Introduction
Uterine leiomyomas are monoclonal tumors in which
interactions of sex steroids, somatic mutations and growth factors
are involved in the neoplastic transformation of the myometrium
(1). Several studies suggest that
ovarian hormones and genetic factors may have a role in leiomyoma
growth (2). The extracellular
matrix (ECM) of the leiomyoma is constituted predominantly of
collagens, proteoglycans and matrix glycoproteins, and their
overproduction is directly involved in leiomyoma volume expansion
(3). The development of the fibroid
is due to the hyper-production of collagen and to the lack of
degradation (4).
The leiomyoma is characterized by cytoskeletal
alterations influencing contractility, cell migration and cell
proliferation (5). TNF-α
upregulates MMP-2 expression and stimulates cell migration through
the extracellular signal-regulated kinase (ERK) pathway in the
leiomyoma (6). Integrin plays a key
role in cytoskeletal remodeling towards activation of RHOA
(transforming protein RhoA), which takes part in cell signaling
leading to cell contractility, ECM stiffening and tumor growth
(7).
RHOA is essential in the regulation of the signal
transduction pathway, and is involved in a microtubule-dependent
signal required for the myosin contractile ring formation during
cell cycle cytokinesis (8). Cell
migration requires cytoskeletal rearrangement and RHOA is crucial
in the regulation of this mechanism: by activating MYL9 (myosin
regulatory light polypeptide 9), it increases the metastatic
potential in cancer cells (9).
Several factors can induce alterations in the
mechanotransduction signal from ECM via the transmembrane receptor
to the interior of the cell (10).
This signal causes the reorganization of the actin cytoskeleton,
mediated by RHOA, inducing cell proliferation (11).
In this study, we present the data of the altered
expression of several cytoskeletal proteins involved in cell
migration in the tissue of the leiomyoma compared to the normal
myometrium.
Materials and methods
Tissue samples were obtained from ten premenopausal
patients who underwent hysterectomy for symptomatic uterine
leiomyomas. The procedures complied with the Declaration of
Helsinki and were approved by the Review Board of the Institute for
Maternal and Child Health - IRCCS 'Burlo Garofolo' (Trieste,
Italy). All subjects signed a written informed consent.
The median age of patients was 46.5 with a minimum
of 42 years and a maximum of 52 years.
Tissue samples
Two samples were collected from each patient: one
from the central area of the leiomyoma and one from the unaffected
myometrium. All the leiomyomas were confirmed histologically as
benign ordinary leiomyomas. Samples were stored at −80°C until
proteomic analysis was performed.
Two-dimensional gel electrophoresis
Clean leiomyoma and myometrium (300 mg each) were
homogenized in 1.2 ml of dissolution TUC buffer [7 M urea, 2 M
thiourea, 4% CHAPS, 40 mM Tris, 65 mM DTT and 0.24% Bio-Lyte
(3–10)] with a protease inhibitor mix (2 mM
PMSF, 1 mM benzamidine, 1 mM EDTA, 1 mM NaF) and a trace of
bromophenol blue. The solutions were vortexed at maximum speed
several times and kept at room temperature for 1 h and centrifuged
at 10,000 × g at 4°C for 30 min. The protein content of the
supernatant was determined using the Bradford assay. Eight hundred
micrograms of proteins from each sample were used for the 2-DE
analysis.
NL IPG Readystrips, 18-cm, pH 3-10 (Bio-Rad,
Hercules, CA, USA) were rehydrated at 50 V for 12 h at 20°C.
Isoelectric focusing (IEF) was performed in a Protein IEF cell
(Bio-Rad) set to 170.000 Vh. After IEF, the IPG strips were
equilibrated by serial incubation (20 min) in an equilibration
buffer (6 M urea, 2% SDS, 50 mM Tris-HCl pH 8.8, 30% glycerol, and
1% DTT) and in an equilibration buffer containing 4% iodoacetamide
instead of DTT. Equilibrated IPG strips were transferred onto a 12%
polyacrylamide gel for the second dimension. After the second
dimension, gels were fixed and stained for 48 h with colloidal
Coomassie Blue, and excess dye was removed with distilled water. On
average, three experimental replicates were performed per sample.
Molecular masses were determined by precision protein standard
markers (Bio-Rad). 2-DE gels were scanned with a Molecular Imager
PharosFX system (Bio-Rad) and the quantitative analysis of the
spots was carried out using the ProteomeWeaver 4 program
(Bio-Rad).
Quantification of spot levels
Spot normalization was automatically performed by
the software and was based on a normalization algorithm intended
for numerical analysis, which did not require any internal
standard. For each gel an intensity factor was computed to ensure
all normalization factors were as close to one as possible. The
matching produced a list of super spots, which represent certain
protein species present in the gel. For the correct matching, each
super spot was manually controlled prior to normalization. For each
matched pair of gels, the quotient between the pair-matched spot
was calculated. The normalization factor was the median of these
quotients (ProteomeWeaver 4 program; Bio-Rad).
