Introduction
Recently Homo sapiens longevity assurance
homolog 2 of yeast LAG (Lass2) has attracted the interest of
researchers since a large amount of evidence has demonstrated that
Lass2 is a potential tumor-suppressor gene. Deficiency of Lass2 is
involved in the tumorigenesis of various types of tumors,
especially hepatocellular carcinoma (HCC) (1–4). It
has been reported that two of three non-specific Lass2-deleted mice
presented liver cancer spontaneously when they were about nine
months old (3). In our recently
published study, using a hepatocellular-specific Lass2-knockout
(KO) animal model, we found that Lass2-KO mice were more
susceptible to the carcinogen DEN, i.e., DEN caused the Lass2-KO
mice to develop liver tumors earlier and the tumors developed more
rapidly (5).
To explore the biological functions of Lass2 and the
related mechanisms it employs to suppress HCC, the structures and
functions of the livers of the Lass2-KO mice and wild-type (WT)
mice were compared. Meanwhile, microarrays of mRNAs and miRNAs of
the livers from the two genotypes were performed and analyzed.
Materials and methods
Animals
The hepatocyte-specific Lass2-KO mice used in the
present study were generated by the crossing of mice (C57BL/6J)
carrying floxed the second exon of Lass2 and Albumin-Cre transgenic
mice (C57BL/6J), as previously reported (5). All protocols for animal care and use
were approved by the Regulations for the Administration of Affairs
Concerning Experimental Animals (The State Science and Technology
Commission of P.R. China, 1988), and the Animal Experimental Center
of Jiangsu University was licensed for animal experiments. All of
the mice were housed in pathogen-free (SPF) animal facilities under
a standard 12-h-light/12-h-dark cycle. Animals received free access
to water and commercial mouse chow throughout the present study.
One-month-old male Lass2-KO and -WT mice were sacrificed by
cervical dislocation. The mice for the experiments that followed
were n=10/group. Liver weight/body weight were measured.
Biochemical estimation
The mice were sacrificed by cervical dislocation,
and blood harvested from the inferior vena cava was centrifuged at
1,600 × g for 10 min at room temperature to obtain serum for
hepatic biochemical estimation. The activities of aspartate
aminotransferase (AST), alanine aminotransferase (ALT) and lactate
dehydrogenase (LDH) in serum were estimated using an AutoAnalyzer
(Hitachi 7600, Japan) at the Affiliated Hospital of Jiangsu
University.
Histological sections and staining
Liver tissues were immediately removed from the
sacrificed mice, partly fixed in AAF (100% alcohol 85 ml, acetic
acid 5 ml, formalin 10 ml) for morphological examination, and
partly stored at −80°C for further use. The tissues were
paraffin-embedded and sectioned (5-μm thick). Periodic acid-Schiff
(PAS) staining was performed using a commercially available kit
(cat. no. 0609A14; Shellfish Gamma Biotechnology Co., Ltd.,
Nanjing, China). Briefly, the sections were incubated in 0.5%
periodic acid solution for 15 min, rinsed with distilled water, and
exposed to Schiff’s reagent for 20 min followed by two 3-min
exposures to 0.6% sodium metabisulfite.
Western blotting
Tissues were lysed in RIPA lysis buffer containing
the protease inhibitor phenylmethanesulfonyl fluoride (cat. nos.
P0013C and ST506; both from Beyotime Institute of Biotechnology).
The cell extracts were centrifuged at 12,000 × g for 20 min at 4°C
in a Beckman Avanti-30 centrifuge, and the supernatants were used
for the experiments. The protein concentrations were determined
with the BCA assay kit (cat. no. P0009; Beyotime Institute of
Biotechnology). The equivalent tissue proteins (10 μg/lane) were
subjected to electrophoresis on a Mini-Protean Tetra
Electrophoresis System (165–8001; Bio-Rad, Hercules, CA, USA) and
transferred onto PVDF membranes (Millipore, Bedford, MA, USA) via a
semi-dry transfer system (Bio-Rad). The membranes were blocked with
5% non-fat milk for 1 h at room temperature in TBST [50 mM (pH 7.5)
Tris, 0.9% NaCl and 0.1% Tween-20] and then incubated with a 1:200
dilution of rabbit anti-mouse/human albumin (cat. no. BS6520;
Bioworld Technology, Inc., Dublin, OH, USA) overnight at 4°C. The
membranes were washed and then incubated with peroxidase goat
anti-rabbit antibody (cat. no. XR-9920; ProSci Inc., Poway, CA USA)
for 1 h at room temperature and developed using the BeyoEcl Plus
kit (cat. no. P0018; Beyotime Institute of Biotechnology) for 1
min, and then scanned by ChampChemi professional (SG2011; Beijing
Sage Creation Science Co., Ltd., Beijing, China).
