Introduction
Epithelial ovarian cancer (EOC), the most common
ovarian malignancy, continues to be the leading cause of death
among gynecological malignancies (1). The poor survival from EOC is due to
the high percentage of patients diagnosed at an advanced stage and
a lack of effective treatment (2).
Currently the standard treatment protocol used in the initial
management of advanced-stage ovarian cancer is primary
cytoreductive surgery, followed by platinum/taxane combination
chemotherapy. Approximately 70% of patients will have a complete
clinical response to this initial therapy, indicated by an absence
of clinically detectable residual disease based on clinical exam,
radiologic imaging, or measurement of serum tumor marker CA125
(3). Unfortunately, the majority of
these patients will eventually develop recurrence or persistent
platinum-resistant disease (4).
Therefore, more effective treatments for advanced ovarian cancer
are urgently required (5).
Pinellia pedatisecta Schott (PPS), a
traditional Chinese medicine (TCM), has been used, based on many
TCM books, to treat several disorders. In the 1970s, Pedate
Pinellia Rhizome was used in the Obstetrics and Gynecology
Hospital of Fudan University to treat 247 cervical cancer patients
with a total effective rate of 81.5%. Following, Li et al
found that a novel lipid-soluble extract (PE) from PPS can induce
apoptosis in human cervical cancer cells with few side effects on
normal cells, suggesting that PPS extract could be used as a
potential drug for cervical cancer treatment (6). In addition, Sun et al found
that total PPS extract can inhibit mouse sarcoma-180 growth in
vivo by direct intraperitoneal injection (7). However, the pharmacological mechanism
of this action has not been established.
In this study, we first examined whether the PPS
extract component could induce apoptosis and exert proliferation
inhibition on ovarian cancer cell line SKOV3. To determine how PPS
extract affects apoptosis and tumor inhibition, we performed high
throughput mRNA sequencing and bioinformatics analysis to determine
transcriptome difference between PPS-treated SKOV3 cells and
controls.
Materials and methods
Cell line
The ovarian cancer cell line SKOV3 was purchased
from Shanghai Institutes for Biological Sciences and maintained as
recommended by the supplier (American Type Culture Collection,
Rockville, MD, USA). Cells were cultured in RPMI-1640 supplemented
with 10% fetal bovine serum (FBS), 1% sodium pyruvate, and 1%
nonessential amino acids. All tissue culture reagents were obtained
from Sigma-Aldrich (St. Louis, Mo, USA).
Preparation of total extract from
PPS
Fresh PPS was collected from Xuchang, Henan
Province, China. It was washed and peeled, and ground to generate a
homogenate with normal saline. The homogenate was centrifuged at
3000 rpm and 4°C for 20 min and the supernatant was retrieved.
Crude extract of PPS was obtained by two times gradient
sedimentation with acetone (V/V). The sediments was dissolved into
0.9% NaCl (m/m) and followed by dialysis in distilled water at 4°C
for 48 h. After high-speed centrifugation, the supernatant was
extracted as the final total extract. Extract protein
concentrations were measured by bicinchoninic acid (BCA) method.
Detection of phosphorus, saccharide and lipid protein in the total
extract was measured by ammonium molybdate spectrophotometry,
Molisch's test and Sudan black B staining method, respectively.
Cytotoxicity assay
SKOV3 cells at logarithmic growth phase were seeded
at 5000 cells per well in 96-well culture plates and cultured for
24 h. The PPS extract protein at increasing concentrations (0.8, 4,
20, 100, 200, 300, 400 µg/ml) was added to the wells. After
72 h, CCK-8 (10% V/V) was added to the culture and incubated for 1
h. With 630 nm wavelength as the reference, the absorption values
at 450 nm were collected for analysis in an automatic ELISA
detection instrument. Medium alone was used as the blank control.
Cells without drug treatment or with cisplatin (DDP) treatment were
used as negative or positive controls, respectively. All the
experiments were replicated six times.
Flow cytometry analysis
SKOV3 cells in logarithmic growth phase were seeded
in 6-well plates at approximately 8×104 cells per well
and cultured for 24 h. Increasing concentrations of the protein
extract of PPS (0, 20, 50, 100, 200, 400 µg/ml) were added
to the wells and cells were cultured for an additional 72 h at
37°C. The cells were trypsinized, washed twice with ice-cold PBS
extract, adjusted to 1×106 cells/ml and fixed with 70%
ethanol. Then, 100 µl cell suspension aliquot was taken to
each labeled tube, followed by adding 10 µl Annexin V-FITC
and 10 µl PI (20 µg/ml) (Annexin V/FITC kit: BD
Biosciences). After 20 min incubation in darkness at room
temperature, 400 µl PBS binding buffer was then added and
cell apoptosis was detected immediately by flow cytometry (BD
Biosciences Clontech, San Jose, CA, USA) (within 30 min after
adding PBS binding buffer). Samples in triplicates were examined.
SKOV3 cells without treatment were used as the reference. Data
analysis was conducted using the Cell Quest software.