Trypsin digestion and MS analysis
Spots from 2-DE were washed 4 times with 50 mM
NH4HCO3 and acetonitrile (ACN; Sigma-Aldrich)
alternatively and dried under vacuum in a SpeedVac system. Three
microliters of 12.5 ng/µl sequencing grade modified trypsin
(Promega, Madison, WI, USA) in 50 mM NH4HCO3
was added to each gel spot and samples were digested overnight at
37°C. Peptides were extracted with three changes of 50% ACN/0.1%
formic acid (FA; Fluka), dried under vacuum and stored at −20°C
until mass spectrometry (MS) analysis was performed.
Samples were dissolved in 10 µl of 0.1%
trifluoroacetic acid (TFA; Riedel-de Haën). One microliter of each
sample was mixed with 1 µl of matrix solution
(α-cyano-4-hydroxycinnamic acid; Fluka). Five mg/ml in 70% ACN/0.1%
TFA) and 0.8 µl of the resulting solution were spotted onto
a stainless steel MALDI target plate for the MS analysis on the
MALDI-TOF/TOF 4800 analyzer (AB Sciex, Framingham, MA, USA). The
analysis was performed in a data dependent mode: a full MS scan was
acquired from each sample, followed by MS/MS spectra of the ten
most intense signals.
Few samples that could not be identified by
MALDI-TOF/TOF analysis were further analyzed by LC-MS/MS on an
LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific,
Rockford, IL, USA), coupled with a nano-HPLC Ultimate 3000 (Dionex
- Thermo Fisher Scientific). Samples were loaded onto an in-house
pico-frit column packed with C18 material (ReproSil, 300Å, 3
µm; Dr. Maisch HPLC GmbH) and separated using a 20-min
linear gradient of ACN/0.1% formic acid (from 3 to 40% ACN), at a
flow rate of 250 nl/min. Capillary voltage was set at 1.3–1.5 kV
and source temperature at 200°C. The analysis was performed in a
data dependent mode, and a full scan at 60,000 resolution on the
Orbitrap was followed by MS/MS fragmentation scans on the ten most
intense ions acquired with CID fragmentation in the linear
trap.
MS and MS/MS spectra obtained from MALDI-TOF/TOF
analysis were converted into MGF (Mascot generic format) files to
be elaborated with Proteome Discoverer 1.4 (Thermo Fisher
Scientific), while raw data files from the LTQ-Orbitrap XL mass
spectrometer were directly analyzed with the software. Proteome
Discoverer was interfaced to a Mascot search engine, version 2.2.4
(Matrix Science, London, UK).
The database used for protein identification was
UniProt Human (version 20140709, 88993 sequences), while enzyme
specificity was set to trypsin with 1 missed cleavage. The mass
tolerance window was set to 10 ppm for parent mass and to 0.6 Da
for fragment ions for the files from LTQ-Orbitrap XL, while the
tolerances were 50 ppm (parent) and 0.3 Da (fragment ions) for the
MALDI-TOF/TOF data. Carbamidomethylation of cysteine residues was
set to 'fixed modification' and methionine oxidation to 'variable
modification'.
Proteome Discoverer calculates a false discovery
rate (FDR) based on the parallel search against a randomized
database. Proteins were considered as positive hits if at least two
independent peptides were identified with medium (95%) or high
(99%) confidence.
STRING 9.0 network analysis and
biological functions
Possible connections among identified cytoskeletal
proteins with significant variations as compared to normal
myometrium versus leiomyoma were analyzed by a protein and gene
network software. For each protein, related gene names were
acquired in UniProtKB and used for network generation by the use of
STRING 9.0 (http://www.string-db.org/).
Differential proteins distributed in the biological
function data were obtained from Gene Ontology (http://amigo.geneon-tology.org/rte).
Western blotting
Western blot analysis was performed as previously
described (12). Protein extracts
(30 µg) used for 2-DE were separated by 10% SDS-PAGE, and
then transferred to a nitrocellulose membrane in a blotting
chamber. The residual binding sites on the membrane were blocked by
treatment with defatted dry milk proteins, and incubated overnight
at 4°C with 1:1,000 diluted primary rabbit polyclonal antibody
against myosin regulatory light polypeptide 9 (Sigma-Aldrich).
After washing, membranes were incubated with an HRP-conjugated
anti-rabbit IgG (Sigma-Aldrich) in a dilution of 1:3,000. At the
end the protein expression was visualized by chemiluminescence
(SuperSignal West Pico Chemiluminescent; Thermo Fisher Scientific)
according to manufacturer's instructions. The intensity of the
signals was quantified by VersaDoc Imaging system (Bio-Rad). The
intensities of the immunostained bands were normalized with the
total protein intensities measured by Coomassie brilliant blue
G-250 from the same blot.
Statistical analyses
Statistical analyses were carried out with the
non-parametric Wilcoxon sign-rank test for paired samples for both
2-DE and western blot data. A p-value <0.05 was considered as
statistically significant. Analyses were conducted with Stata/IC
12.2 for Windows (StataCorp LP, College Station, TX, USA).
