e., the miR-195 mimic significantly enhanced apoptosis of
Adriamycin-induced tumor cells compared to the control cells
(apoptosis rate from 32±4 to 54±6% for MCF-7 cells and from 22±2 to
42±4% for MCF-7/ADR cells) (Fig.
7B). These data suggest that miR-195 expression affected cell
sensitivity to Adriamycin and was mediated by suppression of Raf-1
expression.
Furthermore, to understand the miR-195-mediated
Raf-1 gene pathway, we found that modulation of miR-195 expression
inhibited expression of Raf-1 protein, which in turn affects Bcl-2
and P-glycoprotein expression for changes in cancer cell
sensitivity to Adriamycin treatment. Our data showed the changes in
expression of Bcl-2 and P-glycoprotein in MCF-7 and MCF-7/ADR cells
after transfection with miR-195 mimic and inhibitor or Raf-1-siRNA
(Fig. 7C and D). In particular,
expression of Bcl-2 and P-glycoprotein was significantly lower in
tumor cells transfected with miR-195 mimic than that of the control
vector-transfected tumor cells, whereas there was a slight increase
in the expression of Bcl-2 and P-glycoprotein in the
anti-miR-195-transfected cells. Similarly, Raf-1-siRNA transfection
had such effects (Fig. 7C and D).
These data demonstrated that expression of miR-195 was inversely
associated with Raf-1 expression in these two breast cancer cell
lines and miR-195 expression reduced tumor cells survival but
increased apoptosis by the downregulation, at least in part, of
Raf-1 expression. Meanwhile, expression of Bcl-2 and P-glycoprotein
was affected by miR-195 or Raf-1 siRNA, in association with the
sensitivity of breast cancer cells to Adriamycin treatment.
Discussion
Altered expression of miRNAs plays an important role
in regulating cell activities, including proliferation,
morphogenesis, apoptosis, differentiation as well as cancer
development and progression (14,26).
Dysregulated expression of miRNA was also reported to play a role
in resistance to cancer therapy (27,28).
In particular, previous studies have shown aberrant miR-195
expression in various types of human cancer, such as gastric,
bladder and breast cancer (16,17),
thus, it was reported that miR-195 plays a tumor-suppressor role in
human cancer. For example, overexpression of miR-195 in cancer cell
lines induced apoptosis and inhibited tumor cell invasiveness in
different cancer cell lines (29–31)
and modulated cancer cell sensitivity to chemotherapy by targeting
the Bcl-w proto-oncogene in head and neck and liver cancer
(32,33). Thus, in the present study, we
investigated the effects of miR-195 on the regulation of breast
cancer viability, apoptosis and sensitivity to Adriamycin
treatment, and then explored the underlying mechanisms by targeting
Raf-1 expression and Raf-1-related pathway genes. We found that
miR-195 expression was reduced in breast cancer tissue and cells
compared to the normal breast tissues and cells and that induction
of miR-195 expression promoted apoptosis and inhibited survival of
breast cancer cells. Our data also showed that miR-195 is able to
bind to 3′-UTR of Raf-1 mRNA to suppress Raf-1 expression,
consistent with the finding by Flavin et al(34). Indeed, Subramanian and Yamakawa
(35) recently showed that miR-195
is a direct negative regulator of Raf-1 expression and that
inhibition of Raf-1 and MEK kinases simultaneously increased
apoptosis of colon cancer cells. Other studies revealed that Raf-1
was overexpressed in breast cancer (36,37).
Thus, these findings indicate that suppression of Raf-1 activities
by miR-195 could have profound effects on the suppression of cancer
development or progression. In this study, we observed that Raf-1
was highly expressed in drug-resistant breast cancer tissues and
cells and that miR-195 levels were inversely associated with Raf-1
expression. Moreover, we found that miR-195 expression or Raf-1
knockdown inhibited breast cancer cell survival and promoted them
to apoptosis, and that expression of miR-195 inhibited Raf-1
expression in MCF-7 and MCF-7/ADR cells. In addition, the potential
interaction between Raf-1 and miR-195 was confirmed by reversed
expression of miR-195 and Raf-1 in MCF-7 and MCF-7/ADR cells. For
example, when the miR-195 mimic was transfected into MCF-7 and
MCF-7/ADR cells, miR-195 expression was induced and Raf-1
expression was reduced, followed by reduced cell survival and
apoptosis induction. These phenomena were consistent with Raf-1
siRNA transfection in MCF-7 and MCF-7/ADR cells. However, our data
showed that anti-miR-195 was not able to produce the inverse data,
which may be because these cell lines used expression of low levels
of miR-195. Clinically, miR-195, together with miR-497, was
significantly downregulated in cancer tissues and promoters of
these two miRNAs were regulated by a common CpG methylation
mechanism or high frequency of loss of heterozygosity on chromosome
17p13.1 (38,39). This study, therefore, demonstrated
that antitumor effects of miR-195 were mediated by targeting of
Raf-1 expression in breast cancer cells.