Transcriptome sequencing
SKOV3 cells and those treated with 200 µg/ml
PPS extract for 72 h were subjected to mRNA-Seq library
construction and transcriptome sequencing. Approximately
108 cells from each group were collected after PBS
washes. Oligo(dT) beads were used to capture poly(A) mRNA after
total RNA was extracted. Each poly(A)-enriched RNA sample was
chemically fragmented into small pieces using divalent cations at
94°C for 5 min. The fragmented RNA was reverse-transcribed into
cDNA using random hexamer primers containing a tagging sequence at
the 3′ end and a SuperScript III Double-Stranded cDNA synthesis kit
according to the manufacturer's protocol (Invitrogen, Carlsbad, CA,
USA). The double-stranded cDNA was subjected to end-repair and
further 3′ terminal tagging by the addition of 5′ DNA adaptors and
T4 DNA ligase with overnight incubation at 16°C for 16 h. The
targeted di-tagged cDNA was purified by polyacrylamide gel
electrophoresis and gel excision (200±25 bp). The clean di-tagged
cDNA was enriched by 18 PCR cycles with primer pairs annealed to
the tagging sequences of the di-tagged cDNA. Library purification
by PAGE removed any residual nucleotides, PCR primers and small
amplicons. Two paired-end libraries were sequenced using the
HiSeq2000 platform (Illumina). The initial short reads from this
study are being submitted to the NCBI sequence read archive
database and the accessions will be released soon.
Bioinformatics analysis
The raw Illumina/Solexa reads of SKOV3 cells and PPS
extract-treated cells were subjected to adapter trimming and low
quality filtering using the Trimmomatic program (8). The high quality clean reads were
aligned to the human genome using TopHat (9). Human genome (hg19) sequence and gene
annotation were obtained from the UCSC genome Browser (http://genome.ucsc.edu/). Cuffdiff was used to perform
differential gene expression profiling with default parameters
(10,11). The significantly differentially
expressed transcripts (DETs) between SKOV3 and PPS extract-treated
cell line were selected using the following criteria: i) if the
FPKM value for a certain transcript in both samples was >1, the
difference between them should be at least 3-fold. ii) If the FPKM
value for a certain transcript in one sample was <1, the FPKM
value for this gene in the paired sample should be >3. Mapping
transcripts to the KEGG pathway were conducted using the BLASTX and
followed by custom scripts (12).
Kegg pathway enrichment analysis of the DETs compared with
transcriptome background (Cuffdiff detected transcript status as
'OK') was performed by hypergenometric distribution testing using
the Phyper function of the R software package (http://www.r-project.org/). Bonferroni correction was
used to adjust P-value for each pathway.
Quantitative real-time PCR validation of
mRNA-Seq data
The mRNAs which have been used to perform mRNA-Seq
experiments from two cell lines were used for qPCR validation.
Total RNA from each sample was treated with DNase I before reverse
transcription by SuperScript III double-stranded cDNA synthesis
kit. Ten intron-spanning target gene primers and two endogenous
control (18s rRNA and GAPDH) primers were designed using Primer 5.0
for qPCR experiment. SYBR Green I based qPCR was performed
according to the routine protocols (13) on a 7500 Real-time PCR System
(Applied Biosystems, Foster City, CA, USA) and three technical
replicates were performed for all the genes in each sample. Gene
expression difference between two lines was calculated by the
2−ΔΔCt method (14).
Statistical methods
All data were first subjected to normality test and
expressed as mean ± standard deviation. Independent-sample t-test
was used to explore the apoptosis rate and/or proliferation
inhibition differences between PPS extract-treated group and
control group, cisplatin-treated group and control group, 150
µg/ml PPS extract treatment for 48 and 72 h, and 1
µg/ml cisplatin treatment for 48 and 72 h. Multiple
concentrations of PPS extract-treated SKOV3 cells within the same
group were compared using one-way ANOVA after homogeneity of
variance was tested. LSD method was used to analyze the difference
in apoptosis rate between two samples if homogeneity of variance
existed. Otherwise, Dunnet's T3 testing was adopted. Data analysis
was performed using SPSS software version 16.0.
Results
Effect of PPS extract on the
proliferation of SKOV3 cells
We evaluated the total extract obtained from the
PPS. The absorption value of total extract at 660 nm wavelength was
entirely identical to the blank control (A660=0) indicating that
there was no phosphorus presence. In addition, total extract could
not be stained by Sudan dye suggested the absence of lipoprotein.
Moreover, total extract can react with α-naphthol to generate
purple compound indicating the presence of glycoprotein in the
extract. The productivity of total protein extracted from fresh PPS
was 1% (9.97 g total extract from 1000 g PPS). We found that 20
µg/ml of the total extract of PPS significantly inhibited
the growth of SKOV3 cells in vitro. As the concentration of
the PPS extract increased, the inhibition rate was enhanced,
showing a significant dose-effect relationship. However, 0.8
µg/ml and 4 µg/ml PPS extract had no obvious effect
on cell proliferation inhibition. When the extract protein
concentration reached 100 µg/ml or higher, the inhibitory
effect of PPS on SKOV3 proliferation was significantly different
between the two adjacent concentrations (F=407.084, P<0.001,
Fig. 1A). Cisplatin group also
showed a significant dose-effect relationship. The inhibitory
effect of cisplatin on SKOV3 cells was significantly different
between different concentrations (F=799.621, P<0.001, Fig. 1B).