Results
Proteomic studies
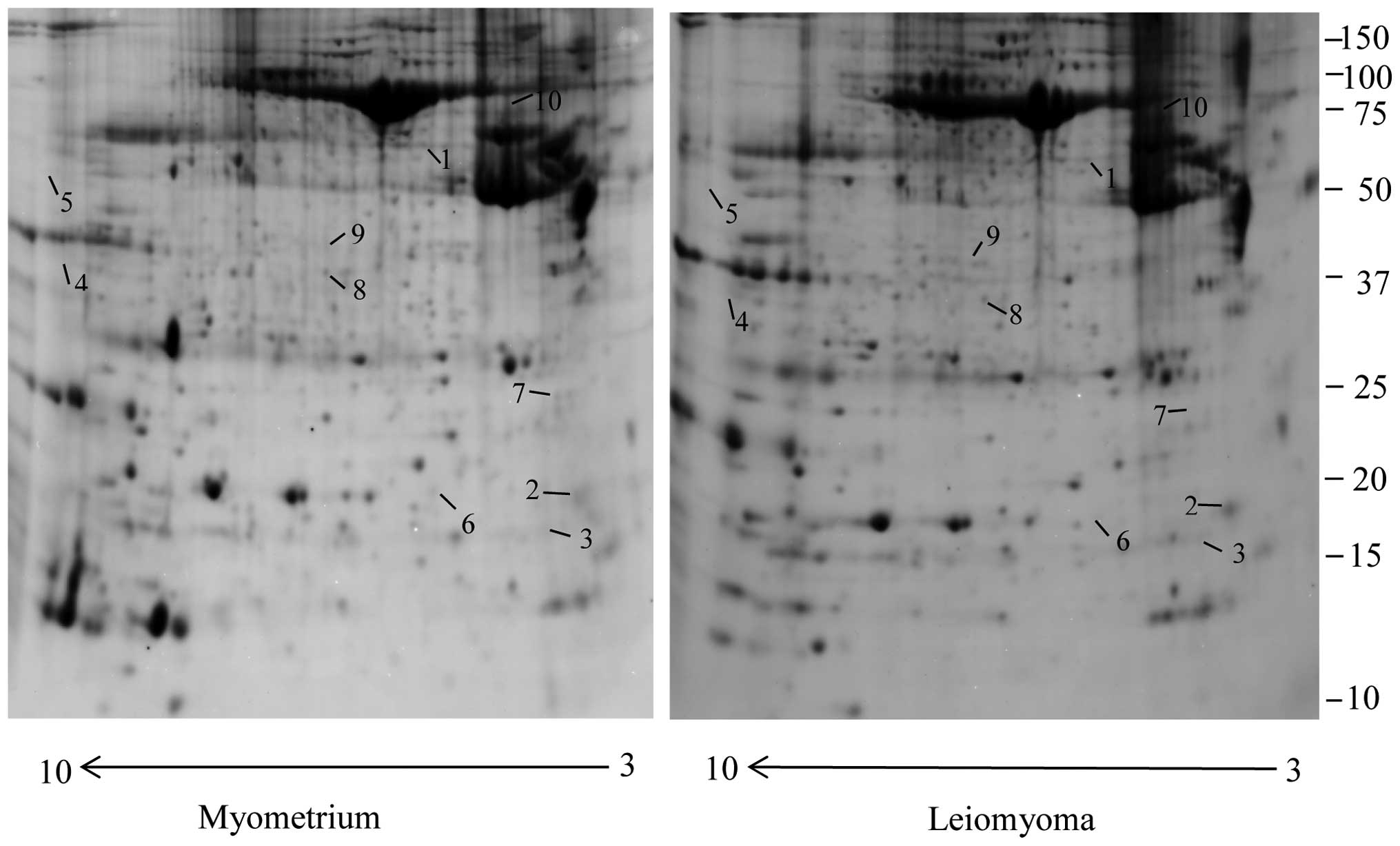
Using the 2-DE coupled with MS, a comparative
proteomics analysis was performed between uterine leiomyoma and
myometrium tissues. Correlation analysis of gel-pairs performed
well, with average matching efficiency of about 75%. An average of
2,200 spots was detected on gels for both types of proteomes. In
this study 10 protein spots, belonging to cytoskeletal proteins
with several biological functions, were found to be significantly
dysregulated in leiomyoma samples compared to the myometrium. Eight
spots were significantly upregulated (>1.5-fold) and two were
significantly downregulated (<0.6 fold) (Fig. 1), and corresponded to 10 proteins
identified by MALDI-TOF/TOF and LTQ-Orbitrap XL by searching the
MS/MS data against the human section of the UniProt database
(Table I). We found a significant
increase in the leiomyoma of tubulin β (TUBB), myosin regulatory
light polypeptide 9 (MYL9), desmin (DES), four and a half LIM
domains protein 1 (FHL1), keratin, type II cytoskeletal 1 (KRT1),
keratin, type I cytoskeletal 9 (KRT9), LIM and SH3 domain protein 1
fragment (LASP1), actin α cardiac muscle 1 (ACTC1). Transgelin
(TAGLN), prelamin A/C (LMNA) were found to be significantly
downregulated in the leiomyoma compared to the myometrium.