Furthermore, to date, Adriamycin is the major drug
used in the clinical treatment of breast cancer. Mechanistically,
Adriamycin induces tumor cells to undergo apoptosis and stimulates
cytokine production as well (40).
However, Adriamycin drug resistance is a significant clinical
problem and to make this drug more effective, this drug resistance
needs to be overcome. In this study, we showed breast cancer MCF-7
cells were sensitive to Adriamycin, whereas MCF-7/ADR cells were
resistant; however, both induction of miR-195 and inhibition of
Raf-1 expression sensitized MCF-7/ADR cells to Adriamycin treatment
by reduced cell viability and induction of apoptosis. This is
consistent with previously reported data that Raf-1 expression was
associated with highly aggressive tumors (such as inflammatory
breast cancer), protected tumor cells from differentiation and
apoptosis, and promoted tumor cell proliferation, invasiveness and
chemoresistance (41,42). Furthermore, since Raf-1 has been
widely accepted as a drug resistance protein, our data showed that
reduced miR-195 expression plays a role in the development of
multiple drug resistance by loss of miR-195-suppression of Raf-1.
In our previous study, we found that reduced miR-195 expression was
significantly associated with advanced clinical stages of breast
cancer and was inversely associated with Raf-1 expression in breast
cancer (38).
In addition, the present study also showed that
expression of Bcl-2 and P-glycoprotein levels was significantly
higher in MCF-7/ADR cells than in HBL-100 and MCF-7 cells.
Expression of miR-195 or inhibition of Raf-1 expression
significantly inhibited expression of Bcl-2 and P-glycoprotein in
breast cancer cells, which was associated with sensitivity of tumor
cells to Adriamycin treatment. In previous studies, it was observed
that an activated Raf-1 was able to upregulate Bcl-2 and
P-glycoprotein levels in breast cancer cells (43,44).
Anti-apoptosis function of Bcl-2 protein contributed to drug
resistance of different types of cancer, while P-glycoprotein
protected breast cancer cells from chemotherapy-induced apoptosis
(45,46). Thus, the elevated levels of miR-195
in miR-195 mimic-transfected MCF-7 and MCF-7/ADR cells not only
downregulated expression of Bcl-2 and P-glycoprotein, but also
increased sensitivity of these cells to Adriamycin.
Our present data demonstrated that the induced
expression of miR-195 in MCF-7 and MCF-7/ADR cell lines suppressed
Raf-1 expression and sensitized tumor cells to Adriamycin
treatment, suggesting that targeting of Raf-1 expression using
miR-195 may have significant implications for prevention and
reversal of breast cancer cell resistance to chemotherapy. However,
there may be multiple underlying mechanisms, as previous studies
demonstrated crosstalk or interactions between the Ras/Raf/MEK/ERK
and Ras/PI3K/PTEN/Akt pathways in the regulation of cell growth,
survival and apoptosis (47,48).
Moreover, numerous miRNAs may participate to modulate certain gene
expressions as part of a complex network for an anti-apoptotic
program in tumor cells resistant to apoptosis-inducing
chemotherapeutic agents, and these miRNAs have shown to be
dysregulated in breast cancer cells resistant to genotoxic agents
(49). Future studies will
investigate the effects of miR-195-regulated Raf-1 expression in
mediating breast cancer sensitivity to Adriamycin treatment in
animal experiments and in clinical trials. Our findings also
contribute to the understanding of ADR regulation in cancer cells.
Additionally, these findings may be beneficial for further research
for predicting ADR in patients and designing personalized therapy
for breast carcinoma patients.
Acknowledgements
This study was supported in part by the National
Science Foundation of China (grant no. 31140035), and the
Foundation of Chongqing Science and Technology Commission (grant
no. CSTC, 2011jjA10060), and was supported by grants from the
National Natural Science Foundation of China (NSFC30872770), the
Natural Science Foundation Project of CQ CSTC (CSTC, 2011BB5131)
and the National Ministry of Education Foundation of China
(KJ120327).
References
|
1
|
Jemal A, Bray F, Center MM, et al: Global
cancer statistics. CA Cancer J Clin. 61:69–90. 2011. View Article : Google Scholar
|
|
2
|
O'Driscoll L and Clynes M: Biomarkers and
multiple drug resistance in breast cancer. Curr Cancer Drug
Targets. 6:365–384. 2006. View Article : Google Scholar
|
|
3
|
Coley HM: Mechanisms and strategies to
overcome chemotherapy resistance in metastatic breast cancer.