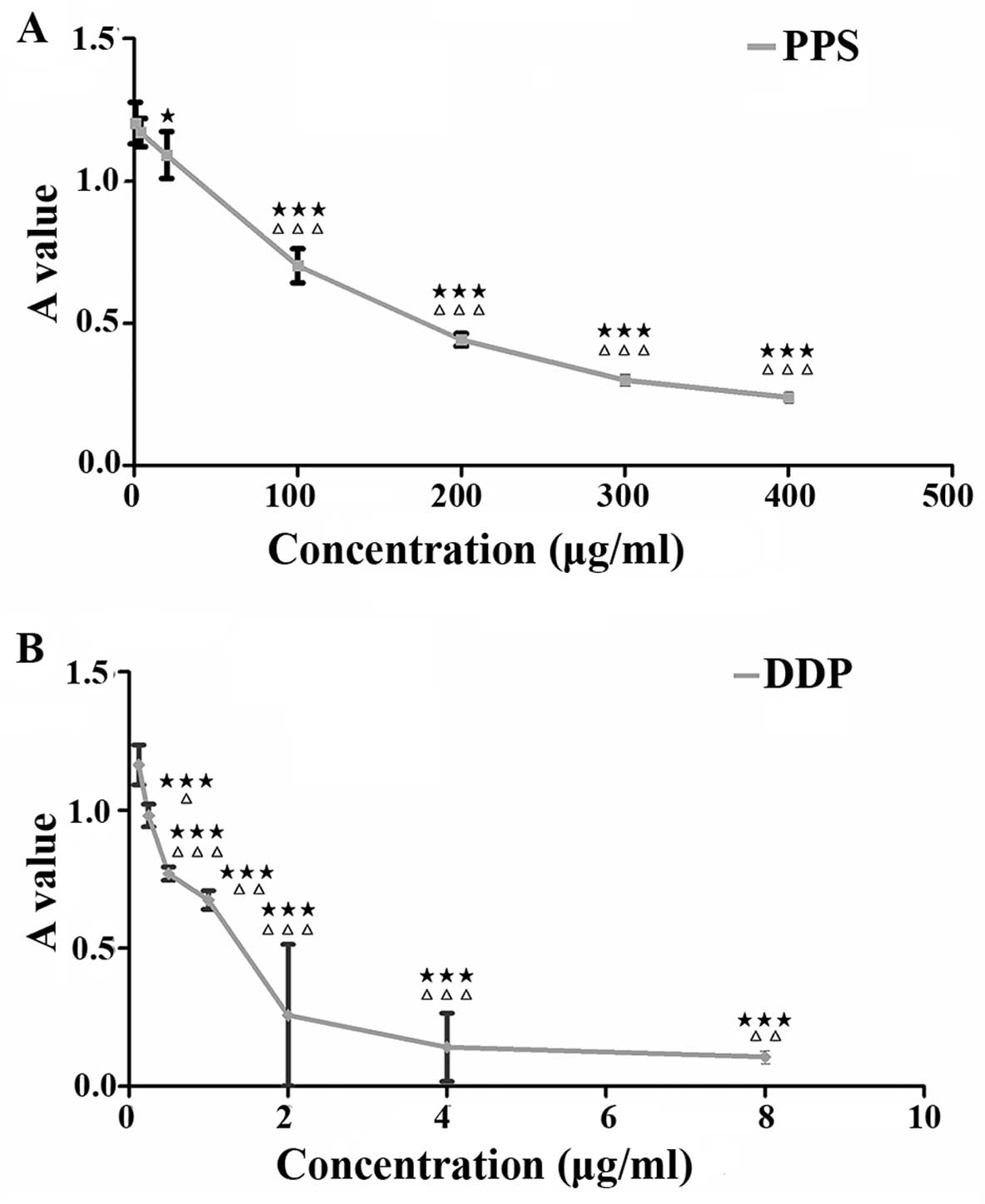 | Figure 1Effect of PPS extract and cisplatin
on the proliferation inhibition of SKOV3 cells. (A) Cells were
cultured in 10% FBS medium and treated with PPS (0.8, 4, 20, 100,
200, 300, 400 µg/ml) for 72 h. (B) Cisplatin (DDP) (0.125,
0.25, 0.5, 1.0, 2.0, 4.0, 8.0 µg/ml) was used as the
positive control. Each group was replicated six times. Absorption
value was expressed as mean ± SD from six independent experiments.
