Introduction
Mutations of EGFR, KRAS,
PIK3CA, and others, and fusion transcripts of ALK, ROS1 and
RET are oncogenic alterations in lung cancers (LCs) and some of
them are therapeutic targets (1–8). For
example, crizotinib, a small molecule inhibitor of ALK, has been
shown to selectively inhibit the growth of ALK-positive LC
(9), meaning that a subclass of LC
patients are likely to benefit clinically from an ALK inhibitor.
Therefore, targetable oncogenic alterations in LCs may have a
significant clinical impact.
Fibroblast growth factor receptor 3 (FGFR3) has been
revealed to be activated by the mutation or fusion of its own gene
in several types of cancer, such as urinary bladder cancer,
glioblastoma, rhabdomyosarcoma and LC (10–17).
Some tumor-specific FGFR3 mutations, including p.R248C and
p.S249C, drive the anchorage-independent growth of NIH3T3 cells and
tumor formation in xenograft models, and cells harboring such
FGFR3 mutations showed an enhanced sensitivity to BGJ398, a
selective FGFR kinase inhibitor (17). Regarding fusions, FGFR3-TACC3 and
FGFR3-BAIAP2L1 oncogenic fusions have thus far been identified
(13–15), and cells harboring the FGFR3-TACC3
fusion showed enhanced sensitivity to three FGFR kinase inhibitors,
including BGJ398 (13). Thus,
oncogenic FGFR3 mutations and fusions in tumors can be
therapeutic targets. To determine which patients can benefit from
FGFR inhibitors in the future, the incidence of FGFR3
mutations or fusions in various tumors derived from patients from
various demographic areas must be determined. However, to date,
only a few studies have been published regarding FGFR3
mutations and fusions in LC and Asian patients with LC have not yet
been analyzed (12,16,17).
Notably, according to previous reports (12,16,17),
the only experimentally proven oncogenic FGFR3 mutations in
LCs are p.R248C and p.S249C, which are located within the first ten
bases of exon 7. Therefore, we considered this region to be a
mutation cluster region in LCs. In the present study, to determine
the status of FGFR3 mutation and fusion in LCs derived from
Japanese patients, we examined LCs from Japanese patients for the
mutations in the mutation cluster region of FGFR3 and the
expression of FGFR3-TACC3 and FGFR3-BAIAP2L1 fusion transcripts and
pathologically and molecularly characterized LCs containing such an
alteration. This is the first published study to describe
FGFR3 mutations in LCs derived from Japanese patients.
Materials and methods
Primary LC
Samples of surgical specimens were obtained from 362
Japanese LC patients who underwent surgery for cancer at Hamamatsu
University Hospital and Mikatahara Seirei General Hospital.
Informed consent was obtained from all the patients, and the study
was approved by the Institutional Review Boards (IRBs) of Hamamatsu
University School of Medicine and Mikatahara Seirei General
Hospital. The clinicopathological profiles of the cases are shown
in Table I. The histological
classification was based on the World Health Organization system.
Among the 362 cases, 214 cases were used in the mutational
analysis, whereas 190 cases were used in the reverse
transcription-polymerase chain reaction (RT-PCR) analysis; 42 cases
were used in both analyses.
 | Table ISummary of the clinicopathological
profiles of the patients. |
Table I
Summary of the clinicopathological
profiles of the patients.
| Numbera |
|---|
|
|
|---|
|
Characteristics | Total | Mutational
analysis | RT-PCR
analysis |
|---|
| No. of
patients | 362 | 214 | 190 |
| Age, years (mean ±
SD) | 66.5±9.6 | 66.4±9.7 | 66.9±9.9 |
| Gender, n (%) |
| Male | 259 (71.5) | 157 (73.4) | 134 (70.5) |
| Female | 103 (28.5) | 57 (26.6) | 56 (29.5) |
| Histology, n
(%) |
|
Adenocarcinoma | 210 (58.0) | 125 (58.4) | 109 (57.4) |
| Squamous cell
carcinoma | 110 (30.4) | 63 (29.4) | 60 (31.6) |
| Large cell
carcinoma | 13 (3.6) | 9 (4.2) | 5 (2.6) |
| Small cell
carcinoma | 12 (3.3) | 11 (5.1) | 3 (1.6) |
| Adenosquamous
carcinoma | 11 (3.0) | 4 (1.9) | 9 (4.7) |
| Pleomorphic
carcinoma | 6 (1.7) | 2 (0.9) | 4 (2.1) |
Search for FGFR3 mutations using
PCR-sequencing
Genomic DNAs were extracted from the lung tissue
samples using a DNeasy kit (Qiagen, Valencia, CA, USA) and were
examined for somatic mutations in the DNA sequences (the first half
of exon 7) covering the mutation cluster region in the FGFR3
gene. PCR was performed in 20-μl reaction mixtures containing
HotStarTaq DNA polymerase (Qiagen) under the following conditions:
30 sec at 94°C, 30 sec at 65°C and 60 sec at 72°C for 45 cycles.