Cancer Treat Rev. 34:378–390. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gonzalez-Angulo AM, Morales-Vasquez F and
Hortobagyi GN: Overview of resistance to systemic therapy in
patients with breast cancer. Adv Exp Med Biol. 608:1–22. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Marquette C and Nabell L:
Chemotherapy-resistant metastatic breast cancer. Curr Treat Options
Oncol. 13:263–275. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lai EC: MicroRNAs are complementary to
3′UTR sequence motifs that mediate negative post-transcriptional
regulation. Nat Genet. 30:363–364. 2002.
|
|
7
|
Guo H, Ingolia NT, Weissman JS and Bartel
DP: Mammalian microRNAs predominantly act to decrease target mRNA
levels. Nature. 466:835–840. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sempere LF, Christensen M, Silahtaroglu A,
et al: Altered MicroRNA expression confined to specific epithelial
cell subpopulations in breast cancer. Cancer Res. 67:11612–11620.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Majumder S and Jacob ST: Emerging role of
microRNAs in drug-resistant breast cancer. Gene Expr. 15:141–151.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kastl L, Brown I and Schofield AC:
miRNA-34a is associated with docetaxel resistance in human breast
cancer cells. Breast Cancer Res Treat. 131:445–454. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhu H, Wu H, Liu X, et al: Role of
MicroRNA miR-27a and miR-451 in the regulation of
MDR1/P-glycoprotein expression in human cancer cells. Biochem
Pharmacol. 76:582–588. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kovalchuk O, Filkowski J, Meservy J, et
al: Involvement of microRNA-451 in resistance of the MCF-7 breast
cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther.
7:2152–2159. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bao L, Hazari S, Mehra S, et al: Increased
expression of P-glycoprotein and doxorubicin chemoresistance of
metastatic breast cancer is regulated by miR-298. Am J Pathol.
180:2490–2503. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ng EK, Wong CL, Ma ES and Kwong A:
MicroRNAs as new players for diagnosis, prognosis, and therapeutic
targets in breast cancer. J Oncol. 2009:3054202009.PubMed/NCBI
|
|
15
|
Iorio MV, Casalini P, Tagliabue E, et al:
MicroRNA profiling as a tool to understand prognosis, therapy
response and resistance in breast cancer. Eur J Cancer.
44:2753–2759. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang QQ, Xu H, Huang MB, et al:
MicroRNA-195 plays a tumor-suppressor role in human glioblastoma
cells by targeting signaling pathways involved in cellular
proliferation and invasion. Neuro Oncol. 14:278–287. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fei X, Qi M, Wu B, et al: MicroRNA-195-5p
suppresses glucose uptake and proliferation of human bladder cancer
T24 cells by regulating GLUT3 expression. FEBS Lett. 586:392–397.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
McCubrey JA, Steelman LS, Chappell WH, et
al: Roles of the Raf/MEK/ERK pathway in cell growth, malignant
transformation and drug resistance. Biochim Biophys Acta.
1773:1263–1284. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cekanova M, Majidy M, Masi T, et al:
Overexpressed Raf-1 and phosphorylated cyclic adenosine
3′-5′-monophosphatate response element-binding protein are early
markers for lung adenocarcinoma. Cancer. 109:1164–1173.
2007.PubMed/NCBI
|
|
20
|
Hoyle PE, Moye PW, Steelman LS, et al:
Differential abilities of the Raf family of protein kinases to
abrogate cytokine dependency and prevent apoptosis in murine
hematopoietic cells by a MEK1-dependent mechanism. Leukemia.
14:642–656. 2000. View Article : Google Scholar
|
|
21
|
Chang F, Steelman LS and McCubrey JA:
Raf-induced cell cycle progression in Human TF-1hematopoietic
cells. Cell Cycle. 1:220–226. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hwang YH, Choi JY, Kim S, et al:
Overexpression of c-raf-1 proto-oncogene in liver cirrhosis and
hepatocellular carcinoma. Hepatol Res. 29:113–121. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Steelman LS, Chappell WH, Abrams SL, et
al: Roles of the Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR pathways in
controlling growth and sensitivity to therapy-implications for
cancer and aging. Aging. 3:192–222. 2011.PubMed/NCBI
|
|
24
|
Meslin F, Hamaï A, Gao P, et al: Silencing
of prion protein sensitizes breast adriamycin-resistant carcinoma
cells to TRAIL-mediated cell death. Cancer Res. 67:10910–10919.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lu C, Shao C, Cobos E, et al:
Chemotherapeutic sensitization of leptomycin B resistant lung
cancer cells by pretreatment with doxorubicin. PLoS One.
7:e328952012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Blenkiron C, Goldstein LD, Thorne NP,
Spiteri I, et al: MicroRNA expression profiling of human breast
cancer identifies new markers of tumor subtype. Genome Biol.
8:R2142007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hannafon BN, Sebastiani P, de las Morenas
A, et al: Expression of microRNA and their gene targets are
dysregulated in preinvasive breast cancer. Breast Cancer Res.