RNA isolation and qPCR analysis
Total RNA including miRNA from liver tissues was
extracted by TRIzol (cat. no. 15596026; Invitrogen Corp., Carlsbad,
CA, USA) or the miRNeasy Mini kit (cat. no. 217004; Qiagen, Hilden,
Germany) according to the manufacturer’s suggestions. For mRNA
qPCR, RNA was transcribed into cDNA using the QuantiTect reverse
transcription kit and QuantiTect SYBR-Green PCR kits (cat. no.
205311 and no. 204243; both from Qiagen) according to the
manufacturer’s protocols. For miRNA qPCR, RNA was transcribed into
cDNA using the miScript Reverse II transcription kit (cat. no.
218161; Qiagen). The reaction component consisted of total RNA 1
μg, miScript HiSpec buffer 4 μl, Nucleics Mix 2 μl, miScript
reverse transcriptase mix 2 μl, RNase-free H2O up to 20
μl. The reaction was carried out at 37°C for 60 min and 95°C for 5
min on the ABI PCR 9700 system (Applied Biosystems, Foster City,
CA, USA). cDNA was diluted in 80 μl nuclease-free H2O
for further application by LightCycler 480 SYBR-Green I master
(Roche, Switzerland; cat. no. 04887352001). The reaction system for
qPCR consisted of: LightCycler 480 SYBR-Green I Master 5 μl,
forward primer 0.2 μl, reverse primer 0.2 μl, cDNA 1 μl,
nuclease-free H2O 3.6 μl. PCR was run on ABI 7500 Fast
(Applied Biosystems) and normalized against the expression of GAPDH
or U6. The program was performed at 95°C for 10 min, 95°C for 10
sec plus 60°C for 30 sec for 40 cycles. For the melting curve
evaluation, the temperature was slowly increased from 60°C to 97°C,
and 5 acquisitions per °C were performed continuously. All samples
were analyzed in triplicate. Relative expression was calculated
using the comparative threshold cycle (Ct) method and was indicated
as n-fold change = −(ΔCtKO − ΔCtWT). The
primers for Tnfaip3, NF-κB and GAPDH were: for Tnfaip3,
5′-CAGCACCTAAG CCAACGAGT-3′ and 5′-TGGACCTGTCAATGTGTTCG-3′; for
NF-κB, 5′-AGCTTATGCCGAACTTCTCG-3′ and 5′-GAC TCCGGGATGGAATGTAA-3′;
for GAPDH, 5′-GCAAGG TCATCCCAGAG-3′ and 5′-AAGTCGCAGGAGACAAC-3′;
for miR-694 and U6, the miRNA specific primers were respectively:
5′-CTGAAAATGTTGCCTGAAG-3′ and 5′-CAAGG ATGACACGCAAATTCG-3′, whereas
the reverse primer was the manufacturer-provided miScript universal
primer.
Microarrays of mRNA and miRNA
Liver tissues from 2 one-month male Lass2-KO mice
and 2 one-month male WT mice were respectively subjected to the
experiments of Mouse OneArray (MOA 2.0 ver.) and Mouse and Rat
miRNA OneArray (MRmiOA 4.0 ver.) using Phalanx microarray platform
by Oebiotech Co. (Shanghai, China). Briefly, RNA was extracted from
liver tissues with TRIzol reagent (Invitrogen), whose quantity and
purity were assessed using a NanoDrop ND-1000 spectrophotometer
(NanoDrop Technologies Inc., Wilmington, DE, USA). The purity and
integrity of each extracted RNA met the requirements:
A260/A280 >1.6,
A260/A230 >1 and RNA integrity number
(RIN) value >5. Small RNA fraction indicated the abundance of
RNA <200 nt compared with the overall RNA area from Agilent RNA
6000 Nano assay and was acceptable for the miRNA assay. Possibility
of genomic DNA contamination was excluded by gel electrophoresis.
Two micrograms of RNA from each group was respectively converted
into cyanine-5 labeled target cRNA, hybridized to either Mouse
OneArray or Mouse and Rat miRNA OneArray by the Affymetrix GeneChip
fluidics station 450, and scanned with an Affymetrix GeneChip
scanner 3000 7G. After normalization, differentially expressed
mRNAs or miRNAs were established at log2 |Fold change|
>1 and P<0.05.