 | Table ICytoskeletal protein expression levels
measured by mass spectrometry in the leiomyoma and in the
myometrium proteome. |
Table I
Cytoskeletal protein expression levels
measured by mass spectrometry in the leiomyoma and in the
myometrium proteome.
| Accession no. | Spot no. | Protein
description | Gene symbol | Total score | Fold changea | Biological
function |
|---|
| P68032 | 10 | Actin, α cardiac
muscle 1 | ACTC1 | 248 | 3.6 | Cell motility |
| Q13642 | 4 | Four and a half LIM
domains protein 1 | FHL1 | 224 | 3.4 | Cell migration |
| P04264 | 5 | Keratin, type II
cytoskeletal 1 | KRT1 | 107 | 3.4 | Regulators of
inflammation |
| Q14847 | 9 | LIM and SH3 domain
protein 1 fragment | LASP1 | 66 | 2 | Cell migration |
| P24844 | 2 | Myosin regulatory
light polypeptide 9 | MYL9 | 100 | 1.8 | Cell migration |
| P17661 | 3 | Desmin | DES | 190 | 1.6 | Main component of
intermediate filaments |
| P35527 | 8 | Keratin, type I
cytoskeletal 9 | KRT9 | 214 | 1.6 | Keratin filament
assembly |
| P07437 | 1 | Tubulin β
chain | TUBB | 317 | 1.6 | Compnent of
microtubules |
| P02545 | 7 | Prelamin-A/C | LMNA | 120 | 0.5 | Nuclear
assembly/chromatin organization |
| Q01995 | 6 | Transgelin | TAGLN | 1,200 | 0.5 | Replicative
senescence |
Three of these proteins are involved in the
promotion of cell migration (FHL1, LASP1 and MYL9), while TAGLN is
involved in replicative senescence.
Validation of myosin regulatory light
polypeptide 9
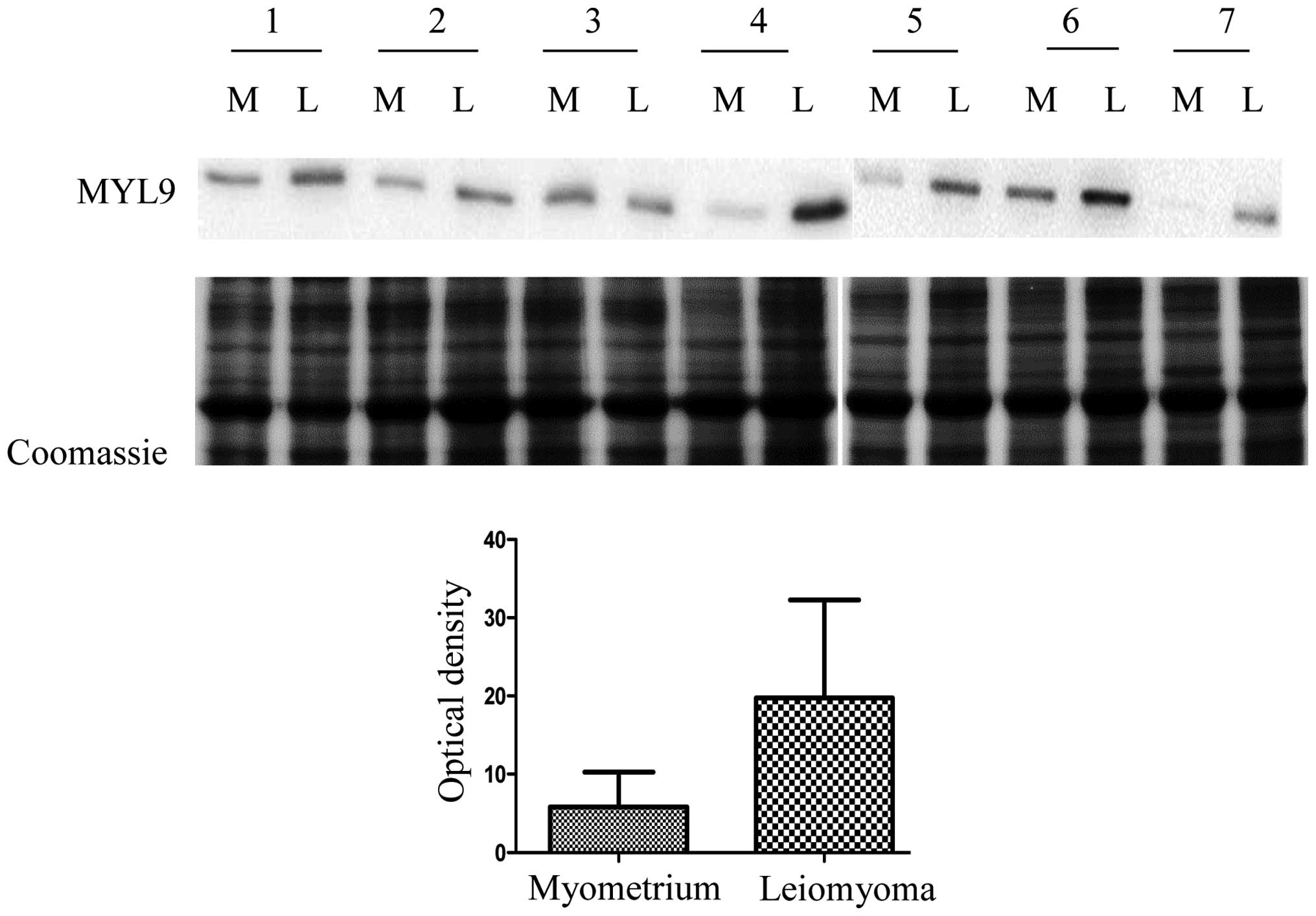