The following set of primers was used: 5′-CTG AGC GTC ATC TGC CCC
C-3′ and 5′-TGG GGC TGT GCG TCA CTG TAC-3′. PCR-amplified products
were purified with ExoSAP-IT (GE Healthcare Bio-Sciences,
Piscataway, NJ, USA) and were sequenced directly using a BigDye
Terminator Cycle Sequencing Reaction kit (Applied Biosystems,
Tokyo, Japan) and the ABI 3130 Genetic Analyzer (Applied
Biosystems).
Search for FGFR3 fusion transcripts using
RT-PCR
Total RNA was extracted from the lung tissue samples
using an RNeasy kit (Qiagen) and was converted to first-strand cDNA
using a SuperScript First-Strand Synthesis System for RT-PCR
(Invitrogen, Carlsbad, CA, USA) according to the supplier’s
protocol. PCR was performed in 20-μl reaction mixtures containing
HotStarTaq DNA polymerase under the following conditions: 30 sec at
94°C, 30 sec at 59°C and 60 sec at 72°C for 45 cycles. The
following reverse PCR primers were used: 5′-CAG CCT CCA CTG GTT TCT
GTA G-3′ for the sequence at exon 4 of TACC3, 5′-TGG TAC ACA ACC
TCT TCG AAC C-3′ for the sequence at exon 12 of TACC3, and 5′-GGA
CAT GTC CCA GTT CAG TTG-3′ for the sequence at exons 3 and 4 of
BAIAP2L1. The forward PCR primer used was the same, i.e., 5′-GAC
CGT GTC CTT ACC GTG AC-3′ for the sequence at exon 18 of FGFR3. The
PCR products were fractionated using electrophoresis on an agarose
gel and were stained with ethidium bromide.
Quantitative RT (qRT)-PCR
The expressions of the FGFR3 mRNA transcripts were
measured using real-time qRT-PCR with a LightCycler instrument
(Roche, Palo Alto, CA, USA). PCR amplification of the FGFR3
transcript and the transcript of the control housekeeping gene
GAPDH was performed with the cDNA and a QuantiTect SYBR
Green PCR kit (Qiagen). The following PCR primers were used: 5′-GCA
CAC ACG ACC TGT ACA TGA TC-3′ and 5′-CCA GGT ACT CGT CGG TGG AC-3′
for the FGFR3 transcript, and 5′-GCT CAG ACA CCA TGG GGA AG-3′ and
5′-TGT AGT TGA GGT CAA TGA AGG GG-3′ for the GAPDH transcript. The
T/N ratios were calculated by dividing the normalized transcript
amounts in the cancerous tissue by the amounts in the non-cancerous
tissue.
Immunohistochemical staining
Sections of formalin-fixed, paraffin-embedded tissue
samples were used for immunohistochemical staining using a
Histofine Simple Stain MAX PO kit (Nichirei, Tokyo, Japan), as
previously described (18). The
primary antibodies were: anti-CK14, anti-thyroid transcription
factor-1 (TTF-1) (both from Novocastra Laboratories, Newcastle, UK)
and anti-p53 (clone DO7; Dako, Tokyo, Japan). Hematoxylin and eosin
(H&E) staining was also performed.
Search for EGFR, KRAS, PIK3CA and p53
mutations using PCR-sequencing
Genomic DNA derived from the lung tissue samples
containing an FGFR3 mutation was examined for somatic
mutations in the DNA sequences of mutation cluster regions in the
EGFR, KRAS, PIK3CA and p53 genes. PCR
amplification was performed as previously described (7). Sequencing was performed as described
in the ‘Search for FGFR3 mutations using PCR-sequencing’
section.