For advanced data analysis, all biological
replicates were pooled and calculated to identify differentially
expressed genes based on the threshold of fold change and the
P-value. The correlation of expression profiles between biological
replicates and treatment conditions was demonstrated by
unsupervised hierarchical clustering analysis. A subset of genes
was selected for clustering analysis. An intensity filter was used
to select genes where the difference between the maximum and
minimum intensity values exceeded 4,000 among all microarrays. For
this microarray project, the number of genes clustered was 272
genes. According to previously selected differentially expressed
gene lists, Gene Ontology (GO) analysis was performed by Oebiotech
Co. Targetscan 5.1 was utilized to predict miR-targeting mRNA.
mRNA-miRNA integration analysis demonstrated potential mRNA targets
with inverse expression alterations as their regulatory miRs
displayed in the mRNA microarray or miRNA microarray.
Statistical analysis
Data were analyzed by the Student’s t-test.
P<0.05 was considered to indicate a statistically significant
difference, and P≤0.01 and P≤0.001 are indicated by relevant
symbols in the figures and legends. qPCR and western blot analyses
were repeated three times.
Results
Structural alterations in the liver
tissues from hepatocyte-specific Lass2-KO mice were noted in the
PAS-stained sections
The average ratio of liver weight/body weight of the
Lass2-KO mice was higher than the ratio in the control WT mice
(Fig. 1A). Compared to the liver
tissues from the WT mice, the hepatocytes of the Lass2-KO displayed
abundant vesicles (Fig. 1B).
Liver functions and the expression of ALB
are altered in Lass2-KO mice compared with the WT control
The production of ALB in the liver was attenuated in
the Lass2-KO mice (Fig. 2A). The
levels of ALT, AST and LDH in serum were respectively elevated in
the Lass2-KO mice, indicating abnormal liver function in the
Lass2-KO mice (Fig. 2B–D), in
accordance with the structural injury of the Lass2-KO
hepatocytes.
Profiles of mRNAs and miRNAs in the liver
tissues are respectively reprogrammed in Lass2-KO mice vs. WT
control
mRNA and miRNA microarray results showed that over
600 mRNA were markedly upregulated and over 700 genes were
downregulated; whereas 13 miRNAs were markedly upregulated and 31
miRNAs were downregulated (Table
I). According to the GO analysis, ‘response to wounding’ and
‘inflammatory response’ were among the top-10 altered pathways
(Table II). miRNA-mRNA integrated
analysis identified 4 upregulated and 3 downregulated miRNAs and
their respective negatively controlled target genes (Table III).
 | Table IAltered miRNAs in the Lass2-KO mouse
liver tissues compared to the WT mouse liver tissues. |
Table I
Altered miRNAs in the Lass2-KO mouse
liver tissues compared to the WT mouse liver tissues.
| Upregulated
miRNAs | Downregulated
miRNAs |
|---|
|
mmu-miR-1198-5p | mmu-miR-1192 |
mmu-miR-467d-3p |
|
mmu-miR-125b-5p |
mmu-miR-1224-5p | mmu-miR-467f |
| mmu-miR-142-3p |
mmu-miR-1247-3p | mmu-miR-483-5p |
|
mmu-miR-199a-3p | mmu-miR-149-3p | mmu-miR-494-3p |
|
mmu-miR-199b-3p |
mmu-miR-1894-3p | mmu-miR-5109 |
|
mmu-miR-199a-5p | mmu-miR-1931 | mmu-miR-5136 |
|
mmu-miR-199b-5p |
mmu-miR-1934-3p |
mmu-miR-669a-3p |
| mmu-miR-2137 |
mmu-miR-30c-1-3p |
mmu-miR-669o-3p |
| mmu-miR-2861 | mmu-miR-320-3p |
mmu-miR-669c-3p |
| mmu-miR-326-5p | mmu-miR-346-3p |
mmu-miR-669p-3p |
| mmu-miR-34a-5p |
mmu-miR-466f-3p |
mmu-miR-694 |
| mmu-miR-491-3p |
mmu-miR-466h-3p | mmu-miR-711 |
| mmu-miR-5130 |
mmu-miR-466i-3p | mmu-miR-744-5p |
| mmu-miR-466q | mmu-miR-762 |
|
mmu-miR-467b-3p | mmu-miR-882 |
| |
mmu-miR-92a-2-5p |
 | Table IITop 10 enrichment pathway terms from
the Gene Ontology (GO) analysis (biological process). |
Table II
Top 10 enrichment pathway terms from
the Gene Ontology (GO) analysis (biological process).
