1. Comprehensive cancer projects
Large-scale collaborative cancer projects generate a
large amount of cancer data. The International Cancer Genome
Consortium (ICGC) (1) and The
Cancer Genome Atlas (TCGA) (2) are
the most prominent examples of such efforts (Table I). ICGC aims to obtain a
comprehensive description of the genomic, transcriptomic and
epigenomic changes in 50 different tumor types and/or subtypes that
are of clinical and social significance (1). The curated data types are: sample
donor IDs, cancer project, simple somatic mutations (SSMs) and
genes with SSMs. The data are complemented by associated
attributes, including the primary site of the tumor at diagnosis
(e.g., brain, skin, blood, bone and prostate), gender, tumor stage
at diagnosis (e.g., 4, M0, M3, 1, M1), available data types (e.g.,
copy number and structural somatic mutations, miRNA expression,
gene expression, DNA methylation and exon junction.
 | Table ICancer databases. |
The ICGC Data Portal (3) provides tools for querying, visualizing
and downloading the data released quarterly by the consortium’s
member projects. The ICGC Data Portal contains data from other
large-scale cancer genome projects, including TCGA, Johns Hopkins
University (Baltimore, MD, USA) (4,5) and
Tumor Sequencing Project (TSP) (6).
The ICGC Data Portal is based on the BioMart data management
platform (7,8), which uses a seamless federated data
model to enable the cross querying of diverse biological databases
in a unified manner. To maintain the uniformity of ICGC datasets,
the same set of data models, ontologies, controlled vocabularies
and references have been applied in all of the ICGC’s member
databases. Three interfaces are available: cancer projects,
advanced search and data repository. The cancer projects interface
contains data available in the 49 ICGC member projects, as well as
additional filters and a selection of attributes. The advanced
search interface contains the complete set of filters and
attributes. The database can be queried interactively using three
main options: donors, genes and mutations. By selecting any of
these options, the results are presented in tabulated form. The
results can be filtered based on several search criteria. The Data
Repository provides access to all ICGC cancer project data,
including uniformly processed and annotated data files. The results
can be downloaded and exported for further analysis.
TCGA is a joint project of the National Cancer
Institute (NCI) and the National Human Genome Research Institute
(NHGRI) (both from Bethesda, MD, USA) that provides a comprehensive
map of the important genomic changes that occur in the major types
and subtypes of cancer (2). It
contains clinical information, genomic characterization data and
high level sequence analysis of the tumor genomes. The TCGA Data
Portal enables investigators to explore, download and analyze
datasets generated by TCGA. The data types stored in TCGA include
gene expression, copy number, somatic mutations, single nucleotide
polymorphisms (SNPs), microRNAs, clinical outcomes and tissue slide
images. Four main methods for downloading data are available: i)
Data Matrix enables users to select and download a subset of data
for a particular cancer type, but does not allow searching and
downloading data across multiple cancer types simultaneously; ii)
Bulk Download facilitates the bulk download of archives of data as
uploaded by the TCGA Centers; iii) File Search allows users to
filter and download data files in a more easily accessible manner;
and iv) Access HTTP Directories enables users to access the HTTP
directories where the data archives are stored. The TCGA Roadmap
(9) engine was developed to index
and annotate the TCGA files and capture file metadata in the TCGA
open-access HTTP by applying third-generation web technologies (Web
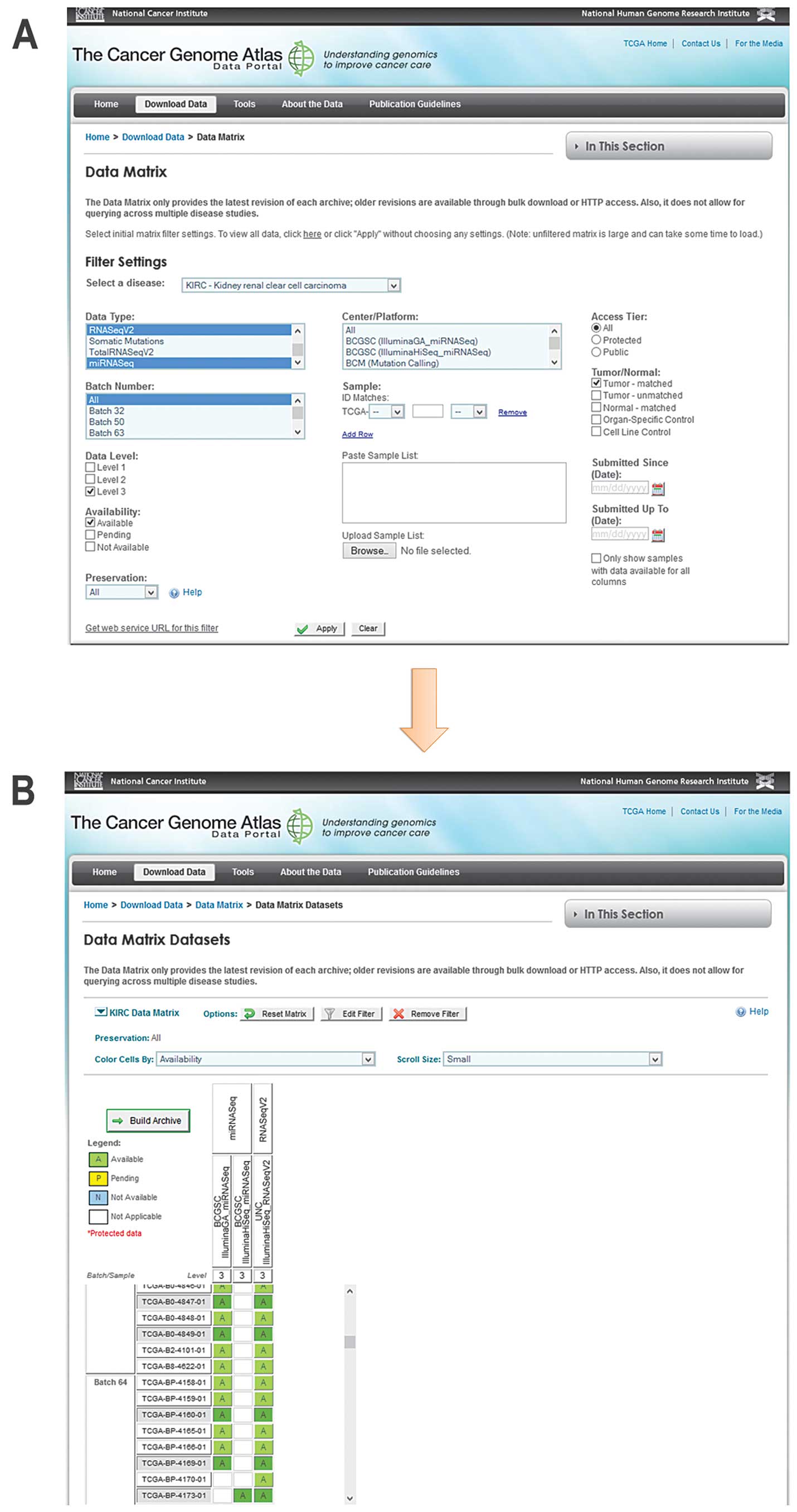
3.0) (10). An example of searching
for and downloading processed data concerning expressed genes and
microRNAs from next generation sequencing (NGS) experiments in
matched tumors is shown in Fig.