Fluorescence in situ hybridization (FISH)
analysis
Paraffin-embedded tissue sections were de-waxed and
re-hydrated, then boiled in 0.01 M citrate buffer (pH 6.0) to
release the closed chromosomal structures. A combination of
Cy3-labeled bacterial artificial chromosome (BAC) clone
(RP11-100B16) for the FGFR1 locus, BAC clone (RP11-245C23
and RP11-355N16) for the PIK3CA locus, or BAC clone
(RP11-275H4) for the SOX2 locus and a SpectrumGreen-labeled
control BAC probe for the near centromere locus on chromosome 3 or
8 were placed on a slide and covered with a coverslip. All the BAC
probes were obtained from Advanced GenoTechs Co. (Tsukuba, Japan).
The slides with the hybridization mixture were denatured on a
digital hot plate (HP-15; As One Corporation, Osaka, Japan) and
then incubated overnight at 42°C. After washing the slide in 50%
formamide/2X SSC, mounting medium containing DAPI (Vector
Laboratories, Burlingame, CA, USA) was used for nuclear
counterstaining. The slides were promptly examined under a
fluorescence microscope (Olympus BX51-FL; Olympus, Tokyo, Japan)
equipped with epifluorescence filters and a photometric CCD camera
(Sensicam; PCO Company, Kelheim, Germany). The images captured were
digitized and stored in the image analysis program (MetaMorph;
Molecular Devices, Palo Alto, CA, USA). The average ratio of the
FGFR1, PIK3CA, or SOX2 signal number to the
control probe signal number was calculated for each cancer. If the
ratio of a cancer was >2.5, the cancer was defined as
amplification-positive.
Results
In the present study, we examined 214 LCs for
mutations in the mutation cluster region of the FGFR3 gene
using a sequencing analysis; we also examined 190 LCs for
FGFR3-TACC3 and FGFR3-BAIAP2L1 fusion transcripts using an RT-PCR
analysis. Although the expression of FGFR3-TACC3 and FGFR3-BAIAP2L1
fusion transcripts was not detected in any of the carcinomas,
FGFR3 mutations were detected in two (0.9%) LCs (Fig. 1A). The mutations that occurred in
the LCs derived from cases no. 57 and no. 183 were the same, i.e.,
a somatic c.743G>A mutation associated with an amino acid
exchange from Arg to His at codon 248. The p.R248H mutation in the
FGFR3 gene has not been previously reported, suggesting it
is a novel mutation. Of note, with regards to codon 248, it was
previously reported that the p.R248C mutation drives cellular
transformation (17), indicating
that the p.R248H mutation may have an oncogenic potential similar
to that of the p.R248C mutation. When we next examined the
expression levels of FGFR3 mRNA transcripts in the two LCs
containing an FGFR3 p.R248H mutation using a qRT-PCR
analysis, the ratio of the level of FGFR3 mRNA expression in the
cancerous tissue to the level in the corresponding non-cancerous
tissue (T/N ratio) was increased in both cases (Fig. 1B), suggesting the involvement of
mutated FGFR3 in LC. Case no. 57 was a 62-year-old man who was a
smoker [Brinkman index (BI)=1,800] and case no. 183 was a
66-year-old man who was also a smoker (BI=1,000). The histological
classification of both LCs was squamous cell carcinoma (Fig. 2A and B). An immunohistochemical
study revealed that the carcinomas were positive for CK14 but
negative for TTF-1 (Fig. 2C and D,
Table II), indicating that the
immunophenotype of the carcinoma was compatible with that of
squamous cell carcinoma. Among the 214 cases used for the
mutational analysis, 63 cases were histologically classified as
squamous cell carcinoma; thus, the incidence of the FGFR3
mutation among the squamous cell carcinoma cases was 3.2% (2/63).
These findings suggest that a subset of LC may carry an
FGFR3 mutation.