| Gene set name | Genes in
overlap | P-value |
|---|
| Response to
wounding | 10 | 0.0407 |
| Humoral immune
response | 3 | 0.0588 |
| Homeostasis of
number of cells | 2 | 0.112 |
| Viral genome
replication | 2 | 0.122 |
| Locomotory
behavior | 5 | 0.122 |
| Inflammatory
response | 6 | 0.148 |
| Development of
primary sexual characteristics | 2 | 0.172 |
| Response to
external stimulus | 12 | 0.172 |
| Jak-Stat
cascade | 2 | 0.225 |
| Viral infectious
cycle | 2 | 0.235 |
 | Table IIImiRNA-mRNA integrated analysis of the
Lass2-KO mouse liver tissues. |
Table III
miRNA-mRNA integrated analysis of the
Lass2-KO mouse liver tissues.
| A, Upregulated
miRNAs and their downregulated predicted target genes |
|---|
|
|---|
| mmu-miR-2861 | mmu-miR-5130 | mmu-miR-142-3p |
mmu-miR-125b-5p |
|---|
| Lrrc41 | Osbpl7 | Atp2a2 | Fam116a |
| Srm | | Baz1a | Ier3ip1 |
| | Rras | Map2k7 |
| | Mastl | Fam78b |
| | Arl15 | Slc17a7 |
| | Lifr | Rasal2 |
| | Tgfb2 | |
| | Myst2 | |
|
| B, Downregulated
miRNAs and their upregulated predicted target genes |
|
| mmu-miR-1192 |
mmu-miR-466f-3p |
mmu-miR-694 |
|
| Tcf4 | Pgm2l1 | Col4a1 | Mef2c |
| Ap3m1 | Tm6sf1 | Junb | Ncoa7 |
| Sgk1 | Angptl2 | Cttnbp2nl | Ska1 |
| Scd2 | Prc1 | 2700081O15Rik | Gng2 |
| Adam23 | Smad7 | Ptgfrn | Abi2 |
| Elavl4 | Klf6 | Ncoa7 | Nipal1 |
| Rbm28 | Trp53inp1 | Zfp532 | Tnfaip3 |
| Phf15 | Tmem65 | Samd4 | Fam120c |
| Arrdc4 | Dgkd | Prrg3 | Srebf1 |
| Slc16a5 | Fam120c | Zmat3 | Man1c1 |
| Arhgef17 | Gpm6b | Ptp4a1 | Vldlr |
| Zfp532 | Nr4a1 | Adam23 | Fbln5 |
| Nrxn2 | Eif4enif1 | Tcf4 | Rai2 |
| Ccna2 | Mef2c | Jun | Prrg3 |
| Fam46a | Pde4d | Fam46a |
| Elavl4 | Tnfaip8 | Plekha6 |
| Odz3 | | E130203B14Rik |
| | | Nup153 |
| | | Trib1 |
| | | Cd63 |
| | | Pde5a |
| | | Serpinh1
(PAI-1) |
| | | Sgms1 |
| | | Elavl4 |
| | | Pde4d |
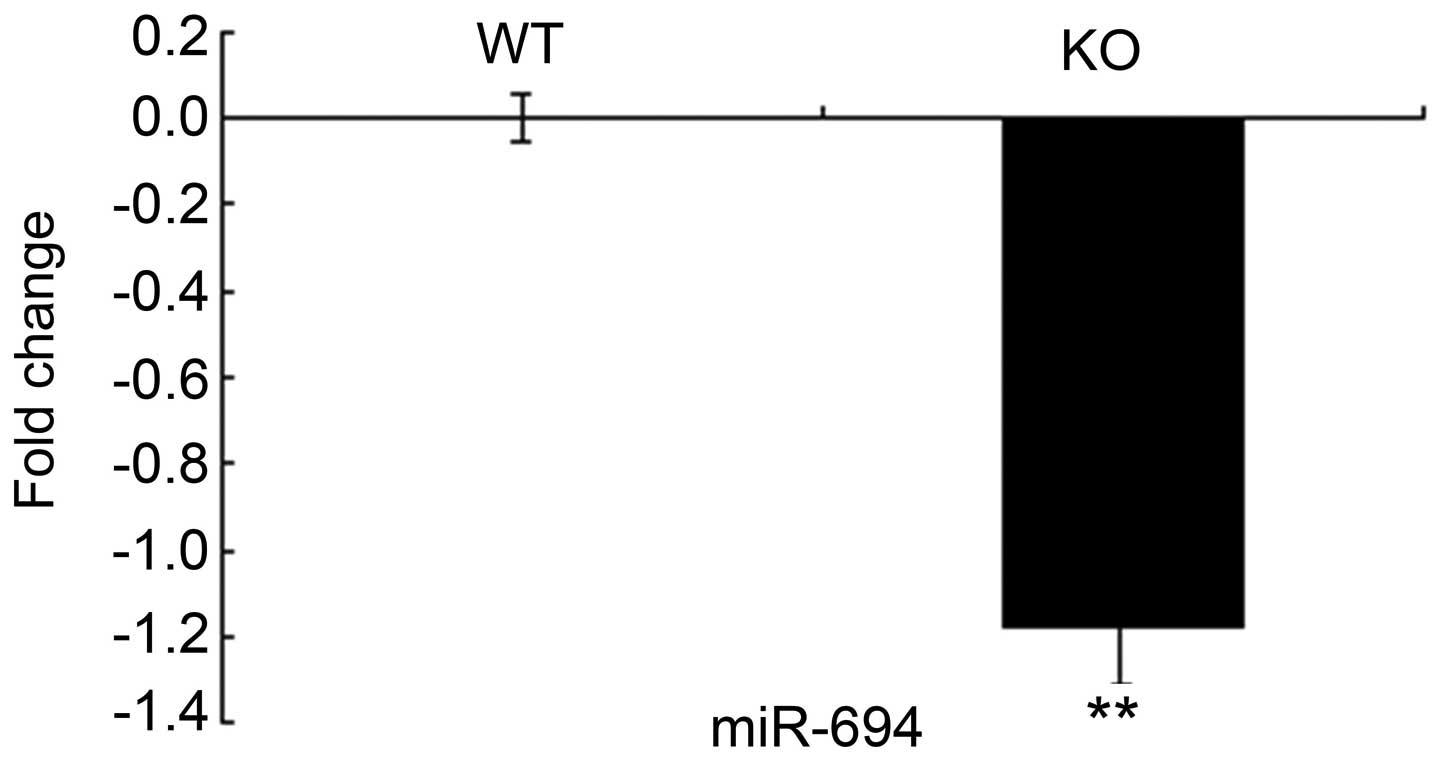
qPCR confirms that miR-694 is markedly
downregulated
The level of miR-694 was downregulated in the
Lass2-KO liver tissue, as confirmed by qPCR (Fig. 3).
Tnfaip3 is markedly upregulated whereas
NF-κB is downregulated