1.
The Clinical Proteomic Tumor Analysis Consortium
(CPTAC) (11,12), launched by NCI, aims at elucidating
the molecular basis of cancer through the application of proteomic
techniques. In particular, CPTAC analyzes cancer samples by mass
spectrometry in order to identify and quantify their constituent
proteins and localize the post-translational protein modifications,
such as phosphorylation. The CPTAC Data Portal is the central
repository for the distribution of proteomic data collected by the
Proteome Characterization Centers (PCCs). The CPTAC Data Portal
uses an Aspera Connect Transfer Server for the transport of large
data files. The Cancer Genome Project (CGP) (13) at the Sanger Institute (Cambridge,
UK), seeks to identify somatic variants/mutations critical in the
etiology and pathogenesis of human cancers by using the sequenced
human genome and high-throughput mutation detection
technologies.
2. Resources
The large volume of data that emerges from these
large-scale programs has resulted in the concomitant development of
new databases for accessing and analyzing cancer data (Table I).
Tools
Specialized web-based tools are available to enable
investigators to query, retrieve and analyze cancer-related data in
a rapid, reliable and efficient manner. The Cancer Genome Anatomy
Project (CGAP) (14) of NCI
includes a number of bioinformatic analysis tools and
interconnected modules that enable users to access CGAP data. These
data include cancer-relevant genes and SNPs, malignant tissues and
chromosomal aberrations in cancer patients. Moreover, CGAP provides
information regarding the differential expression of a given gene
in normal, precancerous and cancerous tissues based on Serial
Analysis of Gene Expression (SAGE), as well as RNA interference
(RNAi) constructs that target cancer-related genes, and diagrams of
biochemical pathways and protein complexes. The UCSC Cancer
Genomics Browser (15,16) is a suite of web-based tools used to
integrate, display and analyze cancer genomics and clinical data.
The browser allows whole-genome views of several different types of
genomics and associated clinical data. Various datasets can be
viewed together as coordinated ‘heatmap tracks’, thus enabling the
user to make comparisons across studies and cancer types. Annotated
biological pathways, collections of genes, genomic or clinical
information can be sorted, filtered, aggregated, classified and
viewed interactively based on any given feature set, including
clinical features, annotated biological pathways and
user-contributed collections of genes. The Cancer Genome Workbench
(CGWB) (17) includes copy number,
mutation, expression and methylation data from various projects,
including TCGA, the Catalogue of Somatic Mutations in Cancer
(COSMIC) (18,19), Johns Hopkins University, and the
Therapeutically Applicable Research To Generate Effective
Treatments (TARGET) initiative. CGWB provides a series of tools for
visualizing genomic and transcription alterations from different
cancer samples. The data in CGWB can be viewed in three different
ways: i) Integrated track, which provides a sample-level view of
genomic alterations from multiple data sources; ii) Heatmap view,
an interactive graphical view of gene expression and copy number
data and their associated clinical features; and iii) Bambino, an
alignment viewer for NGS data.
Cancer driver genes
Several repositories of driver genes or gene
families that play a causal role in carcinogenesis have been
developed. The Tumor Gene Family Databases (TGDBs) (20) contain a broad range of information
regarding genes involved in cancer. Apart from TGDB itself, the
data of two component databases, the Oral Cancer Gene Database
(OrCGDB) (21) and the Breast
Cancer Gene Database (BCGD) (22),
have been merged into TGDBs. Gene information includes gene
aliases, cell location, biochemical function, frequency in various
tumors, oncogenicity, chromosomal location, tumor gene type (either
proto-oncogene or tumor suppressor gene) and the signal
transduction pathways in which the gene of interest is
involved.
The DriverDB database (23) compiles a large amount (>6,000
cases) of exome-sequencing (exome-seq) data, annotation databases
such as dbSNP (24), 1000 Genome
(25) and COSMIC, as well as
various bioinformatics algorithms for the identification of driver
genes or mutations. The database can be queried either by cancer
type, where the driver genes/mutations for a specific cancer type
are estimated, or by gene where the mutation information of a
driver gene in five different aspects is presented. Meta-Analysis,
another option offered in DriverDB, enables users to identify
driver genes in custom-defined samples according to clinical
criteria. The RAS Oncogene Database (RASOnD) (20) integrates large amounts of genomics
and proteomics data derived from publicly available databases such
as NCBI’s GenBank (26), Online
Mendelian Inheritance in Man (OMIM) (27), Universal Protein Resource (UniProt)
(28), Protein Databank (PDB)
(29), Kyoto Encyclopedia of Genes
and Genomes (KEGG) (30) and PubMed
(31). The RASOnD database contains
199,046 entries from 101 species, allowing investigators to
retrieve information regarding RAS oncogene SNPs, chromosomal
positions, disease associations and nucleotide and amino acid
positions.
Genetic variations
Cancer is characterized by abundant genetic
abnormalities in the form of mutations, SNPs, copy number
alterations (CNAs), genomic rearrangements and gene fusions. To
manage the increasing amount of information, public resources have
been implemented to collect, curate, annotate and analyze data
regarding cancer genetic variations. COSMIC (18,19) is
the largest public database that stores and displays information on
somatically acquired mutations involved in cancer and associated
clinical and phenotypic data. Currently, COSMIC contains
information on 28,735 genes, 2,002,811 coding mutations and 10,435
fusion gene mutations reported in 1,029,547 cancer samples. The
data are primarily extracted from published scientific literature
and whole-genome sequencing screens from CGP. To provide a uniform
representation of the data, a histology and tissue ontology has
been created. COSMIC uses the BioMart (32) data mining software that enables
users to filter the available data according to cancer sample,
gene, mutation, tumor site, histology and tumor. The results are
presented in tabulated format (Fig.
2).
The Cancer Gene Census (CGC) (33) lists >1% of all human genes which
bear mutations that causally contribute to carcinogenesis. The Gene
Census data include gene symbol according to HGNC (34), a short description of the gene, gene
chromosomal location, type of mutations (i.e., somatic or
germline), type of tumor and the cancer syndrome in which the
mutated gene is involved. The data are provided in a table and can
be downloaded and exported in several formats. The catalogue is
updated at regular intervals. BioMuta (35) is a curated database of
cancer-related non-synonymous single-nucleotide variations (nsSNVs)
that affect functional sites. The datasets are derived from the
TCGA, COSMIC, ClinVar (36) and
UniProt Knowledgease (UniProtKB) (37) databases. Due to the large amount of
data present in the primary NGS repositories, the High-performance
Integrated Virtual Environment (HIVE) platform (35) has been implemented in BioMuta in
order to store, analyze, compute and curate NGS data and associated
metadata. CaSNP (38) is a
comprehensive collection of CNAs from 11,485 Affymetrix SNP arrays
with raw data from NCBI’s Gene Expression Omnibus (GEO) (39), additional arrays from the TCGA
consortium and a few individual publications covering 34 different
cancer types in 105 studies. The user can query CaSNP by gene,
region or cancer type and retrieve information regarding the
frequencies of copy number aberrations for each study. CaSNP also
provides a heatmap showing CNAs estimated at each SNP marker around
the query region across all studies. CanProVar (40) has been developed to store and
prevent germline and somatic amino acid variations in the human
proteome associated with human tumorigenesis based on published
literature. The CanGEM (41)
database stores clinical information on tumor samples and array
Comparative Genomic Hybridization (aCGH) data to detect gene CNAs
in cancer. Users can create custom datasets for specific clinical
sample characteristics or CNAs of individual genes. The Integrative
Cancer Profiler System (ICPS) (10)
database integrates genomic alterations such asCNA and LOH, with
transcription signatures (SAGE, microarray) in order to study gene
profiles in one or more different types/subtypes of cancer.