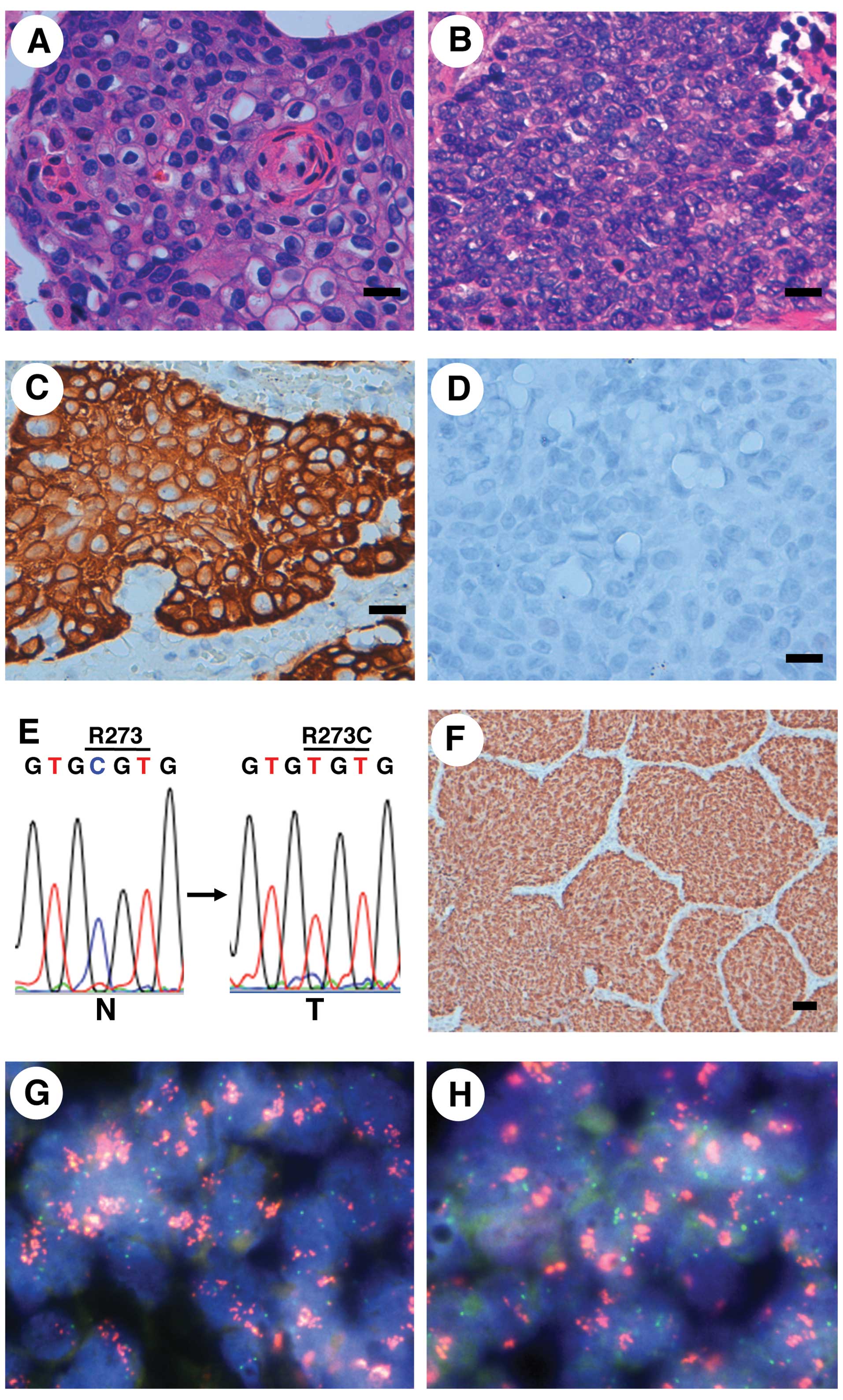 | Figure 2Pathological, immunohistochemical,
mutational and FISH analyses for lung cancers (LCs) containing an
FGFR3 mutation. (A and B) Microscopic image (H&E) of the
squamous cell carcinoma in case no. 57 (A) and case no. 183 (B).
Scale bar, 20 μm. (C and D) The squamous cell carcinoma of case no.
57 was immunohistochemically positive for CK14 (C) and negative for
TTF-1 (D). Scale bar, 20 μm. (E) Detection of a somatic p53
missense mutation in LC from case no. 183. A CGT to TGT mutation associated with
the conversion from Arg to Cys at codon 273 was detected in the LC.