Tnfaip3, one of the markedly upregulated mRNAs in
the Lass2-KO liver tissues, which is also a putative target gene of
miR-694, was confirmed by qPCR (Fig.
4A). Its negatively controlled NF-κB was found to be
downregulated (Fig. 4B).
Discussion
Lass2 is a member of the Lass family, which is
conserved among eukaryotes and is abundantly distributed in the
liver, kidney and brain. The major function of Lass2 is
synthesizing long-chain ceramide-C:24–26 (6). Ceramide serves as the precursor of a
series of more complex sphingolipids. Short-chain ceramides
function as a second messenger in a variety of cellular events,
including apoptosis and differentiation (7,8), and
regulate various cellular processes linked to cancer development,
progression, metastasis and resistance to therapy (9,10). By
comparison, long-chain ceramide, as the important element in
constructing membranous structures, may regulate the cellular
behavior via influencing the property of membranes. For example,
ablation of Lass2 causes morphological alterations of the property
of membranes (11,12). In the present study, numerous
vesicles were noted in the hepatocytes from young Lass2-KO mice
(Fig. 1), in accordance with
previous reports from other researchers. Based on the attenuated
production of albumin and increased hepatic biochemical indices in
the Lass2-KO mice, (Fig. 2), it
appears that the hepatocellular-specific Lass2-KO mice underwent
hepatocellular injury even at an early age.
The microarrays of mRNAs and miRNAs of the Lass2-KO
mice liver tissues vs. control demonstrated that miR-694 was
upregulated, which was confirmed by qPCR. Moreover, the predicted
target gene Tnfaip3 was found to be upregulated, as shown in the
results of either the microarray of mRNA or qPCR. NF-κB, which is
usually commonly negatively controlled by Tnfaip3 was found
downregulated in the Lass2-KO mouse liver tissues. In our previous
study (5), another target gene
Serpinh1 (PAI-1) of miR-694 was found to be upregulated. The data
strongly suggest that Lass2 deletion influences the expression
level of miR-694.