Black pentangle indicates statistical difference between
experimental group and blank control group (★P<0.05,
★★P<0.01, ★★P<0.001). White triangle
indicates statistical difference between certain concentration
group and the next higher concentration group treated with PPS
extract or cisplatin (ΔP<0.05,
ΔΔP<0.01, ΔΔΔP<0.001). |
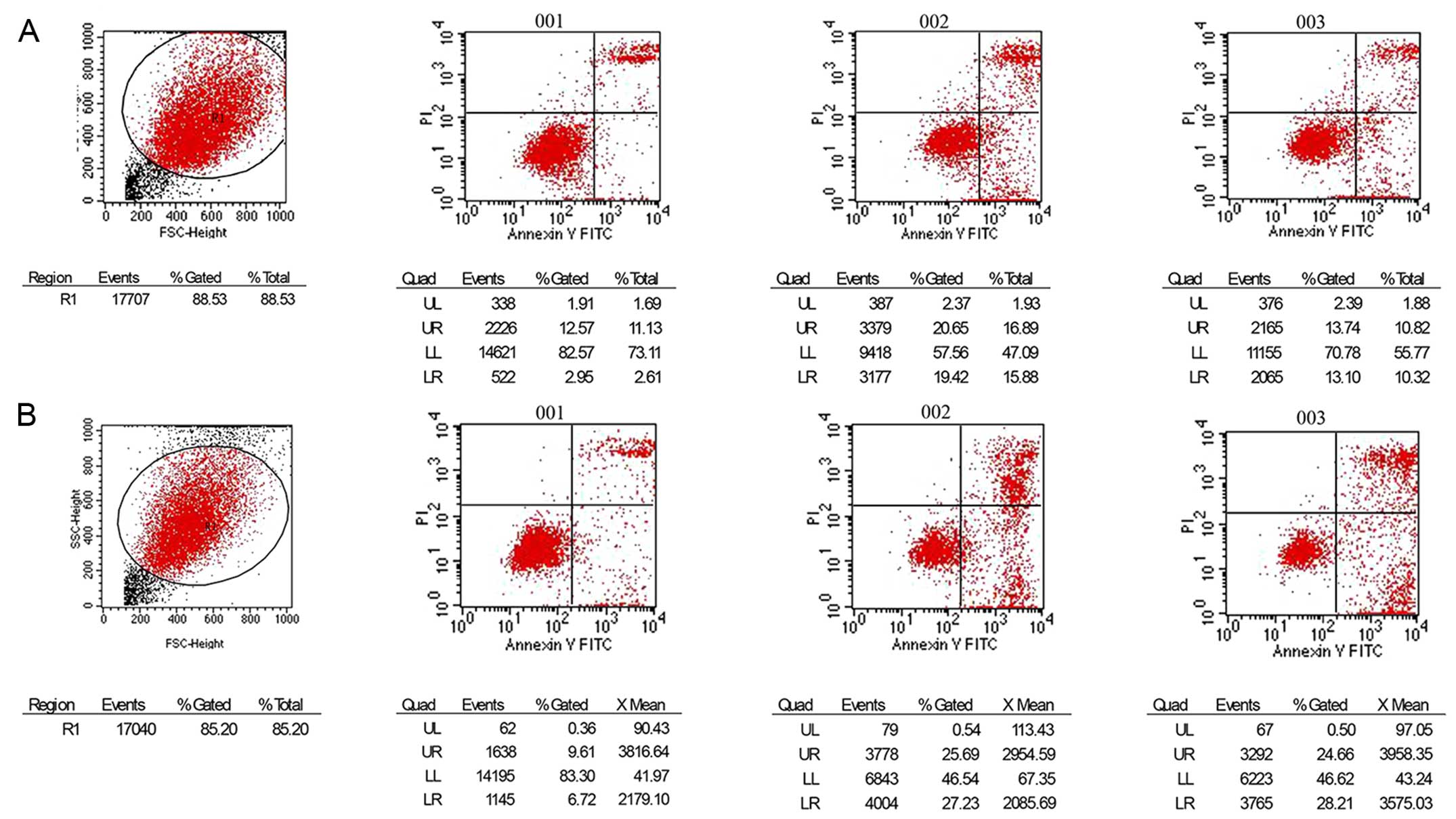
Effect of the different concentrations of
the PPS extract on cell apoptosis rate
We observed that PPS protein extract at higher than
100 µg/ml significantly increased the early apoptosis rate
of SKVO3 cells compared with that of the control group (Fig. 2A). PPS extract induced apoptosis of
SKOV3 cells in a dose-dependent manner. When the protein extract
concentration increased from 20 µg/ml to 50 µg/ml,
the early apoptosis rate was slightly increased. However, this rate
was significantly increased at extract concentration of 100
µg/ml or higher when compared with the next higher
concentration group (Fig. 2A). Only
the high concentrations (200 µg/ml, 400 µg/ml) of PPS
extract had significant effects on late cell apoptosis rate and
necrosis rate, whereas the low concentrations of PSS protein had no
obvious effects (Fig. 2B).
 | Figure 2Effects of PPS extract on early
apoptosis rate, late apoptosis rate and necrosis rate in SKOV3
cells. Cells were cultured in 10% FBS medium and treated with PPS
(0, 20, 50, 100, 200, 400 µg/ml) for 72 h. Apoptosis rates
are expressed as mean ± SD from three independent experiments. (A)
Early apoptosis rate of SKOV3 cells treated with PPS. (B) Late
apoptosis rate and necrosis rate of SKOV3 cells treated with PPS.