N, non-cancerous lung tissue DNA; T, cancerous lung tissue DNA. (F)
Immunohistochemical detection of p53 accumulation in the LC from
case no. 183. Scale bar, 50 μm. (G and H) PIK3CA (G) and
SOX2 (H) amplifications in LC from case no. 57, as shown
using a FISH analysis. Red signals, BAC probe for the PIK3CA
(G) and SOX2 (H) locus; green signals, control probe for the
near centromere locus on chromosome 3. FGFR3, fibroblast growth
factor receptor 3; H&E, hematoxylin and eosin; TTF-1, thyroid
transcription factor-1. |
 | Table IIClinical profiles of the cases with
LC containing an FGFR3 mutation and the pathological,
immunohistochemical, mutational and amplification status of the
LCs. |
Table II
Clinical profiles of the cases with
LC containing an FGFR3 mutation and the pathological,
immunohistochemical, mutational and amplification status of the
LCs.
|
Characteristics | Case No. 57 | Case No. 183 |
|---|
| FGFR3
mutation | p.R248H | p.R248H |
| Age (years) | 62 | 66 |
| Gender | Male | Male |
| Smoking habit | Smoker
(BI=1,800) | Smoker
(BI=1,000) |
| Histology | Squamous cell
carcinoma | Squamous cell
carcinoma |
| Stage | III | II |
| Lymph node
metastasis | Positive | Positive |
| CK14
expression | Positive | Positive |
| TTF-1
expression | Negative | Negative |
| EGFR
mutation (exons 19 and 21) | Wild-type | Wild-type |
| KRAS
mutation (exon 2) | Wild-type | Wild-type |
| PIK3CA
mutation (exons 9 and 20) | Wild-type | Wild-type |
| p53 mutation
(exons 4–9) | Wild-type | p.R273C
mutation |
| FGFR1
amplification | Amplification
(−) | Amplification
(−) |
| PIK3CA
amplification | Amplification
(+) | Amplification
(−) |
| SOX2
amplification | Amplification
(+) | Amplification
(−) |
We next examined whether the LC cases containing the
FGFR3 p.R248H mutation also contained mutations in other
genes that are often mutated in LC (3,8,12,19,20).
Mutation cluster regions for EGFR, KRAS,
PIK3CA and p53 (3,8,12,19,20)
were searched for somatic mutations. No somatic mutations in exon 2
of KRAS, exons 19 and 21 of EGFR, or exons 9 and 20
of PIK3CA were detected; however, a somatic c.817C>T
mutation associated with an amino acid exchange from Arg to Cys at
codon 273 (p.R273C) was detected in the p53 gene in case no.
183 (Fig. 2E, Table II). The fact that the missense
mutation was detected in the DNA binding region of the p53 protein
suggested that the mutant p53 protein was stable in the cancer
cells. We therefore performed immunohistochemical staining for p53
in case no. 183 and the results showed the nuclear accumulation of
p53 exclusively in the cancer cells (Fig. 2F). These results suggested that a
somatic p53 mutation, but not EGFR, KRAS or
PIK3CA mutations, may occur in a subset of LCs containing an
FGFR3 mutation.
We also examined whether the LC cases containing the
FGFR3 p.R248H mutation also contained gene amplifications,
which are frequently observed in LC (12,21,22).
The amplification status of the FGFR1, PIK3CA and
SOX2 genes was examined using a FISH analysis. Although
FGFR1 amplification was not detected in either case, the
amplification of the PIK3CA and SOX2 genes was
detected in case no. 57 (Fig. 2G and
H, Table II). These results
suggested that PIK3CA and SOX2 gene amplification,
but not FGFR1, may occur in a subset of LCs containing an
FGFR3 mutation.
Discussion
In the present study, FGFR3 mutations were
found in two (0.9%) of the 214 LCs that were examined, but the
expression of FGFR3-TACC3 and FGFR3-BAIAP2L1 fusion transcripts was
not detected in any of the 190 LCs that were examined. Both LCs
containing an FGFR3 mutation were histologically diagnosed
as squamous cell carcinoma, and squamous cell carcinoma accounted
for 63 out of the 214 LCs examined in the mutational analysis,
resulting in an incidence of FGFR3 mutation among squamous
cell carcinoma cases of 3.2% (2/63). Both of the mutations were
p.R248H, which was a novel mutation located in the same codon as
the oncogenic p.R248C mutation. Both LCs exhibited an increased
FGFR3 expression, suggesting the involvement of mutated FGFR3 in
LC. Regarding the co-occurring genetic abnormalities, one case
exhibited a p53 p.R273C mutation, while the other case
exhibited PIK3CA and SOX2 amplifications. No somatic
mutations were detected in the mutation cluster regions of the
EGFR, KRAS and PIK3CA genes in either case.
These results suggested that an FGFR3 p.R248H mutation is
involved in the carcinogenesis of a subset of LCs.