However, the function of miR-694 is uncertain.
According to miRNA-mRNA integrated analysis in this study, the
down-regulated Lass2-related miR-694 elevated 25 mRNAs including
Tnfaip3. Tnfaip3 is commonly considered as an inflammation
suppressor (13) by inactivation of
lymphocytes of suppressing NF-κB (14,15).
Although it is commonly considered a tumor suppressor,
overexpression of Tnfaip3 has also been reported in several
non-lymphoma solid cancers, including HCC (16–18).
Our data suggest that Tnfaip3/NF-κB might play a role in inhibiting
the inflammation and protecting injured hepatocytes caused by
deletion of Lass2. In another report, Lass2-KO mice displayed
resistance to LPS-induced liver injury (19), which might also be explained by the
inhibitory immunity mediated by Tnfaip3/NF-κB.
The functions of other predicted target genes of
miR-694 are listed in Table IV.
According to published reports, miR-694 target genes are involved
it the regulation of various important cellular events, including
transcription, cell signal transduction, metabolism of lipid and
glucose and trafficking, suggesting a diversity of regulatory
functions of miR-694. miRNAs act as highly effective regulators of
intracellular events. Moreover, via endocytosis or exocytosis of
miRNA-containing vesicles, miRNAs modulate the extracellular milieu
including stromal cells and extracellular matrix (20). For example, altered Tnfaip3/NF-κB
may influence the immunocytes in the liver, or PAI-1 which was
found to be upregulated, may regulate the synthesis of ECM in the
liver (5), either of which is a
predicted target gene of miR-694. The actual functions of miR-694
may likely be beyond what is listed in Table IV.
 | Table IVFunctions of the miR-694 predicted
target genes. |
Table IV
Functions of the miR-694 predicted
target genes.
| Function | miR-694 target gene
(ref.) |
|---|
| Transcription | Mef2c (21) |
| Ncoa7 (22) |
| Tnfaip3 (13,14) |
| Elavl4 (23,24) |
| Cell cycle | Ska1 (25) |
| Nup153 (26) |
| Cell signal
transduction | Gng2 (27) |
| Abi2 (28,29) |
| Pde5a (30) |
| Pde4d (31) |
| Metabolism
regulator | Trib1 (32–34) |
| Sgms1 (35–37) |
| Srebf1 (38) |
| Vldlr (39) |
| Trafficking | CD63 (40,41) |
| Man1c1 (42) |
| Nup153 (26) |
| Sgms1 (43) |
| ECM deposition | Fbln5 (44,45) |
| PAI-1 (46) |
| Immunity
regulation | Trib1 (47,48) |
| Tnfaip3 (13,14) |
Overall, the present study first reports the
attenuated expression level of miR-694 in Lass2-KO mouse liver
tissue and its alteration of the Tnfaip3/NF-κB pathway. Our data
strongly suggest that miR-694 functions in maintaining homeostasis
of the liver and provide the basis to explore the functions of
Lass2-related microRNAs.
Acknowledgements
This study was supported by the Grant from the State
Key Laboratory of Oncogenes and Related Genes (no. 90-10-02, to
X.L.) and Clinical Medicine Science & Technology Project of
Jiangsu Province of China (no. BL2013024).
Abbreviations:
|
Lass2
|
longevity assurance gene 2
|
|
KO
|
knockout
|
|
miR or miRNA
|
microRNA
|
|
Tnfaip3
|
tumor necrosis factor α-induced
protein 3
|
|
NF-κB
|
nuclear transcription factor-κB
|
|
qPCR
|
real-time quantitative PCR
|
|
HCC
|
hepatocellular carcinoma
|
References
|
1
|
Pewzner-Jung Y, Ben-Dor S and Futerman AH:
When do Lasses (longevity assurance genes) become CerS (ceramide
synthases)? Insights into the regulation of ceramide synthesis. J
Biol Chem. 281:25001–25005. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang H, Wang J, Zuo Y, et al: Expression
and prognostic significance of a new tumor metastasis suppressor
gene LASS2 in human bladder carcinoma. Med Oncol. 29:1921–1927.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fan S, Niu Y, Tan N, et al: LASS2 enhances
chemosensitivity of breast cancer by counteracting acidic tumor
microenvironment through inhibiting activity of V-ATPase proton
pump. Oncogene. 32:1682–1690. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Imgrund S, Hartmann D, Farwanah H, et al:
Adult ceramide synthase 2 (CERS2)-deficient mice exhibit myelin
sheath defects, cerebellar degeneration, and hepatocarcinomas. J
Biol Chem. 284:33549–33560. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen L, Lu X, Zeng T, et al: Enhancement
of DEN-induced liver tumourigenesis in hepatocyte-specific
Lass2-knockout mice coincident with upregulation of the
TGF-β1-Smad4-PAI-1 axis. Oncol Rep. 31:885–893. 2014.PubMed/NCBI
|
|
6
|
Teufel A, Maass T, Galle PR, et al: The
longevity assurance homologue of yeast lag1 (Lass) gene family
(Review). Int J Mol Med. 23:135–140. 2009.PubMed/NCBI
|
|
7
|
Morad SAF and Cabot MC:
Ceramide-orchestrated signalling in cancer cells. Nat Rev Cancer.