Black pentangle indicates statistical difference between
experimental group and blank control group (★P<0.05,
★★P<0.01, ★★★P<0.001). White triangle
indicates statistical difference between certain concentration
group and the next higher concentration group treated with PPS
extract (ΔP<0.05, ΔΔP<0.01,
ΔΔΔP<0.001). |
Time-dependent effect of PPS extract on
cell apoptosis rate
After the treatment with a relatively high
concentration of the PPS extract, SKOV3 cells showed obvious
apoptosis, and the apoptosis rate increased with time extension
(Fig. 3). The early apoptosis rate
in the cells treated with 150 µg/ml PPS protein extract for
48 h was 14.860±1.590%, significantly different from the control
group (t=14.224, P<0.001). Similarly, the early apoptosis rate
reached 24.426±4.149% after 72 h treatment with the same extract
concentration and the difference between the control group and the
treated group was also statistically significant (t=8.461,
P=0.001). The apoptosis rates of cells treated with 150
µg/ml PPS extract were significantly different between the
48 and 72 h treatment groups (t=3.729, P=0.02).
As a classical chemotherapeutical drug, cisplatin
has been widely used to treat various cancers. After the treatment
with 1 μg/ml cisplatin for 48 and 72 h, the early apoptosis rates
of SKOV3 cells reached 10.033±2.834% and 22.470±6.913%,
respectively, both significantly different from the blank control
group (t=5.116, P=0.007; t=4.629, P=0.01, respectively). The
difference between 48 and 72 h treatment was also statistically
significant (t=2.877, P=0.045). These data indicated that PPS
extract plays a significant role in apoptosis induction of ovarian
cancer cell line SKOV3 (Fig.
3).
Transcriptome profiling of SKOV3 and PPS
extract-treated SKOV3 and qPCR validation
To reveal how the PPS extract alters the gene
expression of SKOV3 cells after 72 h, the transcriptomes of SKOV3
cells and PPS extract-treated SKOV3 cells were sequenced using
Illumia/Solexa technology. In total, 44,576,818 and 52,295,034 raw
reads with 100 bp in length were obtained from SKOV3 and PPS
extract-treated cell lines, respectively. After filtering to
exclude low quality, adaptor contamination and ambiguous base
containing reads, 43,071,352 and 50,530,232 high quality clean
reads remained, representing approximately 4.3 and 5.0 Giga base.
We mapped the reads from two cell lines to hg19 genome for
quantitative measurement of gene expression and found that 17,217
transcripts were detected as expression. Differential gene
expression analysis between two samples showed that 1,754
transcripts were significantly differentially expressed, of which
840 were upregulated and 914 were downregulated in PPS
extract-treated group as compared with the untreated group.
To determine in which biochemical pathways these
differentially expressed transcripts (DETs) were enrich, total
expressed transcripts (17,217) were mapped to the KEGG pathway.
Transcripts (14,135) were assigned to 283 pathways representing
23,308 functional occurrences, of which 1,341 annotated transcripts
corresponding to 2,365 frequency of use in our indentified DETs.
Enrichment analyses were performed using hypergeometric
distribution testing followed by multiple hypothesis testing.
Result showed that DETs associated with steroid biosynthesis
(q<0.01) were over-represented as 11 of 2,365 (0.47%) DETs could
be mapped to this metabolism pathway, whereas the counterparts in
the transcriptome background were 32 and 23,308 (0.14%),
respectively. Table I showed the