In this study we decided to validate myosin
regulatory light polypeptide 9 (MYL9) because it is considered to
be a cell migration marker, in several cancers and could play a key
role in leiomyoma development. MYL9 expression in seven leiomyomas
was compared to the expression in matched normal myometrial tissue
samples by western blot analysis. MYL9 expression was significantly
higher in the leiomyoma with respect to the myometrium, confirming
results obtained from the 2-DE analysis. In Fig. 2 we report the quantitative analysis
of MYL9 expression (P=0.031). We opted for a Coomassie staining
protocol because, as described by Lv et al (13), and according to our results, the two
housekeeping proteins, β-actin and tubulin, are upregulated in
leiomyoma, and thus not adequate as controls. We do not have
information on the expression of the other housekeeping
proteins.
Protein-protein interaction analysis
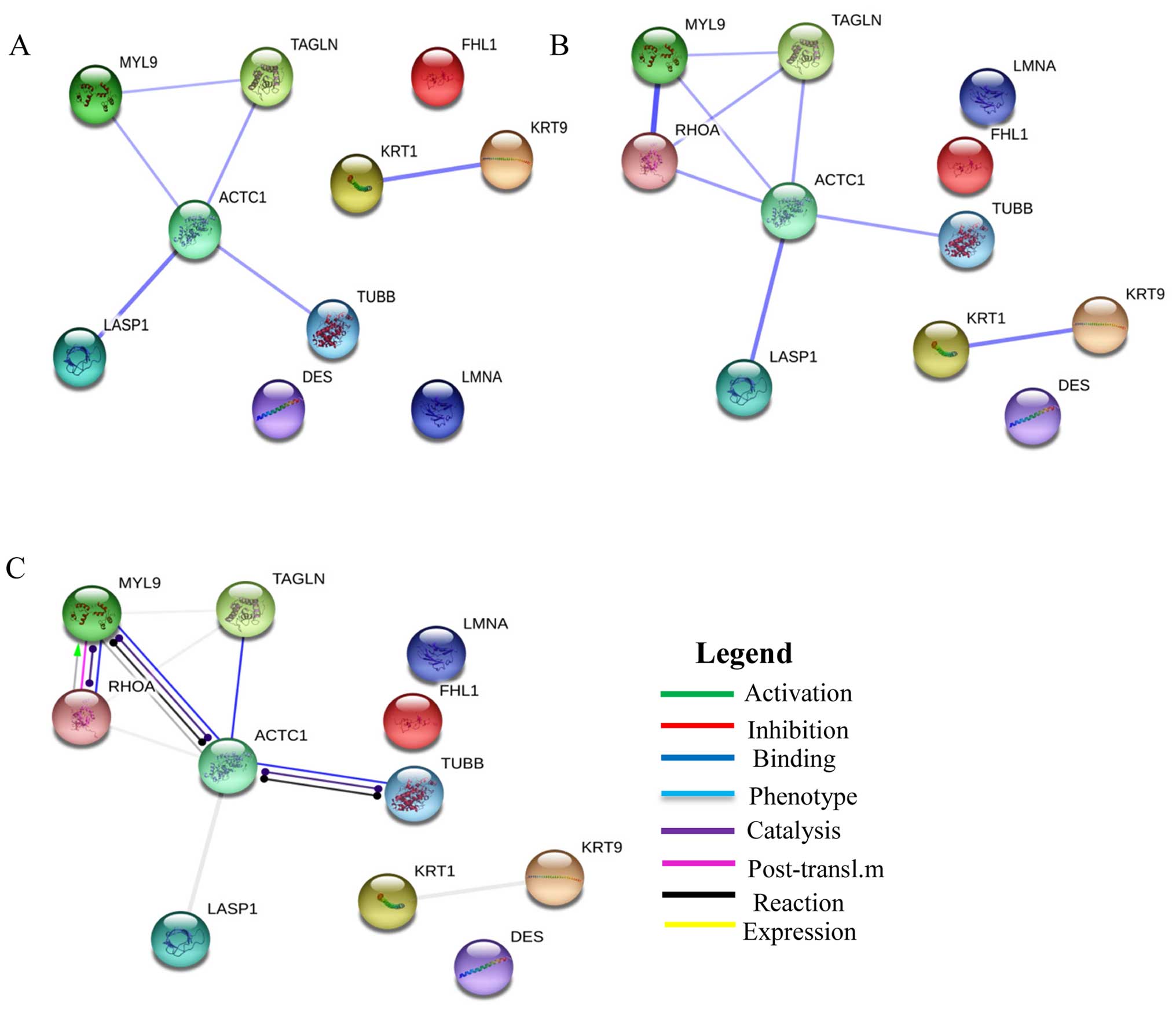
The dysregulated cytoskeletal related proteins
identified in this study were loaded on STRING 9.0 to generate a
prediction of protein-protein interaction networks. The strongest
interactions were between: LASP1 and ACTC1 (combined score, 0.774),
and KRT1 and KRT9 (combined score, 0.76) (Fig. 3A). We further analyzed the
interaction between transforming protein RhoA (a key protein in the
regulation of the signal transduction pathway involved in leiomyoma
growth) and the proteins identified in our study (Fig. 3B).