13:51–65. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Stiban J, Tidhar R and Futerman AH:
Ceramide synthases: roles in cell physiology and signaling. Adv Exp
Med Biol. 688:60–71. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Saddoughi SA and Ogretmen B: Diverse
functions of ceramide in cancer cell death and proliferation. Adv
Cancer Res. 117:37–58. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu J, Beckman BS and Foroozesh M: A
review of ceramide analogs as potential anticancer agents. Future
Med Chem. 5:1405–1421. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Park JW, Park WJ, Kuperman Y, Boura-Halfon
S, Pewzner-Jung Y and Futerman AH: Ablation of very long acyl chain
sphingolipids causes hepatic insulin resistance in mice due to
altered detergent-resistant membranes. Hepatology. 57:525–532.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Silva LC, Ben David O, Pewzner-Jung Y, et
al: Ablation of ceramide synthase 2 strongly affects biophysical
properties of membranes. J Lipid Res. 53:430–436. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ma A and Malynn BA: A20: linking a complex
regulator of ubiquitylation to immunity and human disease. Nat Rev
Immunol. 12:774–785. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pujari R, Hunte R, Khan W and Shembade N:
A20-mediated negative regulation of canonical NF-κB signaling
pathway. Immunol Res. 57:166–171. 2013.PubMed/NCBI
|
|
15
|
Zhang F, Yang L and Li Y: The role of A20
in the pathogenesis of lymphocytic malignancy. Cancer Cell Int.
12:442012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang M and Li S: Bladder polypoid
cystitis-derived A20 associates with tumorigenesis. Cell Biochem
Biophys. 67:669–673. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hjelmeland AB, Wu Q, Wickman S, et al:
Targeting A20 decreases glioma stem cell survival and tumor growth.
PLoS Biol. 8:e10003192010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang CM, Wang Y, Fan CG, et al: miR-29c
targets TNFAIP3, inhibits cell proliferation and induces apoptosis
in hepatitis B virus-related hepatocellular carcinoma. Biochem
Biophys Res Commun. 411:586–592. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ali M, Fritsch J, Zigdon H, Pewzner-Jung
Y, Schütze S and Futerman AH: Altering the sphingolipid acyl chain
composition prevents LPS/GLN-mediated hepatic failure in mice by
disrupting TNFR1 internalization. Cell Death Dis. 4:e9292013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Su Y, Li X, Ji W, et al: Small molecule
with big role: MicroRNAs in cancer metastatic microenvironments.
Cancer Lett. 344:147–156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cante-Barrett K, Pieters R and Meijerink
JPP: Myocyte enhancer factor 2C in hematopoiesis and leukemia.
Oncogene. 33:403–410. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Higginbotham KS, Breyer JP, Bradley KM, et
al: A multistage association study identifies a breast cancer
genetic locus at NCOA7. Cancer Res. 71:3881–3888. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bronicki LM and Jasmin BJ: Emerging
complexity of the HuD/ELAVl4 gene; implications for neuronal
development, function, and dysfunction. RNA. 19:1019–1037. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lee EK, Kim W, Tominaga K, et al:
RNA-binding protein HuD controls insulin translation. Mol Cell.
45:826–835. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ye AA and Maresca TJ: Cell division:
kinetochores SKAdaddle. Curr Biol. 23:R122–R124. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ullman KS, Shah S, Powers MA and Forbes
DJ: The nucleoporin nup153 plays a critical role in multiple types
of nuclear export. Mol Biol Cell. 10:649–664. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Matsuda T, Hashimoto Y, Ueda H, et al:
Specific isoprenyl group linked to transducin gamma-subunit is a
determinant of its unique signaling properties among G-proteins.