Currently, ICPS contains five different data types and 23,375
experiments covering 11 major cancer types. Apart from public data,
ICPS also supports in-house data of users.
Given that ~50% of human tumors harbor TP53 gene
mutations (42), a UMD TP53
(43) database was created to
provide detailed information on TP53 mutants such as the molecular
and cell properties of each TP53 mutant and localization or various
gains of functions. The UMD TP53 database contains >110,000
entries. The International Agency for Research on Cancer (IARC)
TP53 (44,45) is a comprehensive resource that
compiles all TP53 gene variations in human cancers derived from
scientific publications. The datasets available in the resource
are: TP53 somatic and germline mutations, validated common TP53
polymorphisms identified in human population and their functional
and clinical impact, TP53 gene status (i.e., wild-type, mutant,
null) in various human cell lines, mouse models with engineered
TP53 constructs and experimentally induced TP53 mutations.
Epigenetic modifications
Epigenetic modifications, such as DNA methylation
and chromatin-modifying factors, play a critical role in
carcinogenesis by regulating tumor-suppressor gene silencing,
proto-oncogene activation and chromosomal instability (46). MethyCancer (47), the database of human DNA methylation
and cancer, was developed to study the association of DNA
methylation, gene expression and cancer. It contains data of DNA
methylation, cancer-relevant genes and CpG Island (CGI) clones
derived from high-throughput sequencing. The MethyView option
allows the graphical presentation of CGI information of >30,000
genes. PubMeth (48) is a cancer
methylation database that includes information of genes that have
been reported in the literature to be methylated in various cancer
types. The information is extracted from PubMed abstracts using a
text-mining approach, GoldMine, followed by manual annotation.
There are two options for searching the database, the
‘gene-centric’ (the cancer types/subtypes where the genes of the
interest are reported to be methylated) and the ‘cancer-centric’
(the genes reported to be methylated in a particular cancer
type/subtype). In ChromoHub V2 (49), chemical, structural and biological
data extracted from public repositories, such as TCGA and ICGC, are
mapped on phylogenetic trees of protein families involved in
chromatin-mediated signaling.
OncomiRs
OncomiRs, microRNAs that are associated with diverse
cancer-related processes, play a significant role in the epigenetic
regulation of cancer. The miRCancer database (50) provides a comprehensive collection of
microRNA expression profiles in various human malignancies that are
automatically extracted from publications in PubMed. OncomiRDB
(51) is a database developed for
annotating the experimentally validated oncomiRs from literature.
The database includes 2,259 entries of oncomiR regulations,
covering 328 miRNAs and 829 target genes in 25 cancer tissues
extracted from published literature. The user is able to search by
miRNA, tissue, tumor, target gene and function (e.g.,
proliferation, apoptosis, migration).
Transcriptomics
Databases have been designated to extract, store and
interpret data from large-scale and genome-wide expression studies.
Oncomine (52) is a cancer
microarray database that collects and curates 715 gene expression
data-sets and 86,733 samples and associated clinical data from most
major types of data. Oncomine allows a user to contact a
gene-centric search to retrieve the differential expression
analyses of a gene of interest across all available datasets; in a
study-centric search, the genes that are differentially expressed
in the selected study are provided. To facilitate data mining, the
current version of Oncomine enables multi-gene search, gene
ontology-based filtering and integration of Oncomine concepts.
Integrated Tumor Transcriptome Array and Clinical data Analysis
(ITTACA) (53) is a central
repository of transcriptome microarray and associated clinical data
from breast carcinoma, bladder carcinoma and uveal melanoma. A web
interface offers different options for class comparison analyses,
such as the comparison of profiles of expression distribution and
patient survival analyses. The user is able to analyze the
differential expression of one or more gene between two groups of
samples with different phenotypes, and, conversely, the genes
differentially expressed between two groups of samples (Fig. 3).
The Cancer Gene Expression Database (CGED) (54) contains cancer gene expression
profiles and related clinical information. The expression data are
obtained by adaptor-tagged competitive PCR from breast, colorectal,
esophageal, gastric, hepatocellular, lung and thyroid cancers and
glioma. The database can be queried either using gene identifiers
or by functional categories. Mosaic plots are used for the
visualization of gene expression data and comparison of the
expression patterns of various genes. CancerMA (55) is an integrated bioinformatic
pipeline used for the automated identification of novel candidate
cancer biomarkers by analyzing the expression profiles of a
user-defined gene list across public cancer microarray [GEO,
ArrayExpress (56)] experimentally
verified datasets. A total of 80 microarray datasets covering 13
types of cancer are available.
Proteins
Differentially expressed proteins (DEPs) that
contribute to the onset and progression of cancer have been
identified. The first database of DEPs in human cancers, dbDEPC
(57,58), currently contains 4,029 DEPs,
curated from 331 mass spectrometry experiments across 20 types of
human cancer. This resource enables the users to investigate
whether a protein of interest has altered in particular cancers
and, to create an association network of query proteins. Moreover,
dbDEPC shows a heatmap that represents the expression profiles of a
certain protein across various cancer types. An example of how to
query dbDEPC is shown in Fig.
4.
Phosphorylation
The genes encoding protein kinases, enzymes that
phosphorylate proteins, are among the most commonly mutated genes
in human cancers. The MoKCa database (59) provides a collection of the mutations
present in protein kinases involved in cancer, along with
structural and functional annotation and, wherever possible,
prediction of the impact of these mutations in the structure and
function of kinases. The user can select from a pull-down list the
gene that codes for a protein kinase: information is available for
the types of mutations (e.g., missense, silent) found in tumor cell
lines and, the mutated amino acid residues are mapped onto the
tertiary structures of the affected protein kinase domains.
Cell lines
Cancers are thought to be initiated and maintained
by a subpopulation of stem or stem-like cells with tumorigenic
potential (60). SCDE (61,62) is
an integrated repository of curated tissue and cancer stem cell
data from blood, brain and intestine. The datasets are homogenized
with regard to structure, formatting and annotation and stored in
the Investigation/Study/Assay-Tab (ISA-Tab) format. SCDE is linked
to the Galaxy framework which provides a series of analytical tools
to compare those data to genes, molecular signatures and pathways.