gene expression differences between two groups of cell involved in
the steroid biosynthesis.
 | Table ITranscript expression difference
between SKOV3 and PPS extract-treated SKOV3 cells involved in
steroid biosynthesis identified by mRNA-Seq. |
Table I
Transcript expression difference
between SKOV3 and PPS extract-treated SKOV3 cells involved in
steroid biosynthesis identified by mRNA-Seq.
| Transcript
accession | Protein symbol | Enzyme code | FPKM-SKOV3 | FPKM-PPS | Fold change |
|---|
| NM_005891 | ACAT2 | EC:2.3.1.9 | 9.06 | 119.50 | 13.19 |
| NM_002130 | HMGCS1 | EC:2.3.3.10 | 10.94 | 67.12 | 6.13 |
| NM_000859 | HMGCR | EC:1.1.1.34 | 7.56 | 18.69 | 2.47 |
| NM_000431 | MVK | EC:2.7.1.36 | 5.69 | 17.43 | 3.06 |
| NM_006556 | PMVK | EC:2.7.4.2 | 32.00 | 35.38 | 1.11 |
| NM_002461 | MVD | EC:4.1.1.33 | 12.85 | 37.67 | 2.93 |
| NM_001135822 | FDPS | EC:2.5.1.10 | 39.68 | 128.57 | 3.24 |
| NM_001287748 | FDFT1 | EC:2.5.1.21 | 2.68 | 70.79 | 26.38 |
| NM_003129 | SQLE | EC:1.14.99.7 | 17.99 | 68.63 | 3.82 |
| NM_002340 | LSS | EC:5.4.99.7 | 0.0025 | 10.95 | >1000 |
| NM_000786 | CYP51A1 | EC:1.14.13.70 | 4.39 | 18.04 | 4.11 |
| NM_015922 | NSDHL | EC:1.1.1.170 | 26.82 | 46.66 | 1.74 |
| NM_001017369 | MSMO1 | EC:1.14.13.72 | 5.13 | 25.47 | 4.97 |
| NM_006745 | MSMO1 | EC:1.14.13.72 | 18.84 | 65.15 | 3.46 |
| NM_001129765 | NSDHL | EC:1.1.1.170 | 1.36 | 4.70 | 3.46 |
| NM_016371 | HSD17B7 | EC:1.1.1.270 | 3.35 | 7.55 | 2.25 |
| NM_006579 | EBP | EC:5.3.3.5 | 10.77 | 3.36 | 0.31 |
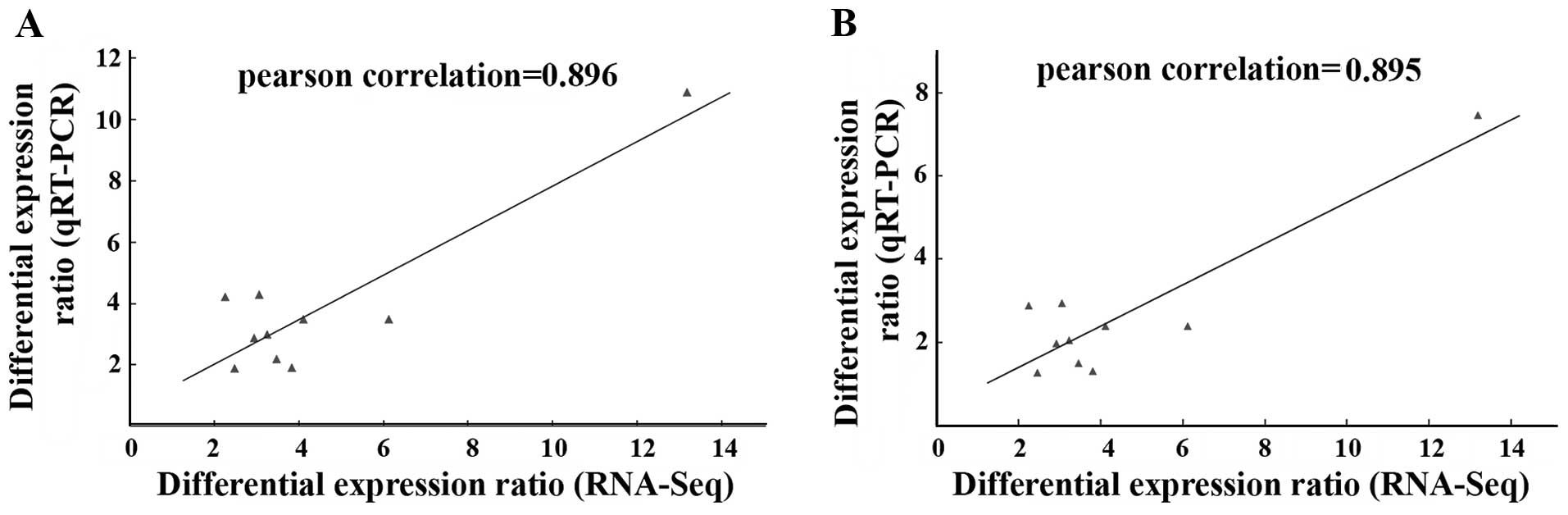
To evaluate high throughput sequencing data, ten
DETs involved in the steroid biosynthesis pathway were selected for
qPCR assay. Table II summarized
the gene primers used in this qPCR assay. 18s rRNA and GAPDH were
used as internal reference to normalize gene expression between two
samples. We found that all genes were upregulated in PPS
extract-treated cell line compared with untreated SKOV3, which is
consistent with the mRNA-Seq data although the exact fold changes
for these genes between the two techniques within two cell lines
are slightly different (Table
III). The correlation coefficients (Pearson) of differential
expression ratio between two techniques were 0.895 and 0.896 when
18s rRNA and GAPDH were used as internal control for qPCR analysis,
respectively (Fig. 4).
 | Table IIPrimers used in the qPCR. |
Table II
Primers used in the qPCR.
| Gene | Primer name | Sequence
(5′→3′) |
|---|
| 18S RRNA | 18S rRNA_F |
GGAGCCTGCGGCTTAATTT |
| 18S rRNA_R |
CAACTAAGAACGGCCATGCA |
| GAPDH | GAPDH_F |
CAAATTCCATGGCACCGTCA |
| GAPDH_R |
GGCAGAGATGATGACCCTTT |
| ACAT2 | ACAT2_F |
CGGCAGGAGAAGCAAGATGA |
| ACAT2_R |
GCTGCCAAGACATGTCCAAA |
| HMGCS1 | HMGCS1_F |
GCTGCTGTCTTCAATGCTGT |
| HMGCS1_R |
TTTGGCCCAATTAGCAGAGC |
| FDPS | FDPS_F |
AACTACTCGACCCACAGAGC |
| FDPS_R |
CTCAGTCAGCACCCTAACGA |
| SQLE | SQLE_F |
GTTCGCCCTCTTCTCGGATA |
| SQLE_R |
TGTTGATGTACAGGCAGCTG |
| LSS | LSS_F |
CCAGCCGGATACAGAGAAGA |
| LSS_R |
CCACAGCACCACCTTTCTTG |
| HSD17B7 | HSD17B7_F |
CGTTTGCTTCACTGCTTGGA |
| HSD17B7_R |
GCTTCTGCCTTGCTCATGTT |
| MVK | MVK_F |
CCTGAAGTACAACGCCTCCT |
| MVK_R |
TGCTACCTTGCCATGTACCA |
| MVD | MVD_F |
CGATGAAGAGCTGGTTCTGC |
| MVD_R |
CCATCCCGTGAGTTCCTCC |
| CYP51A1 | CYP51A1_F |
ACCCTCAGCCTGGTCTACC |
| CYP51A1_R |
GCATCACTCCCCAGAAGGTA |
| MSM01 | MSM01_F |
TGGAACACCTGGCGAGTC |
| MSM01_R |
TCCCCATGTTGCAATCTGGA |
 | Table IIIqPCR validation of ten DETs involved
in the steroid biosynthesis pathway. |
Table III
qPCR validation of ten DETs involved
in the steroid biosynthesis pathway.