Biochemistry. 37:9843–9850. 1998. View Article : Google Scholar
|
|
28
|
Grove M, Demyanenko G, Echarri A, et al:
ABI2-deficient mice exhibit defective cell migration, aberrant
dendritic spine morphogenesis, and deficits in learning and memory.
Mol Cell Biol. 24:10905–10922. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dai Z and Pendergast AM: Abi-2, a novel
SH3-containing protein interacts with the c-Abl tyrosine kinase and
modulates c-Abl transforming activity. Genes Dev. 9:2569–2582.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kulkarni SK and Patil CS:
Phosphodiesterase 5 enzyme and its inhibitors: update on
pharmacological and therapeutical aspects. Methods Find Exp Clin
Pharmacol. 26:789–799. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lynch MJ, Baillie GS and Houslay MD:
cAMP-specific phosphodiesterase-4D5 (PDE4D5) provides a paradigm
for understanding the unique non-redundant roles that PDE4 isoforms
play in shaping compartmentalized cAMP cell signalling. Biochem Soc
Trans. 35:938–941. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ishizuka Y, Nakayama K, Ogawa A, et al:
TRIB1 downregulates hepatic lipogenesis and glycogenesis via
multiple molecular interactions. J Mol Endocrinol. 52:145–158.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Angyal A and Kiss-Toth E: The tribbles
gene family and lipoprotein metabolism. Curr Opin Lipidol.
23:122–126. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dugast E, Kiss-Toth E, Soulillou JP,
Brouard S and Ashton-Chess J: The Tribbles-1 protein in humans:
roles and functions in health and disease. Curr Mol Med. 13:80–85.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Vacaru AM, Tafesse FG, Ternes P, et al:
Sphingomyelin synthase-related protein SMSr controls ceramide
homeostasis in the ER. J Cell Biol. 185:1013–1027. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu J, Zhang H, Li Z, et al: Sphingomyelin
synthase 2 is one of the determinants for plasma and liver
sphingomyelin levels in mice. Arterioscler Thromb Vasc Biol.
29:850–856. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Separovic D, Hanada K, Awad Maitah MIY, et
al: Sphingomyelin synthase 1 suppresses ceramide production and
apoptosis post-photodamage. Biochem Biophys Res Commun.
358:196–202. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ruiz R, Jideonwo V, Ahn M, et al: Sterol
regulatory element-binding protein-1 (SREBP-1) is required to
regulate glycogen synthesis and gluconeogenic gene expression in
mouse liver. J Biol Chem. 289:5510–5517. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Go GW and Mani A: Low-density lipoprotein
receptor (LDLR) family orchestrates cholesterol homeostasis. Yale J
Biol Med. 85:19–28. 2012.PubMed/NCBI
|
|
40
|
Pols MS and Klumperman J: Trafficking and
function of the tetraspanin CD63. Exp Cell Res. 315:1584–1592.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Maecker HT, Todd SC and Levy S: The
tetraspanin superfamily: molecular facilitators. FASEB J.
11:428–442. 1997.PubMed/NCBI
|
|
42
|
Scott DW and Patel RP: Endothelial
heterogeneity and adhesion molecules N-glycosylation: implications
in leukocyte trafficking in inflammation. Glycobiology. 23:622–633.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Subathra M, Qureshi A and Luberto C:
Sphingomyelin synthases regulate protein trafficking and secretion.
PLoS One. 6:e236442011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kapustin A, Stepanova V, Aniol N, et al:
Fibulin-5 binds urokinase-type plasminogen activator and mediates
urokinase-stimulated β1-integrin-dependent cell migration. Biochem
J. 443:491–503. 2012.PubMed/NCBI
|
|
45
|
Choi J, Bergdahl A, Zheng Q, Starcher B,
Yanagisawa H and Davis EC: Analysis of dermal elastic fibers in the
absence of fibulin-5 reveals potential roles for fibulin-5 in
elastic fiber assembly. Matrix Biol. 28:211–220. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Declerck PJ and Gils A: Three decades of
research on plasminogen activator inhibitor-1: a multifaceted
serpin. Semin Thromb Hemost. 39:356–364. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Satoh T, Kidoya H, Naito H, et al:
Critical role of Trib1 in differentiation of tissue-resident
M2-like macrophages. Nature. 495:524–528. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Dugast E, Kiss-Toth E, Docherty L, et al:
Identification of Tribbles-1 as a novel binding partner of Foxp3 in
regulatory T cells. J Biol Chem. 288:10051–10060. 2013. View Article : Google Scholar : PubMed/NCBI
|