13:R242011. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Waters PS, McDermott AM, Wall D, et al:
Relationship between circulating and tissue microRNAs in a murine
model of breast cancer. PLoS One. 7:e504592012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu L, Chen L, Xu Y, et al: microRNA-195
promotes apoptosis and suppresses tumorigenicity of human
colorectal cancer cells. Biochem Biophys Res Commun. 400:236–240.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu T, Zhu Y, Xiong Y, et al: MicroRNA-195
suppresses tumorigenicity and regulates G1/S transition of human
hepatocellular carcinoma cells. Hepatology. 50:113–121. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dai Y, Xie CH, Neis JP, et al: MicroRNA
expression profiles of head and neck squamous cell carcinoma with
docetaxel-induced multidrug resistance. Head Neck. 33:786–791.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang X, Yin J, Yu J, et al: miRNA-195
sensitizes human hepatocellular carcinoma cells to 5-FU by
targeting BCL-w. Oncol Rep. 27:250–257. 2012.PubMed/NCBI
|
|
34
|
Flavin RJ, Smyth PC, Laios A, et al:
Potentially important microRNA cluster on chromosome 17p13.1 in
primary peritoneal carcinoma. Mod Pathol. 22:197–205. 2009.
View Article : Google Scholar
|
|
35
|
Subramanian RR and Yamakawa A: Combination
therapy targeting Raf-1 and MEK causes apoptosis of HCT116 colon
cancer cells. Int J Oncol. 41:1855–1862. 2012.PubMed/NCBI
|
|
36
|
Monazzam A, Razifar P, Ide S, et al:
Evaluation of the Hsp90 inhibitor NVP-AUY922 in multicellular
tumour spheroids with respect to effects on growth and PET tracer
uptake. Nucl Med Biol. 36:335–342. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Leicht DT, Balan V, Kaplun A, et al: Raf
kinases: function, regulation and role in human cancer. Biochim
Biophys Acta. 1773:1196–1212. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li D, Zhao Y, Liu C, et al: Analysis of
MiR-195 and MiR-497 expression, regulation and role in breast
cancer. Clin Cancer Res. 17:1722–1730. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bandera CA, Muto MG, Welch WR, et al:
Genetic imbalance on chromosome 17 in papillary serous carcinoma of
the peritoneum. Oncogene. 16:3455–3459. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ghebeh H, Lehe C, Barhoush E, et al:
Doxorubicin downregulates cell surface B7-H1 expression and
upregulates its nuclear expression in breast cancer cells: role of
B7-H1 as an anti-apoptotic molecule. Breast Cancer Res. 12:R482010.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang X, Thomson SR, Starkey JD, et al:
Transforming growth factor beta1 is upregulated by activated Raf in
skeletal myoblasts but does not contribute to the
differentiation-defective phenotype. J Biol Chem. 279:2528–2534.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Leontovich AA, Zhang S, Quatraro C, et al:
Raf-1 oncogenic signaling is linked to activation of mesenchymal to
epithelial transition pathway in metastatic breast cancer cells.
Int J Oncol. 40:1858–1864. 2012.PubMed/NCBI
|
|
43
|
Weinstein-Oppenheimer CR, Henriquez-Roldan
CF, Davis JM, et al: Role of the Raf signal transduction cascade in
the in vitro resistance to the anticancer drug doxorubicin. Clin
Cancer Res. 7:2898–2907. 2001.PubMed/NCBI
|
|
44
|
Anderson LR, Sutherland RL and Butt AJ:
BAG-1 overexpression attenuates luminal apoptosis in MCF-10A
mammary epithelial cells through enhanced RAF-1 activation.
Oncogene. 29:527–538. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bodur C and Basaga H: Bcl-2 inhibitors:
emerging drugs in cancer therapy. Curr Med Chem. 19:1804–1820.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Shen Y, Chu Y, Yang Y and Wang Z:
Mitochondrial localization of P-glycoprotein in human breast cancer
cell line MCF-7/ADM and its functional characterization. Oncol Rep.
27:1535–1540. 2012.PubMed/NCBI
|
|
47
|
Gollob JA, Wilhelm S, Carter C and Kelley
SL: Role of Raf kinase in cancer: therapeutic potential of
targeting the Raf/MEK/ERK signal transduction pathway. Semin Oncol.
33:392–406. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Singh R and Saini N: Downregulation of
BCL2 by miRNAs augments drug-induced apoptosis - a combined
computational and experimental approach. J Cell Sci. 125:1568–1578.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Neelakandan K, Babu P and Nair S: Emerging
roles for modulation of microRNA signatures in cancer
chemoprevention. Curr Cancer Drug Targets. 12:716–740. 2012.
View Article : Google Scholar : PubMed/NCBI
|