The strongest evidence was the interaction between
RHOA and MYL9 (combined score, 0.99). Another interesting result
based on action prediction was the activation of MYL9 by RHOA
(Fig. 3C).
Discussion
Several studies have highlighted the role of
extracellular forces in the reorganization of the cytoskeleton in
leiomyoma cells. Increases in mechanical stress induce an excessive
ECM deposition and tumorigenesis promotion (11,14).
Cytoskeleton integrity plays an important role in cell cycle
progression, death and differentiation, and abnormal cytoskeleton
functions are often observed in leiomyoma cells (15).
To the best of our knowledge, this is the first
proteomics study presenting data on the expression of several
cytoskeletal proteins involved in cell migration.
Actins and tubulin are two major components of the
cytoskeleton, with key roles in cell morphology (16). Disassembly of actin and microtubule
dynamic instability may be associated to leiomyoma growth (17). A wide variety of anticancer drugs
are able to bind to TUBB, interfering with the microtubule dynamics
such as paclitaxel (18). The
uterine leiomyoma consists mainly of smooth muscle cells with
abundant expression of DES (19).
The overexpression of desmin inside and outside of the cell
certainly indicates its involvement in the mechanotransduction and
in tumor development (10).
FHL1 regulates cytoskeleton-associated proteins
(20) and its overexpression in
myoblasts inhibits cell adhesion and promotes cell migration
(21). Studies on FHL1 gene
expression have shown that FHL1 knockdown induces inhibition of
cell proliferation (21). These
studies can help understanding of the possible involvement of FHL1
in leiomyoma cell proliferation and growth.
In the leiomyoma, integrin activation takes place by
cell contractility and by ECM stiffening. Integrin leads to the
activation of RHOA, which activates myosin light chain kinase.
Phosphorylation of MYL9 by myosin light chain kinase plays an
important role in the regulation of smooth muscle cell contractile
activity and is implicated in cytokinesis and cell locomotion
(4,22).
MYL9 is considered to be a cell migration marker in
breast cancer and its expression is necessary for cytoskeletal
dynamics and experimental metastasis (23,24).
For the first time, in this study we report the validation of MYL9
expression by western blot, confirming the 2-DE result. The STRING
interaction network shows the activation of MYL9 by RHOA, and this
result is corroborated by the literature.
Keratins constitute the intermediate filament of the
cytoskeleton in the epithelia and KRT1 is one of the major
constituents of the cytoskeleton of keratinocytes (25). KRT1 plays an essential role in
preserving the integrity of the cell from mechanical stress, and in
the protection of the cell from inflammation (25). KRT9 is another constituent of the
inter-mediate filament cytoskeleton identified in our study
revealing overexpression of this protein in the leiomyoma with
respect to the normal myometrium.
LASP1, an actin-binding protein, plays a role in the
organization of the cytoskeleton, is involved in a signaling
pathway and in the regulation of cell migration and proliferation
(26). Our STRING network data
confirm the interaction between LASP1 and actin. Specific
phosphorylation of LASP1 at serine/threonine and tyrosine regulates
the function and the localization of the protein. LASP1 is
overexpressed in breast and ovarian cancer, promoting cell
migration and proliferation (27).
This evidence supports our results regarding the overexpression of
LAPS1 in the leiomyoma compared to the normal myometrium.
ACT1 belongs to the actin isoforms α, which are
found in muscle tissues and are a major constituent of the
contractile apparatus (28). This
protein is recruited at early stages of cell adhesion (29). Previously published studies have
found an interaction between α-actinin and actin, and have
suggested a possible role of α-actinin in adhesion maturation
(29). In the leiomyoma, the
overexpression of ACTC1 can lead to the dysregulation of the
cytoskeleton, inducing proliferation and growth (4,22).
Finally, we found that LAMN A/C and TAGLN are
downregulated in the leiomyoma proteome. Our results are in line
with our previous study on interstitial fluid (10).
Lamins constitute a class of intermediate filament
with structural and functional roles, involved in dynamic chromatin
organization and gene transcription (30). Increasing of mechanical forces in
ECM may induce a downregulation on lamin, inducing chromatin
remodeling and dysregulation of gene expression involved in cell
proliferation and adhesion (31).