A novel p.R248H mutation in the FGFR3 gene
was detected in two lung squamous cell carcinomas derived from
Japanese patients with a smoking habit. The detection of the
p.R248H mutation in two LCs suggests a recurrent mutation in LC.
The p.R248C mutation occurred in the same codon as the p.R248H
mutation and was previously shown to drive cellular transformation;
additionally, the transformation was shown to be reversed by a
small molecule FGFR inhibitor (17). Moreover, the p.R248C mutation was
detected not only in LC, but also in urinary bladder cancer and
multiple myeloma (10,11,23).
Regarding the amino acid conversion from Arg to His, p.R175H and
p.R273H in the p53 gene are hotspot mutations and
gain-of-function mutations (24,25),
suggesting the effect of the amino acid exchange on carcinogenesis.
In addition, the screening for non-acceptable polymorphisms (SNAP)
program, which predicts the effect of single amino acid
substitutions on protein function (http://www.rostlab.org/services/SNAP) (26), predicted that the FGFR3
p.R248H mutation was non-neutral. Finally, in our qRT-PCR
experiment, an increased FGFR3 expression was detected in both LCs
containing a p.R248H mutation. Thus, the FGFR3 p.R248H
mutation may play an important role in the genesis and development
of LCs. In the future, a precise functional investigation may aid
in clarifying the role of the FGFR3 p.R248H mutation.
There have been two previous reports investigating
FGFR3 mutations. Activating FGFR3 mutations were
detected at a frequency of 2.2% (4/178 squamous cell carcinomas) in
one study (12) and 4.2% (2/48
squamous cell carcinomas) in another report (16). In the present study, the incidence
of FGFR3 mutation among squamous cell carcinomas derived
from Japanese patients was 3.2%, although functional
characterization of the mutant was not performed. These results
suggested that FGFR3 mutation is a recurrent event in lung
squamous cell carcinomas in multiple populations.
FGFR3 fusion transcripts were previously found in
one study at a frequency of 4.2% (2/48 squamous cell carcinomas)
(16). On the other hand, no FGFR3
fusion transcripts were detected in our cases, suggesting that
FGFR3 fusion may be rare in the Japanese population. However, since
the number of analyzed squamous cell carcinoma cases in our study
was relatively small (n=60) and FGFR3 may be fused with proteins
other than TACC3 and BAIAP2L1, a future large study examining this
issue is required to examine the incidence of FGFR3 fusion in LCs
derived from Japanese patients.
In our FGFR3 mutation-positive cases, the
co-occurrence of p53 mutation in one case and the
co-occurrence of PIK3CA and SOX2 amplification in the
other case were observed. Both p53 mutation and
amplification of the PIK3CA and SOX2 genes are
frequent in lung squamous cell carcinoma (12,21,22).
These results suggest that a combination of FGFR3 mutation
and such alterations may favor lung squamous cell carcinoma and
FGFR3 mutation is not mutually exclusive with such
alterations in lung squamous cell carcinoma.
In conclusion, our FGFR3 mutation-positive
LCs in conjunction with previously detected FGFR3
mutation-positive LCs suggested that an FGFR3 mutation is
involved in the carcinogenesis of a subset of LCs, especially lung
squamous cell carcinomas, and may aid in elucidating the
characteristics of FGFR3 mutation-positive LCs in the
future.
Acknowledgements
The authors wish to acknowledge Mr. T. Kamo
(Hamamatsu University School of Medicine) for his technical
assistance. The present study was supported in part by a
grant-in-aid from the Ministry of Health, Labour and Welfare
(21-1), a grant-in-aid from the Japan Society for the Promotion of
Science (25460476), a grant-in-aid from the Ministry of Education,
Culture, Sports, Science and Technology (221S0001), and the Smoking
Research Foundation.
References
|
1
|
Soda M, Choi YL, Enomoto M, Takada S,
Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K,
Hatanaka H, Bando M, Ohno S, Ishikawa Y, Aburatani H, Niki T,
Sohara Y, Sugiyama Y and Mano H: Identification of the transforming
EML4-ALK fusion gene in non-small-cell lung cancer. Nature.