| Endogenous
control |
Relative
fold difference of transcript abundance between SKOV3 and PPS
protein-treated SKOV3
|
|---|
| ACAT2 | HMGCS1 | FDPS | SQLE | LSS | HSD17B7 | MVK | MVD | CYP51A1 | MSM01 |
|---|
| 18s rRNA | 7.46 | 2.40 | 2.05 | 1.31 | 1.97 | 2.89 | 2.95 | 1.97 | 2.40 | 1.51 |
| GAPDh | 10.90 | 3.50 | 2.99 | 1.92 | 2.87 | 4.22 | 4.30 | 2.88 | 3.50 | 2.20 |
Discussion
Pedate Pinellia Rhizome, the rhizome of PPS,
is a common TCM found in the mid-region of China. It has been
reported in many TCM books to possess efficacy in dispelling wind,
relieving convulsion, drying dampness to eliminate phlegm,
eliminating stagnation, and has long been used to cure
thanatophidia bite, nameless swelling and toxicum. It has also been
shown to have antitumor properties without apparent toxic effects
on noncancerous cells (6,15). In this study, total extract obtained
from PPS was used to investigate apoptosis induction and
proliferation inhibition ability against ovarian cancer cell line
SKOV3. Cisplatin, the most commonly used chemotherapy drug in
ovarian cancer, was used as the positive control. Results showed
that only low concentrations of PPS protein extract (20
µg/ml) had obvious inhibitory effect on cell proliferation,
whereas high concentrations of PPS extract (≥100 µg/ml)
significantly increased the early apoptosis rate of SKOV3 cells. In
addition, the early apoptosis rate was also significantly increased
when the treatment time was extended from 48 to 72 h under the same
extract concentration (150 µg/ml). These results indicated
that the extract of PPS could induce apoptosis in ovarian cancer
cell SKOV3 in a dose- and time-dependent manner.
Transcriptome sequencing and quantitative gene
expression analysis identified 1,754 DETs between SKOV3 and PPS
extract-treated cells. These DETs were significantly enriched in
the steroid biosynthesis pathway. In this pathway, we found that
most of the enzymes responsible for the synthesis from
farnesyl-diphosphate to 3-keto-4-methylzymosterol were
significantly upregulated in response to PPS treatment (Tables I and III). Of note, many genes involved in the
synthesis of farnesyl-diphosphate, the original substrate of the
steroid biosynthesis, were significantly upregulated, although not
enriched, in the terpenoid backbone biosynthesis pathway. These
upregulated DETs constituted a relatively intact reaction chain
associated with the synthesis of acetyl-COA to
3-keto-4-methylzymosterol, indicative of increased accumulation of
zymosterol in the PPS extract-treated cells. Validation of
transcriptome sequencing data by using qPCR also showed that genes
associated with the zymosterol biosynthesis were significantly
upregulated after treated with PPS extract.
In addition, the cholestenol delta-isomerase, which
is enzymatically involved in the catalysis of zymosterol to
5alpha-Cholesta-7,24-dien-3beta-ol, was downregulated, further
suggesting the retained accumulation of zymosterol in PPS
extract-treated cells (Fig. 5).
Furthermore, the expression of genes encoding the down-stream
enzymes in the steroid biosynthesis pathway were not significantly
differential, suggesting that the final product for majority of
farnesyl-diphosphate may not be cholesterol or other secosteroid,
such as vitamine D2, but zymosterol or its precursor. Zymosterol,
an intermediate precursor of cholesterol, was located in the plasma
membrane of human cells and has been considered to play an
important role in regulating membrane fluidity and permeability
(16–18). Previous studies have demonstrated
that compounds derived from the isoprenoid/cholesterol biosynthetic
pathway possessed novel biological activities, such as regulating
transcriptional and post-transcriptional events that in turn affect
lipid synthesis, meiosis, apoptosis, developmental patterning,
protein cleavage, as well as protein degradation (19). The upregulated expression of enzymes
encoding genes involved in the biosynthesis of zymosterol may be
closely related to the increase of cell apoptosis rate after PPS
extract treatment.