The CellLineNavigator (63)
database contains gene expression profiles (generated uniformly) of
>300 human cancer lines. On the basis of their phenotypic
attributes, these cell lines were further classified into 28
tissues of origin and 57 different disease states. The database is
also linked to advanced tools of bioinformatics analyses. The
database can be searched for i) differentially expressed genes; ii)
pathological or physiological states; and iii) gene names or
functional characteristics, such as Gene Ontologies (GOs) (64) and KEGG pathway maps. A combination
of all query options is also possible.
Cytogenetics
Chromosomal aberrations, such as translocations and
their corresponding gene fusions and duplications or deletions and
generated gene gains or losses are known to play an important role
in the onset of tumorigenesis (65). The Mitelman Database of Chromosome
Aberrations and Gene Fusions in Cancer (65) is a repository of chromosomal
rearrangements such as translocations and their resulting gene
fusions that are associated with tumor characteristics. The data
contained in the database have been manually extracted from
literature. The current version of the database includes a total of
64,679 cases and 2,094 gene fusions. The Mitelman Database can be
queried by several different search options. In particular, the
user can search for individual patient cases or associations of
cases, such as concurrent chromosomal aberrations and clinical
associations of cytogenetic abnormalities, or references
themselves.
Immunomics
Tumor-associated antigens (TAAs) have been applied
extensively in the clinical diagnosis and treatment of human
cancers. The publicly available Human Potential Tumor Associated
Antigen (HPtaa) (66) database
contains potential TTAs identified by in silico computing.
HPtaa incorporates publicly available microarray expression data,
GEO’s SAGE data and Unigene (67)
expression data, as well as other relevant knowledge bases such as
CGAP. Currently, a total of 3,518 potential targets are included in
the database. A web query interface enables users to search for
potential TAAs overexpressed in several cancer types with
particular gene features including chromosome (X and Y or
euchromosome), coding capacity (protein-coding genes or else) and
subcellular location (membrane or secretory proteins). The human
immune response (humoral and cellular) to an increasing number of
TAAs has also been well documented. The Academy of Cancer
Immunology supported by the Ludwig Institute (both from New York,
NY, USA) for Cancer Research have established the Cancer Immunome
Database (CID) (68) which provides
information on all the gene products against which an immunome
response has been reported in cancer patients. The user can access
information regarding the genes that encode the cancer antigens.
Given that a gene can yield multiple antigenic epitopes, the
frequency with which the antigenic epitopes are recognized by sera
or cells from healthy and diseased individuals is reported.
Wherever appropriate, access is provided to experimental evidence
(serological results, microscope images and cytotoxic assays) or
patients’ information (the type of cancer from which they are
suffering, the disease stage, and time the samples were obtained).
CTdatabase (69) is a curated
repository of annotated and computationally predicted cancer-testis
(CT) antigens. The CT antigens are broadly classified according to
their expression pattern in human healthy tissue. CTdatabase also
provides information on genes, the verified splice variants,
genomic locations, gene duplications and bibliographical
references.
Anticancer agents
Knowledge bases dedicated to cancer translational
research and identification of drugs and compounds that inhibit
cancer-related target genes are also available. CanSAR (70) is a public resource that supports
cancer translational research and finding of drug through the
integration of biological, chemical, pharmacological and disease
data, structural biology and cellular networks. The user, through a
single portal, is able to access information regarding genes,
protein families, cell lines and compounds, as well as approved
drugs and clinical candidates associates with cancer.
CancerResource (71) is a
comprehensive knowledge base that integrates cancer-relevant
relationships of compounds/drugs and targets deduced from the text
mining of >19 million PubMed abstracts and external resources
such as Therapeutic Target Database (TTD) (72), Comparative Toxicogenomics Database
(CTD) (73), Pharmacogenomics
Knowledge Base (PharmGKB) (74) and
DrugBank (75). CancerResource can
be queried by Cancer (the user can view the genes expressed in
specific cancer tissues, and also browse cancer-related KEGG
pathways), Drug (the user can search by compound or drug and obtain
information about the cancer relevance of the query drug/compound
and its interactions with targets) and Target (drugs interacting
with targets). The Anticancer Agent Mechanism Database (76,77)
contains a list of 122 compounds with anticancer activity
classified by their mechanism of action into alkylating agents,
topoisomerase I/II inhibitors, RNA/DNA antimetabolites, and
antimitotic agents. This set is generated by neural networks able
to predict the mechanism of action of a drug based on its pattern
of activity against a diverse panel of human tumor cell lines in
the NCI drug screening program.
Drug resistance
A major obstacle in cancer therapies is the
development of drug resistance based on mutations in drug targets.
Therefore, it is important to identify mutations in drug targets
responsible for drug resistance. CancerDR (78) provides information of 148 anticancer
drugs and their pharmacological profiling across ~1,000 cancer cell
lines. Pharmacological profiling information of these anticancer
drugs was collected from the Cancer Cell Line Encyclopedia (CCLE)
(79) and COSMIC databases.
CancerDR provides information about each drug target (cancer genes)
that corresponds to these anti-cancer drugs, such as gene sequences
in respective cancer cell lines, mutations, function and structure.
This database allows users to search for drug targets, drugs, cell
lines and structure. Clustering of cell lines on the basis of their
drug sensitivity towards a drug target allows users to identify
groups of cell lines, which are resistance to a particular
anticancer drug, as well as multipotent drugs effective against a
wide range of cancer cell lines. The clustering of sequences of a
drug target is important to identify mutants/variants against the
corresponding drug target.
Integrative resources
IntOGen (80) is an
integrative resource of high-throughput data associated with
genomic, transcriptional, mutational alterations and modules (e.g.,
GO terms, KEGG pathways) involved in carcinogenesis. IntOGen
collects data from various resources such as COSMIC, GEO,
ArrayExpress, Progenetix (81),
TCGA and CGP. Tumor samples in IntOGen are annotated with terms
from the International Classification of Diseases for Oncology
(ICD-O) (82) where the tumors are
classified based on their topography (location in the human body)
and histology (morphology). IntOGen can be queried by Genes,
Projects, Cancer sites and Pathways. The Biomart portal (83) enables more complicated queries and
the bulk download of all analysis results. The interface has a
number of filters and attributes. IntOGen Biomart can be queried
based on i) IntOGen Experiments, where the user can query gene or
module (e.g., GO terms, KEGG pathways) information; ii) IntOGen
Combinations, where the user is allowed to query a combination of
experiments annotated with the same ICD-O term; and iii)
IntOGenOncomodules which enables the user to search for
combinations and experiments (Fig.
5). NCG 4.0 (84,85) is the current version of the Network
of Cancer Genes, a repository of systems-level properties of cancer
genes and oncomiRs (cancer-related microRNAs). It compiles
information on 2,000 cancer genes that have been reported in
literature to be mutated in 23 different types of cancer collected
from 3,460 whole-exome and -genome screenings of cancer samples.
NCG 4.0 reports information on the duplicability, functional
annotation, evolutionary origin and interactions with other human
proteins and microRNAs.
3. Cancer type-specific databases
Databases that focus on certain types or subtypes of
cancer are also available (Table
I).