448:561–566. 2007.PubMed/NCBI
|
|
2
|
Shinmura K, Kageyama S, Tao H, Bunai T,
Suzuki M, Kamo T, Takamochi K, Suzuki K, Tanahashi M, Niwa H, Ogawa
H and Sugimura H: EML4-ALK fusion transcripts, but no NPM-, TPM3-,
CLTC-, ATIC-, or TFG-ALK fusion transcripts, in non-small cell lung
carcinomas. Lung Cancer. 61:163–169. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schmid K, Oehl N, Wrba F, Pirker R, Pirker
C and Filipits M: EGFR/KRAS/BRAF mutations in primary lung
adenocarcinomas and corresponding locoregional lymph node
metastases. Clin Cancer Res. 15:4554–4560. 2009. View Article : Google Scholar
|
|
4
|
Kohno T, Ichikawa H, Totoki Y, Yasuda K,
Hiramoto M, Nammo T, Sakamoto H, Tsuta K, Furuta K, Shimada Y,
Iwakawa R, Ogiwara H, Oike T, Enari M, Schetter AJ, Okayama H,
Haugen A, Skaug V, Chiku S, Yamanaka I, Arai Y, Watanabe S, Sekine
I, Ogawa S, Harris CC, Tsuda H, Yoshida T, Yokota J and Shibata T:
KIF5B-RET fusions in lung adenocarcinoma. Nat Med.
18:375–377. 2012. View
Article : Google Scholar
|
|
5
|
Takeuchi K, Soda M, Togashi Y, Suzuki R,
Sakata S, Hatano S, Asaka R, Hamanaka W, Ninomiya H, Uehara H, Lim
Choi Y, Satoh Y, Okumura S, Nakagawa K, Mano H and Ishikawa Y: RET,
ROS1 and ALK fusions in lung cancer. Nat Med. 18:378–381. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lipson D, Capelletti M, Yelensky R, Otto
G, Parker A, Jarosz M, Curran JA, Balasubramanian S, Bloom T,
Brennan KW, Donahue A, Downing SR, Frampton GM, Garcia L, Juhn F,
Mitchell KC, White E, White J, Zwirko Z, Peretz T, Nechushtan H,
Soussan-Gutman L, Kim J, Sasaki H, Kim HR, Park SI, Ercan D,
Sheehan CE, Ross JS, Cronin MT, Jänne PA and Stephens PJ:
Identification of new ALK and RET gene fusions from
colorectal and lung cancer biopsies. Nat Med. 18:382–384. 2012.
|
|
7
|
Matsuura S, Shinmura K, Kamo T, Igarashi
H, Maruyama K, Tajima M, Ogawa H, Tanahashi M, Niwa H, Funai K,
Kohno T, Suda T and Sugimura H: CD74-ROS1 fusion transcripts in
resected non-small cell lung carcinoma. Oncol Rep. 30:1675–1680.
2013.PubMed/NCBI
|
|
8
|
Oxnard GR, Binder A and Jänne PA: New
targetable oncogenes in non-small-cell lung cancer. J Clin Oncol.
31:1097–1104. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Casaluce F, Sgambato A, Maione P, Rossi A,
Ferrara C, Napolitano A, Palazzolo G, Ciardiello F and Gridelli C:
ALK inhibitors: a new targeted therapy in the treatment of advanced
NSCLC. Target Oncol. 8:55–67. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cappellen D, De Oliveira C, Ricol D, de
Medina S, Bourdin J, Sastre-Garau X, Chopin D, Thiery JP and
Radvanyi F: Frequent activating mutations of FGFR3 in human bladder
and cervix carcinomas. Nat Genet. 23:18–20. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Greulich H and Pollock PM: Targeting
mutant fibroblast growth factor receptors in cancer. Trends Mol
Med. 17:283–292. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cancer Genome Atlas Research Network.
Comprehensive genomic characterization of squamous cell lung
cancers. Nature. 489:519–525. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Singh D, Chan JM, Zoppoli P, Niola F,
Sullivan R, Castano A, Liu EM, Reichel J, Porrati P, Pellegatta S,
Qiu K, Gao Z, Ceccarelli M, Riccardi R, Brat DJ, Guha A, Aldape K,
Golfinos JG, Zagzag D, Mikkelsen T, Finocchiaro G, Lasorella A,
Rabadan R and Iavarone A: Transforming fusions of FGFR and
TACC genes in human glioblastoma. Science. 337:1231–1235.
2012.PubMed/NCBI
|
|
14
|
Williams SV, Hurst CD and Knowles MA:
Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet.