Transgelin is an abundant protein of smooth muscle
cells involved in the stabilization of actin filaments and is
directly and indirectly involved in many cancer-related processes
such as proliferation, migration, differentiation and apoptosis
(32). Evidence shows how
transgelin acts as a tumor suppressor (33) and its downregulation in the
leiomyoma promotes cell proliferation, and is consequently
associated with the growth of the fibroid tumor (34).
In conclusion, the 2-DE approach remains the
technique of choice for comparative proteomic.
Our data demonstrate significant alterations in the
expression of cytoskeletal proteins in the leiomyoma tissue
compared to the normal myometrium. These proteins are involved in
cell migration, tumor growth and cell proliferation. All the
dysregulated cytoskeletal proteins may be directly involved in
signaling pathway, contributing to the development of the
leiomyoma. A better understanding of the involvement of
cytoskeletal proteins in leiomyoma growth may help identify new
therapeutic targets for the development of new pharmacological
approaches.
References
|
1
|
Rein MS: Advances in uterine leiomyoma
research: The progesterone hypothesis. Environ Health Perspect.
108(Suppl 5): 791–793. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Catherino WH and Malik M: Uterine
leiomyomas express a molecular pattern that lowers retinoic acid
exposure. Fertil Steril. 87:1388–1398. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Arslan AA, Gold LI, Mittal K, Suen TC,
Belitskaya-Levy I, Tang MS and Toniolo P: Gene expression studies
provide clues to the pathogenesis of uterine leiomyoma: New
evidence and a systematic review. Hum Reprod. 20:852–863. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Leppert PC, Jayes FL and Segars JH: The
extracellular matrix contributes to mechanotransduction in uterine
fibroids. Obstet Gynecol Int. 2014:7832892014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Davis BJ, Risinger JI, Chandramouli GV,
Bushel PR, Baird DD and Peddada SD: Gene expression in uterine
leiomyoma from tumors likely to be growing (from black women over
35) and tumors likely to be non-growing (from white women over 35).
PLoS One. 8:e639092013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang Y, Feng G, Wang J, Zhou Y, Liu Y, Shi
Y, Zhu Y, Lin W, Xu Y and Li Z: Differential effects of tumor
necrosis factor-α on matrix metalloproteinase-2 expression in human
myometrial and uterine leiomyoma smooth muscle cells. Hum Reprod.
30:61–70. 2015. View Article : Google Scholar
|
|
7
|
Elosegui-Artola A, Bazellières E, Allen
MD, Andreu I, Oria R, Sunyer R, Gomm JJ, Marshall JF, Jones JL,
Trepat X, et al: Rigidity sensing and adaptation through regulation
of integrin types. Nat Mater. 13:631–637. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Vincent S and Settleman J: The PRK2 kinase
is a potential effector target of both Rho and Rac GTPases and
regulates actin cytoskeletal organization. Mol Cell Biol.
17:2247–2256. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Luo XG, Zhang CL, Zhao WW, Liu ZP, Liu L,
Mu A, Guo S, Wang N, Zhou H and Zhang TC: Histone methyltransferase
SMYD3 promotes MRTF-A-mediated transactivation of MYL9 and
migration of MCF-7 breast cancer cells. Cancer Lett. 344:129–137.
2014. View Article : Google Scholar
|
|
10
|
Ura B, Scrimin F, Zanconati F, Arrigoni G,
Monasta L, Romano A, Banco R, Zweyer M, Milani D and Ricci G:
Two-dimensional gel electrophoresis analysis of the leiomyoma
interstitial fluid reveals altered protein expression with a
possible involvement in pathogenesis. Oncol Rep. 33:2219–2226.
2015.PubMed/NCBI
|
|
11
|
Norian JM, Owen CM, Taboas J, Korecki C,
Tuan R, Malik M, Catherino WH and Segars JH: Characterization of
tissue biomechanics and mechanical signaling in uterine leiomyoma.
Matrix Biol. 31:57–65. 2012. View Article : Google Scholar
|
|
12
|
Mischiati C, Ura B, Roncoroni L, Elli L,
Cervellati C, Squerzanti M, Conte D, Doneda L, Polverino de Laureto
P, de Franceschi G, et al: Changes in protein expression in two
cholangiocarcinoma cell lines undergoing formation of
multi-cellular tumor spheroids in vitro. PLoS One. 10:e01189062015.
View Article : Google Scholar
|
|
13
|
Lv J, Zhu X, Dong K, Lin Y, Hu Y and Zhu
C: Reduced expression of 14-3-3 γ in uterine leiomyoma as
identified by proteomics. Fertil Steril. 90:1892–1898. 2008.
View Article : Google Scholar
|
|
14
|
Mouw JK, Yui Y, Damiano L, Bainer RO,
Lakins JN, Acerbi I, Ou G, Wijekoon AC, Levental KR, Gilbert PM, et
al: Tissue mechanics modulate microRNA-dependent PTEN expression to
regulate malignant progression. Nat Med. 20:360–367. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Butcher DT, Alliston T and Weaver VM: A
tense situation: Forcing tumour progression. Nat Rev Cancer.