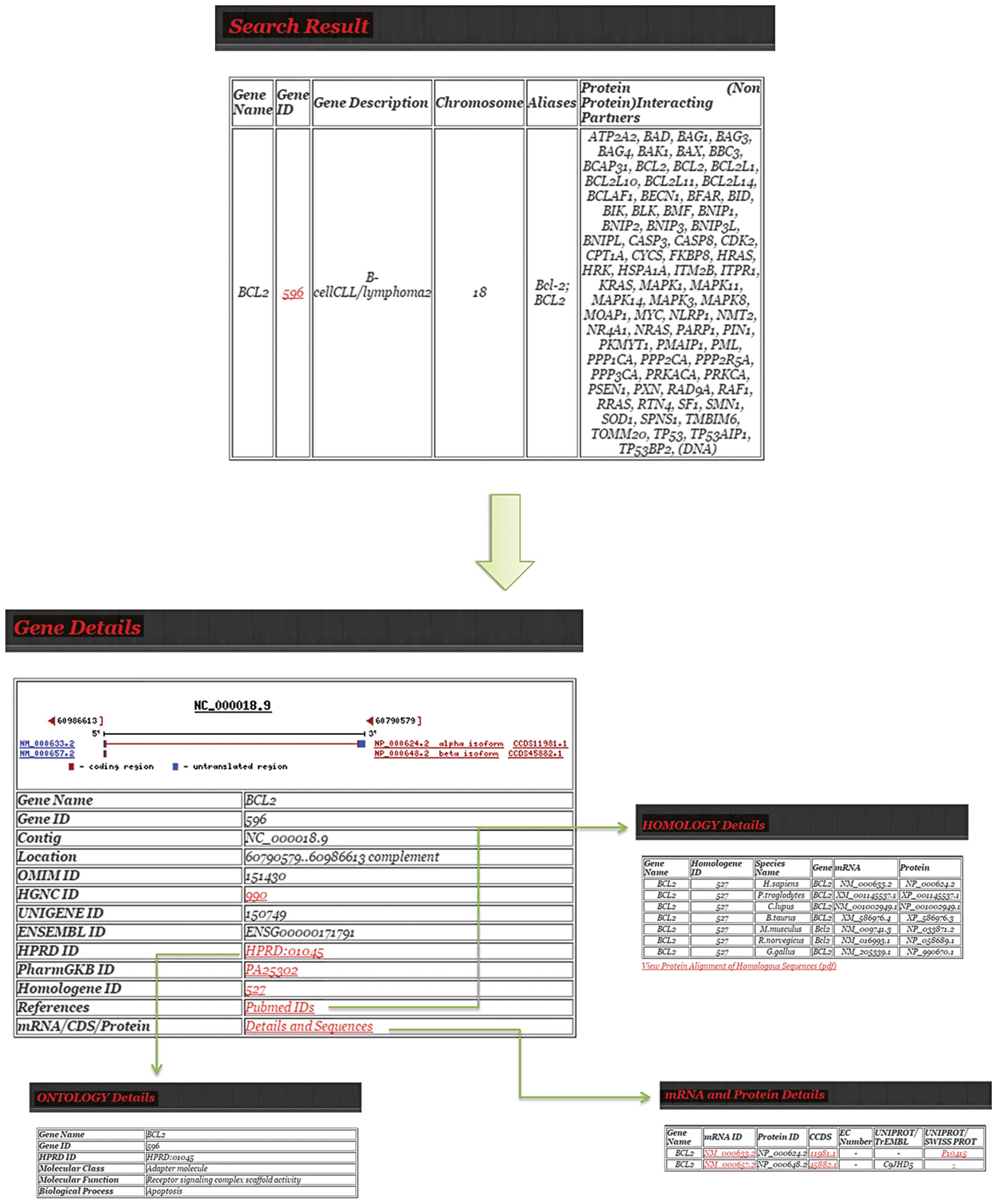
The Cervical Cancer gene DataBase (CCDB) (86) contains a manually curated list of
experimentally validated genes reported to be involved in different
aspects of cervical carcinogenesis. Each record includes
information concerning the gene of interest such as gene structure,
chromosomal location, homology, ontology, and mRNA/CDC/protein
sequences for each isoform encoded by the gene, as well as links to
the original PubMed references and external databases [e.g., HGNC,
Human Protein Reference Database (HPRD) (87), Homologene (67), PharmGKB, PDB]. The database can be
queried by i) Gene name (the user can obtain information pertinent
to the query gene); ii) Category, where the genes are grouped into
categories; and iii) Chromosome number to view all cervical
cancer-related genes present in a particular chromosome (Fig. 6).
The Dragon Database of Genes associated with
Prostate Cancer (DDPC) (88) is an
integrated resource of genes that have been experimentally
confirmed to be involved in Prostate cancer. DDPC provides
information about each gene such as experimental evidence,
associated pathways, orthologous genes, gene ontologies, and
related proteins. The user can select a gene from a pull-down list
or search the database for genes using a combination of one or more
options, including Anatomical System, Cell Line, KEGG Pathways, and
Gene Ontology. DDPC also contains a list of the predicted
transcription factor-binding sites on the promoters of genes
included in the database. Moreover, the database contains DrugBank
drugs reported to be associated with prostate cancer.
The curatedOvarianData (89) resource provides gene expression data
and documented clinical annotations from 2,970 ovarian cancer
patients from 23 studies with ovarian cancer across 11 microarray
platforms. The data are made available as ExpressionSet objects for
R/Bioconductor (90). The gene
expression datasets are obtained from public databases, processed
in a uniform manner and mapped to standard HGNC gene symbols
(34).
The Genes-to-Systems Breast Cancer (G2SBC) Database
(91) is an integrated resource of
genes, transcripts and proteins reported in the literature to be
dysregulated in breast cancer. Moreover, in G2SBC, the analysis is
performed at different levels: the molecular components level,
where the analysis is performed at the level of genes, transcripts
and proteins; the molecular systems level, an analysis based on
biological processes and protein-protein interaction networks; and
the cellular systems level where the user can browse and simulate
mathematical models of carcinogenesis, tumor growth and response to
treatments. An ontology-based query system is also available for
annotations associated with particular ontologies.
The HLungDB (92) is
an integrated resource of lung cancer-related genes, proteins and
microRNAs and pertinent clinical information extracted manually
from the scientific literature. Each entry in the database
describes the relationships between genes and lung cancer,
containing detailed information of the gene, the expression pattern
of the relevant gene (up- or downregulated) in the patient,
experimentally verified information (e.g., transcription factor
binding sites in the promoter of the gene) and protein-protein
interaction networks. The database includes miRNAs that are
differentially expressed in lung cancer or reported to be
associated with lung cancer along with their experimentally
verified identified targets. HLungDB is cross-linked to relevant
external resources, including PubMed, HPRD, HUGO, IPI, EBI and
KEGG. The lung cancer-related genes can be viewed either from a
pull-down list where the genes are sorted by alphabetical order or
by chromosome where the user can view all cancer-related genes
located in the selected chromosome.