22:795–803. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu YM, Su F, Kalyana-Sundaram S, Khazanov
N, Ateeq B, Cao X, Lonigro RJ, Vats P, Wang R, Lin SF, Cheng AJ,
Kunju LP, Siddiqui J, Tomlins SA, Wyngaard P, Sadis S, Roychowdhury
S, Hussain MH, Feng FY, Zalupski MM, Talpaz M, Pienta KJ, Rhodes
DR, Robinson DR and Chinnaiyan AM: Identification of targetable
FGFR gene fusions in diverse cancers. Cancer Discov. 3:636–647.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Majewski IJ, Mittempergher L, Davidson NM,
Bosma A, Willems SM, Horlings HM, de Rink I, Greger L, Hooijer GK,
Peters D, Nederlof PM, Hofland I, de Jong J, Wesseling J, Kluin RJ,
Brugman W, Kerkhoven R, Nieboer F, Roepman P, Broeks A, Muley TR,
Jassem J, Niklinski J, van Zandwijk N, Brazma A, Oshlack A, van den
Heuvel M and Bernards R: Identification of recurrent FGFR3
fusion genes in lung cancer through kinome-centred RNA sequencing.
J Pathol. 230:270–276. 2013.
|
|
17
|
Liao RG, Jung J, Tchaicha J, Wilkerson MD,
Sivachenko A, Beauchamp EM, Liu Q, Pugh TJ, Pedamallu CS, Hayes DN,
Gray NS, Getz G, Wong KK, Haddad RI, Meyerson M and Hammerman PS:
Inhibitor-sensitive FGFR2 and FGFR3 mutations in lung squamous cell
carcinoma. Cancer Res. 73:5195–5205. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shinmura K, Goto M, Suzuki M, Tao H,
Yamada H, Igarashi H, Matsuura S, Maeda M, Konno H, Matsuda T and
Sugimura H: Reduced expression of MUTYH with suppressive activity
against mutations caused by 8-hydroxyguanine is a novel predictor
of a poor prognosis in human gastric cancer. J Pathol. 225:414–423.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pao W and Girard N: New driver mutations
in non-small-cell lung cancer. Lancet Oncol. 12:175–180. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ulivi P, Romagnoli M, Chiadini E, Casoni
GL, Capelli L, Gurioli C, Zoli W, Saragoni L, Dubini A, Tesei A,
Amadori D and Poletti V: Assessment of EGFR and K-ras
mutations in fixed and fresh specimens from transesophageal
ultrasound-guided fine needle aspiration in non-small cell lung
cancer patients. Int J Oncol. 41:147–152. 2012.
|
|
21
|
Mantripragada K and Khurshid H: Targeting
genomic alterations in squamous cell lung cancer. Front Oncol.
3:1952013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pros E, Lantuejoul S, Sanchez-Verde L,
Castillo SD, Bonastre E, Suarez-Gauthier A, Conde E, Cigudosa JC,
Lopez-Rios F, Torres-Lanzas J, Castellví J, Ramon y Cajal S,
Brambilla E and Sanchez-Cespedes M: Determining the profiles and
parameters for gene amplification testing of growth factor
receptors in lung cancer. Int J Cancer. 133:898–907. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Intini D, Baldini L, Fabris S, Lombardi L,
Ciceri G, Maiolo AT and Neri A: Analysis of FGFR3 gene
mutations in multiple myeloma patients with t(4;14). Br J Haematol.
114:362–364. 2001.
|
|
24
|
Petitjean A, Mathe E, Kato S, Ishioka C,
Tavtigian SV, Hainaut P and Olivier M: Impact of mutant p53
functional properties on TP53 mutation patterns and tumor
phenotype: lessons from recent developments in the IARC TP53
database. Hum Mutat. 28:622–629. 2007.PubMed/NCBI
|
|
25
|
Yeudall WA, Vaughan CA, Miyazaki H,
Ramamoorthy M, Choi MY, Chapman CG, Wang H, Black E, Bulysheva AA,
Deb SP, Windle B and Deb S: Gain-of-function mutant p53 upregulates
CXC chemokines and enhances cell migration. Carcinogenesis.
33:442–451. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bromberg Y and Rost B: SNAP: predict
effect of non-synonymous polymorphisms on function. Nucleic Acids
Res. 35:3823–3835. 2007. View Article : Google Scholar : PubMed/NCBI
|
