9:108–122. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cooper JA: The role of actin
polymerization in cell motility. Annu Rev Physiol. 53:585–605.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Salama SA, Kamel MW, Botting S, Salih SM,
Borahay MA, Hamed AA, Kilic GS, Saeed M, Williams MY and
Diaz-Arrastia CR: Catechol-o-methyltransferase expression and
2-methoxyestradiol affect microtubule dynamics and modify steroid
receptor signaling in leiomyoma cells. PLoS One. 4:e73562009.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ganguly A, Yang H and Cabral F: Class III
β-tubulin counteracts the ability of paclitaxel to inhibit cell
migration. Oncotarget. 2:368–377. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Roeder HA, Cramer SF and Leppert PC: A
look at uterine wound healing through a histopathological study of
uterine scars. Reprod Sci. 19:463–473. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Brown S, McGrath MJ, Ooms LM, Gurung R,
Maimone MM and Mitchell CA: Characterization of two isoforms of the
skeletal muscle LIM protein 1, SLIM1. Localization of SLIM1 at
focal adhesions and the isoform slimmer in the nucleus of myoblasts
and cytoplasm of myotubes suggests distinct roles in the
cytoskeleton and in nuclear-cytoplasmic communication. J Biol Chem.
274:27083–27091. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Robinson PA, Brown S, McGrath MJ, Coghill
ID, Gurung R and Mitchell CA: Skeletal muscle LIM protein 1
regulates integrin-mediated myoblast adhesion, spreading, and
migration. Am J Physiol Cell Physiol. 284:C681–C695. 2003.
View Article : Google Scholar
|
|
22
|
Huveneers S and Danen EH: Adhesion
signaling - crosstalk between integrins, Src and Rho. J Cell Sci.
122:1059–1069. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liao XH, Wang N, Liu LY, Zheng L, Xing WJ,
Zhao DW, Sun XG, Hu P, Dong J and Zhang TC: MRTF-A and STAT3
synergistically promote breast cancer cell migration. Cell Signal.
26:2370–2380. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Medjkane S, Perez-Sanchez C, Gaggioli C,
Sahai E and Treisman R: Myocardin-related transcription factors and
SRF are required for cytoskeletal dynamics and experimental
metastasis. Nat Cell Biol. 11:257–268. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Roth W, Kumar V, Beer HD, Richter M,
Wohlenberg C, Reuter U, Thiering S, Staratschek-Jox A, Hofmann A,
Kreusch F, et al: Keratin 1 maintains skin integrity and
participates in an inflammatory network in skin through
interleukin-18. J Cell Sci. 125:5269–5279. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dandapani SV, Sugimoto H, Matthews BD,
Kolb RJ, Sinha S, Gerszten RE, Zhou J, Ingber DE, Kalluri R and
Pollak MR: α-actinin-4 is required for normal podocyte adhesion. J
Biol Chem. 282:467–477. 2007. View Article : Google Scholar
|
|
27
|
Grunewald TG and Butt E: The LIM and SH3
domain protein family: Structural proteins or signal transducers or
both? Mol Cancer. 7:312008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Matsson H, Eason J, Bookwalter CS, Klar J,
Gustavsson P, Sunnegårdh J, Enell H, Jonzon A, Vikkula M, Gutierrez
I, et al: α-cardiac actin mutations produce atrial septal defects.
Hum Mol Genet. 17:256–265. 2008. View Article : Google Scholar
|
|
29
|
Choi CK, Vicente-Manzanares M, Zareno J,
Whitmore LA, Mogilner A and Horwitz AR: Actin and α-actinin
orchestrate the assembly and maturation of nascent adhesions in a
myosin II motor-independent manner. Nat Cell Biol. 10:1039–1050.
2008. View
Article : Google Scholar
|
|
30
|
Dechat T, Adam SA, Taimen P, Shimi T and
Goldman RD: Nuclear lamins. Cold Spring Harb Perspect Biol.
2:a0005472010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Iyer KV, Pulford S, Mogilner A and
Shivashankar GV: Mechanical activation of cells induces chromatin
remodeling preceding MKL nuclear transport. Biophys J.
103:1416–1428. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dvorakova M, Nenutil R and Bouchal P:
Transgelins, cytoskeletal proteins implicated in different aspects
of cancer development. Expert Rev Proteomics. 11:149–165. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang Z, Chang YJ, Miyamoto H, Ni J, Niu Y,
Chen Z, Chen YL, Yao JL, di Sant'Agnese PA and Chang C: Transgelin
functions as a suppressor via inhibition of ARA54-enhanced androgen
receptor transactivation and prostate cancer cell growth. Mol
Endocrinol. 21:343–358. 2007. View Article : Google Scholar
|
|
34
|
Li Q, Shi R, Wang Y and Niu X: TAGLN
suppresses proliferation and invasion, and induces apoptosis of
colorectal carcinoma cells. Tumour Biol. 34:505–513. 2013.
View Article : Google Scholar
|