The Osteosarcoma Database (93) is a repository of osteosarcoma
(OS)-relevant genes and microRNAs. The data stored in database are
extracted from PubMed using an automated dictionary-based gene and
microRNA recognition procedure, manual review and annotation.
Currently, the database contains 911 protein-coding genes and 81
microRNAs deduced from 1,331 abstracts. The user is able to search
by Gene or microRNA. Each entry is linked to PubMed.
The Pancreatic Expression Database (PED) (94), powered by the BioMart software, is a
comprehensive resource of pancreatic cancer data from the
literature obtained using a range of technologies, including
genomics, transcriptomics, proteomics and miRNA. PED includes tools
for mining data by using a combination of queries (e.g., gene
expression and CNAs). The use of BioMart facilitates
interoperability with other BioMart-compliant cancer resources,
which allows users to expand their investigations to a number of
relevant resources, such as Reactome, PRIDE and COSMIC.
The Renal Cancer Gene Database (RCDB) (95) is a manually curated repository of
protein-coding genes and miRNAs associated with various forms of
renal cell carcinomas (RCC). The protein-coding genes have been
classified into six categories according to the type of alteration
observed in RCC: i) methylation; ii) overexpression; iii)
downregulation; iv) mutation; v) translocation; and vi)
unclassified. RCDB also includes the miRNAs dysregulated in RCC.
Users are able to query the protein-coding genes and miRNAs using
keyword, category or, in the case of genes, chromosome. The
ViroBLAST (96) tool is used to
query a user-defined sequence against the sequences available in
RCDB.
References
|
1
|
International Cancer Genome Consortium.
Hudson TJ, Anderson W, Artez A, et al: International network of
cancer genome projects. Nature. 464:993–998. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cancer Genome Atlas Research Network.
Weinstein JN, Collisson EA, Mills GB, et al: The Cancer Genome
Atlas Pan-Cancer analysis project. Nat Genet. 45:1113–1120. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang J, Baran J, Cros A, et al:
International Cancer Genome Consortium Data Portal - a one-stop
shop for cancer genomics data. Database (Oxford) 2011.
bar0262011.
|
|
4
|
Parsons DW, Jones S, Zhang X, et al: An
integrated genomic analysis of human glioblastoma multiforme.
Science. 321:1807–1812. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jones S, Zhang X, Parsons DW, et al: Core
signaling pathways in human pancreatic cancers revealed by global
genomic analyses. Science. 321:1801–1806. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ding L, Getz G, Wheeler DA, et al: Somatic
mutations affect key pathways in lung adenocarcinoma. Nature.
455:1069–1075. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guberman JM, Ai J, Arnaiz O, et al:
BioMart Central Portal: an open database network for the biological
community. Database (Oxford) 2011. bar0412011.
|
|
8
|
Haider S, Ballester B, Smedley D, Zhang J,
Rice P and Kasprzyk A: BioMart Central Portal - unified access to
biological data. Nucleic Acids Res. 37:W23–W27. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Robbins DE, Grüneberg A, Deus HF, Tanik MM
and Almeida JS: A self-updating road map of The Cancer Genome
Atlas. Bioinformatics. 29:1333–1340. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang XY, Shi L, Liu Y, et al: ICPS: an
integrative cancer profiler system. Bioinformatics. 26:2649–2650.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ellis MJ, Gillette M, Carr SA, et al:
Connecting genomic alterations to cancer biology with proteomics:
the NCI Clinical Proteomic Tumor Analysis Consortium. Cancer
Discov. 3:1108–1112. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang B, Wang J, Wang X, et al:
Proteogenomic characterization of human colon and rectal cancer.
Nature. 513:382–387. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pleasance ED, Cheetham RK, Stephens PJ, et
al: A comprehensive catalogue of somatic mutations from a human
cancer genome. Nature. 463:191–196. 2010. View Article : Google Scholar
|
|
14
|
Hess JL: The Cancer Genome Anatomy
Project: power tools for cancer biologists. Cancer Invest.
21:325–326. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Goldman M, Craft B, Swatloski T, et al:
The UCSC Cancer Genomics Browser: update 2013. Nucleic Acids Res.
41:D949–D954. 2013. View Article : Google Scholar :
|
|
16
|
Zhu J, Sanborn JZ, Benz S, et al: The UCSC
Cancer Genomics Browser. Nat Methods. 6:239–240. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang J, Finney RP, Rowe W, et al:
Systematic analysis of genetic alterations in tumors using Cancer
Genome WorkBench (CGWB). Genome Res. 17:1111–1117. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Forbes SA, Bindal N, Bamford S, et al:
COSMIC: mining complete cancer genomes in the Catalogue of Somatic
Mutations in Cancer. Nucleic Acids Res. 39:D945–D950. 2011.
View Article : Google Scholar :
|
|
19
|
Forbes SA, Tang G, Bindal N, et al: COSMIC
(the Catalogue of Somatic Mutations in Cancer): a resource to
investigate acquired mutations in human cancer. Nucleic Acids Res.
38:D652–D657. 2010. View Article : Google Scholar :
|
|
20
|
Kulsum U, Singh V, Sharma S, Srinivasan A,
Singh TP and Kaur P: RASOnD-a comprehensive resource and search
tool for RAS superfamily oncogenes from various species. BMC
Genomics. 12:3412011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Levine AE and Steffen DL: OrCGDB: a
database of genes involved in oral cancer. Nucleic Acids Res.
29:300–302. 2001. View Article : Google Scholar :
|
|
22
|
Baasiri RA, Glasser SR, Steffen DL and
Wheeler DA: The breast cancer gene database: a collaborative
information resource. Oncogene. 18:7958–7965. 1999. View Article : Google Scholar
|
|
23
|
Cheng WC, Chung IF, Chen CY, et al:
DriverDB: an exome sequencing database for cancer driver gene
identification. Nucleic Acids Res. 42:D1048–D1054. 2014. View Article : Google Scholar :
|
|
24
|
Sherry ST, Ward MH, Kholodov M, et al:
dbSNP: the NCBI database of genetic variation. Nucleic Acids Res.
29:308–311. 2001. View Article : Google Scholar :
|
|
25
|
1000 Genomes Project Consortium. Abecasis
GR, Auton A, Brooks LD, et al: An integrated map of genetic
variation from 1,092 human genomes. Nature. 491:56–65. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Benson DA, Clark K, Karsch-Mizrachi I,
Lipman DJ, Ostell J and Sayers EW: GenBank. Nucleic Acids Res.
42:D32–D37. 2014. View Article : Google Scholar :
|
|
27
|
Amberger J, Bocchini CA, Scott AF and
Hamosh A: McKusick’s Online Mendelian Inheritance in Man (OMIM).
Nucleic Acids Res. 37:D793–D796. 2009. View Article : Google Scholar
|
|
28
|
UniProt Consortium. Activities at the
Universal Protein Resource (UniProt). Nucleic Acids Res.
42:D191–D198. 2014. View Article : Google Scholar :
|
|
29
|
Rose PW, Bi C, Bluhm WF, et al: The RCSB
Protein Data Bank: new resources for research and education.
Nucleic Acids Res. 41:D475–D482. 2013. View Article : Google Scholar :
|
|
30
|
Du J, Yuan Z, Ma Z, Song J, Xie X and Chen
Y: KEGG-PATH: Kyoto encyclopedia of genes and genomes-based pathway
analysis using a path analysis model. Mol Biosyst. 10:2441–2447.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
McEntyre J and Lipman D: PubMed: bridging
the information gap. CMAJ. 164:1317–1319. 2001.PubMed/NCBI
|
|
32
|
Shepherd R, Forbes SA, Beare D, et al:
Data mining using the Catalogue of Somatic Mutations in Cancer
BioMart. Database (Oxford) 2011. bar0182011.
|
|
33
|
Futreal PA, Coin L, Marshall M, et al: A
census of human cancer genes. Nat Rev Cancer. 4:177–183. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gray KA, Daugherty LC, Gordon SM, Seal RL,
Wright MW and Bruford EA: Genenames.org: the HGNC resources in
2013. Nucleic Acids Res. 41:D545–D552. 2013. View Article : Google Scholar :
|
|
35
|
Wu TJ, Shamsaddini A, Pan Y, et al: A
framework for organizing cancer-related variations from existing
databases, publications and NGS data using a High-performance
Integrated Virtual Environment (HIVE). Database (Oxford) 2014.
bau0222014. View Article : Google Scholar
|
|
36
|
Landrum MJ, Lee JM, Riley GR, et al:
ClinVar: public archive of relationships among sequence variation
and human phenotype. Nucleic Acids Res. 42:D980–D985. 2014.
View Article : Google Scholar :
|
|
37
|
Magrane M and Consortium U: UniProt
Knowledgebase: a hub of integrated protein data. Database (Oxford)
2011. bar0092011.
|
|
38
|
Cao Q, Zhou M, Wang X, et al: CaSNP: a
database for interrogating copy number alterations of cancer genome
from SNP array data. Nucleic Acids Res. 39:D968–D974. 2011.
View Article : Google Scholar :
|
|
39
|
Barrett T, Wilhite SE, Ledoux P, et al:
NCBI GEO: archive for functional genomics data sets - update.
Nucleic Acids Res. 41:D991–D995. 2013. View Article : Google Scholar
|
|
40
|
Li J, Duncan DT and Zhang B: CanProVar: a
human cancer proteome variation database. Hum Mutat. 31:219–228.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Scheinin I, Myllykangas S, Borze I,
Böhling T, Knuutila S and Saharinen J: CanGEM: mining gene copy
number changes in cancer. Nucleic Acids Res. 36:D830–D835. 2008.
View Article : Google Scholar :
|
|
42
|
Levine AJ and Oren M: The first 30 years
of p53: growing ever more complex. Nat Rev Cancer. 9:749–758. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Leroy B, Anderson M and Soussi T: TP53
mutations in human cancer: database reassessment and prospects for
the next decade. Hum Mutat. 35:672–688. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hainaut P, Hernandez T, Robinson A, et al:
IARC. Database of p53 gene mutations in human tumors and cell
lines: updated compilation, revised formats and new visualisation
tools. Nucleic Acids Res. 26:205–213. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Petitjean A, Mathe E, Kato S, et al:
Impact of mutant p53 functional properties on TP53 mutation
patterns and tumor phenotype: lessons from recent developments in
the IARC TP53 database. Hum Mutat. 28:622–629. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Pavlopoulou A and Kossida S: Cytosine
methyltransferases as tumor markers. Curr Genomics. 11:568–577.
2010. View Article : Google Scholar
|
|
47
|
He X, Chang S, Zhang J, et al:
MethyCancer: the database of human DNA methylation and cancer.
Nucleic Acids Res. 36:D836–D841. 2008. View Article : Google Scholar :
|
|
48
|
Ongenaert M, Van Neste L, De Meyer T,
Menschaert G, Bekaert S and Van Criekinge W: PubMeth: a cancer
methylation database combining text-mining and expert annotation.
Nucleic Acids Res. 36:D842–D846. 2008. View Article : Google Scholar :
|
|
49
|
Liu L, Zhen XT, Denton E, Marsden BD and
Schapira M: ChromoHub: a data hub for navigators of
chromatin-mediated signalling. Bioinformatics. 28:2205–2206. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Xie B, Ding Q, Han H and Wu D: miRCancer:
a microRNA-cancer association database constructed by text mining
on literature. Bioinformatics. 29:638–644. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang D, Gu J, Wang T and Ding Z:
OncomiRDB: a database for the experimentally verified oncogenic and
tumor-suppressive microRNAs. Bioinformatics. 30:2237–2238. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Rhodes DR, Yu J, Shanker K, et al:
ONCOMINE: a cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Elfilali A, Lair S, Verbeke C, La Rosa P,
Radvanyi F and Barillot E: ITTACA: a new database for integrated
tumor transcriptome array and clinical data analysis. Nucleic Acids
Res. 34:D613–D616. 2006. View Article : Google Scholar :
|
|
54
|
Kato K, Yamashita R, Matoba R, et al:
Cancer gene expression database (CGED): a database for gene
expression profiling with accompanying clinical information of
human cancer tissues. Nucleic Acids Res. 33:D533–D536. 2005.
View Article : Google Scholar :
|
|
55
|
Feichtinger J, McFarlane RJ and Larcombe
LD: CancerMA: a web-based tool for automatic meta-analysis of
public cancer microarray data. Database (Oxford) 2012.
bas0552012.
|
|
56
|
Rustici G, Kolesnikov N, Brandizi M, et
al: ArrayExpress update - trends in database growth and links to
data analysis tools. Nucleic Acids Res. 41:D987–D990. 2013.
View Article : Google Scholar
|
|
57
|
He Y, Zhang M, Ju Y, et al: dbDEPC 2.0:
updated database of differentially expressed proteins in human
cancers. Nucleic Acids Res. 40:D964–D971. 2012. View Article : Google Scholar :
|
|
58
|
Li H, He Y, Ding G, Wang C, Xie L and Li
Y: dbDEPC: a database of differentially expressed proteins in human
cancers. Nucleic Acids Res. 38:D658–D664. 2010. View Article : Google Scholar :
|
|
59
|
Richardson CJ, Gao Q, Mitsopoulous C,
Zvelebil M, Pearl LH and Pearl FM: MoKCa database - mutations of
kinases in cancer. Nucleic Acids Res. 37:D824–D831. 2009.
View Article : Google Scholar
|
|
60
|
Reya T, Morrison SJ, Clarke MF and
Weissman IL: Stem cells, cancer, and cancer stem cells. Nature.
414:105–111. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Ho Sui SJ, Begley K, Reilly D, et al: The
Stem Cell Discovery Engine: an integrated repository and analysis
system for cancer stem cell comparisons. Nucleic Acids Res.
40:D984–D991. 2012. View Article : Google Scholar :
|
|
62
|
Sansone SA, Rocca-Serra P, Field D, et al:
Toward interoperable bioscience data. Nat Genet. 44:121–126. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Krupp M, Itzel T, Maass T, Hildebrandt A,
Galle PR and Teufel A: CellLineNavigator: a workbench for cancer
cell line analysis. Nucleic Acids Res. 41:D942–D948. 2013.
View Article : Google Scholar :
|
|
64
|
Gene Ontology Consortium. Blake JA, Dolan
M, Drabkin H, et al: Gene Ontology annotations and resources.
Nucleic Acids Res. 41:D530–D535. 2013. View Article : Google Scholar
|
|
65
|
Mitelman F, Johansson B and Mertens F: The
impact of translocations and gene fusions on cancer causation. Nat
Rev Cancer. 7:233–245. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Wang X, Zhao H, Xu Q, et al: HPtaa
database-potential target genes for clinical diagnosis and
immunotherapy of human carcinoma. Nucleic Acids Res. 34:D607–D612.
2006. View Article : Google Scholar :
|
|
67
|
NCBI Resource Coordinators. Database
resources of the National Center for Biotechnology Information.
Nucleic Acids Res. 42:D7–D17. 2014. View Article : Google Scholar :
|
|
68
|
Jongeneel V: Towards a cancer immunome
database. Cancer Immun. 1:32001.
|
|
69
|
Almeida LG, Sakabe NJ, deOliveira AR, et
al: CTdatabase: a knowledge-base of high-throughput and curated
data on cancer-testis antigens. Nucleic Acids Res. 37:D816–D819.
2009. View Article : Google Scholar :
|
|
70
|
Bulusu KC, Tym JE, Coker EA, Schierz AC
and Al-Lazikani B: canSAR: updated cancer research and drug
discovery knowledgebase. Nucleic Acids Res. 42:D1040–D1047. 2014.
View Article : Google Scholar :
|
|
71
|
Ahmed J, Meinel T, Dunkel M, et al:
CancerResource: a comprehensive database of cancer-relevant
proteins and compound interactions supported by experimental
knowledge. Nucleic Acids Res. 39:D960–D967. 2011. View Article : Google Scholar :
|
|
72
|
Qin C, Zhang C, Zhu F, et al: Therapeutic
target database update 2014: a resource for targeted therapeutics.
Nucleic Acids Res. 42:D1118–D1123. 2014. View Article : Google Scholar :
|
|
73
|
Davis AP, Murphy CG, Johnson R, et al: The
Comparative Toxicogenomics Database: update 2013. Nucleic Acids
Res. 41:D1104–D1114. 2013. View Article : Google Scholar :
|
|
74
|
Thorn CF, Klein TE and Altman RB:
PharmGKB: the Pharmacogenomics Knowledge Base. Methods Mol Biol.
1015:311–320. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Law V, Knox C, Djoumbou Y, et al: DrugBank
4.0: shedding new light on drug metabolism. Nucleic Acids Res.
42:D1091–D1097. 2014. View Article : Google Scholar :
|
|
76
|
Weinstein JN, Kohn KW, Grever MR, et al:
Neural computing in cancer drug development: predicting mechanism
of action. Science. 258:447–451. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Monks A, Scudiero D, Skehan P, et al:
Feasibility of a high-flux anticancer drug screen using a diverse
panel of cultured human tumor cell lines. J Natl Cancer Inst.
83:757–766. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Kumar R, Chaudhary K, Gupta S, et al:
CancerDR: cancer drug resistance database. Sci Rep. 3:14452013.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Barretina J, Caponigro G, Stransky N, et
al: The Cancer Cell Line Encyclopedia enables predictive modelling
of anticancer drug sensitivity. Nature. 483:603–607. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Gundem G, Perez-Llamas C, Jene-Sanz A, et
al: IntOGen: integration and data mining of multidimensional
oncogenomic data. Nat Methods. 7:92–93. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Cai H, Kumar N, Ai N, Gupta S, Rath P and
Baudis M: Progenetix: 12 years of oncogenomic data curation.
Nucleic Acids Res. 42:D1055–D1062. 2014. View Article : Google Scholar :
|
|
82
|
Percy C, Van Holten V and Muir C:
International Classification of Diseases for Oncology. 2nd edition.
World Health Organization; Geneva: 1990
|
|
83
|
Perez-Llamas C, Gundem G and Lopez-Bigas
N: Integrative cancer genomics (IntOGen) in Biomart. Database
(Oxford) 2011. bar0392011.
|
|
84
|
An O, Pendino V, D’Antonio M, Ratti E,
Gentilini M and Ciccarelli FD: NCG 40: the network of cancer genes
in the era of massive mutational screenings of cancer genomes.
Database (Oxford) 2014. bau0152014. View Article : Google Scholar
|
|
85
|
D’Antonio M, Pendino V, Sinha S and
Ciccarelli FD: Network of Cancer Genes (NCG 3.0): integration and
analysis of genetic and network properties of cancer genes. Nucleic
Acids Res. 40:D978–D983. 2012. View Article : Google Scholar :
|
|
86
|
Agarwal SM, Raghav D, Singh H and Raghava
GP: CCDB: a curated database of genes involved in cervix cancer.
Nucleic Acids Res. 39:D975–D979. 2011. View Article : Google Scholar :
|
|
87
|
Keshava Prasad TS, Goel R, Kandasamy K, et
al: Human Protein Reference Database - 2009 update. Nucleic Acids
Res. 37:D767–D772. 2009. View Article : Google Scholar
|
|
88
|
Maqungo M, Kaur M, Kwofie SK, et al: DDPC:
Dragon Database of Genes associated with Prostate Cancer. Nucleic
Acids Res. 39:D980–D985. 2011. View Article : Google Scholar :
|
|
89
|
Ganzfried BF, Riester M, Haibe-Kains B, et
al: curatedOvarianData: clinically annotated data for the ovarian
cancer transcriptome. Database (Oxford) 2013. bat0132013.
|
|
90
|
Gentleman RC, Carey VJ, Bates DM, et al:
Bioconductor: open software development for computational biology
and bioinformatics. Genome Biol. 5:R802004. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Mosca E, Alfieri R, Merelli I, Viti F,
Calabria A and Milanesi L: A multilevel data integration resource
for breast cancer study. BMC Syst Biol. 4:762010. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Wang L, Xiong Y, Sun Y, et al: HLungDB: an
integrated database of human lung cancer research. Nucleic Acids
Res. 38:D665–D669. 2010. View Article : Google Scholar :
|
|
93
|
Poos K, Smida J, Nathrath M, et al:
Structuring osteosarcoma knowledge: an osteosarcoma-gene
association database based on literature mining and manual
annotation. Database (Oxford) 2014. bau0422014. View Article : Google Scholar
|
|
94
|
Cutts RJ, Gadaleta E, Lemoine NR and
Chelala C: Using BioMart as a framework to manage and query
pancreatic cancer data. Database (Oxford) 2011. bar0242011.
|
|
95
|
Ramana J: RCDB: Renal Cancer Gene
Database. BMC Res Notes. 5:2462012. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Deng W, Nickle DC, Learn GH, Maust B and
Mullins JI: ViroBLAST: a stand-alone BLAST web server for flexible
queries of multiple databases and user’s datasets. Bioinformatics.
23:2334–2336. 2007. View Article : Google Scholar : PubMed/NCBI
|