To date, two classical apoptosis signaling pathways
(the extrinsic and intrinsic pathways) have been well established
(20,21). The extrinsic pathway is initiated by
the extracellular hormones or agonists that can be recognized by
the cell surface death receptors (22). The activation of intrinsic pathway
can be initiated by the release of cytochrome c or SMAC/DIABLO from
mitochondria (21). Programmed cell
death (PCD) from both signaling pathways can be finally ascribed to
caspase activation, including the initiator, such as caspase-8, -9,
-10, and the executioner, such as caspase-3 and -7. Since treatment
of SKOV3 cells with 100 µg/ml or higher PPS extract
significantly increased the apoptosis rate, it is possible that the
expression level of genes participated in PCD might have changed.
As expected, the expression of the caspase family members,
including caspase-3β, -7a, -8c, -9α and caspase-10 variant 2, were
significantly upregulated in PPS extract-treated group, except for
caspase-7e, which was downregulated (Table IV). These upregulated caspases may
well explain the increase of cell apoptosis rates.
 | Table IVTranscript expression difference
between SKOV3 and PPS extract-treated SKOV3 cells involved in
apoptosis identified by mRNA-Seq. |
Table IV
Transcript expression difference
between SKOV3 and PPS extract-treated SKOV3 cells involved in
apoptosis identified by mRNA-Seq.
| Transcript
accession | Protein symbol | FPKM-SKOV3 | FPKM-PPS | Fold change |
|---|
| NM_032991 | CASP3 variant
β | 2.7 | 8.85 | 3.27 |
| NM_033356 | CASP8 variant
C | 0 | 3.11 | Inf |
| NM_001229 | CASP9 variant
α | 0.0008 | 3.23 | >1000 |
| NM_001267056 | CASP7 variant
e | 14.77 | 4.18 | 0.28 |
| NM_001227 | CASP7 variant
a | 0.66 | 6.58 | 9.96 |
| NM_032974 | CASP10 variant
2 | 0.36 | 4.53 | 12.72 |
| NM_001204401 | XIAP variant 2 | 0.0022 | 4.14 | >1000 |
| NM_001012270 | BIRC5 variant
2 | 1.54 | 5.42 | 3.52 |
| NM_001171625 | VEGFA variant
3 | 8.71 | 0.09 | 0.01 |
| NM_001243733 | VEGFB-167 | 12.6 | 0 | 0 |
| NM_198156 | VHL variant 2 | 0.13 | 3.65 | 28.1 |
| NM_004465 | FGF10 | 6.11 | 0 | 0 |
Controversially, the inhibitors of apoptosis (IAP)
protein, such as BIRC5 (Survivin) and xIAP, which inhibit caspase
activity by directly binding to the enzyme, were also significantly
upregulated (23,24). The overexpressed IAP family members
in PPS group might also result from the cell anti-apoptotic
response, which in turn antagonized the upregulated caspase
activity. However, the enhanced xIAP activity in PPS-treated cells
might not be antagonized by the second mitochondrial-derived
activator of caspase (SMAC/DIABLO), because the expression of
SMAC-encoded transcript variants was not upregulated. In addition,
few cytosolic pro-apoptotic Bcl-2 family members that can promote
cytochrome c release from mitochondria to cytosol, such as Bid,
Bax, Bak, Bad, were upregulated. Taken together, the PPS
extract-induced PCD in SKOV3 cells may initially be mediated by the
extrinsic apoptosis signaling pathway.
Vascular endothelial growth factor (VEGF) has been
suggested to be a key regulator of angiogenesis in many types of
cancer, including ovarian cancer (25,26).
Due to alternative splicing events, a number of VEGF mRNA variants
have been detected and all variants are derived from the same locus
in human genome (27). VEGF induces
endothelial cell proliferation presumably through binding to the
endothelial cell receptor. Elevated VEGF expression across a wide
range of cancers has been observed, such as breast cancer and
ovarian cancer (28,29). We found that VEGFA variant 3 and
VegFB-167 were moderately expressed in SKOV3 cell line. However,
the expression of these two variants was extremely downregulated in
PPS extract-treated cell line, where FPKM values were almost
undetectable (Table IV). This may
be due to the enhanced von Hippel-Lindau tumor suppressor (VHL)
expression in PPS extract-treated group. VHL possesses ubiquitin
ligase E3 activity and is a well-characterized VEGF transcriptional
repressor, which inhibits VEGF expression through direct
ubiquitination and subsequent degradation of its transcription
factor (30,31), hypoxia-inducible-factor (HIF). For
other hormones, FGF10 was moderately expressed in SKOV3 cells,
whereas its expression was not detected in PPS-treated cells. FGF10
has been proven to play an important role in tumor growth and
invasion in a variety of cancers, such as breast cancer and
pancreatic cancer (32,33). The downregulated FGF10 expression
was consistent with the low proliferation ability in response to
PPS extract treatment.
In conclusion, total extract obtained from PPS can
inhibit epithelial derived ovarian cancer cell line SKOV3
proliferation and further induce apoptosis of these cells in a
dose- and time-dependent manner, indicative of PPS extract
component as a potential drug for ovarian cancer treatment in the
future. Transcriptome sequencing and differential gene expression
analysis revealed that the expression of a number of caspase family
members were upregulated after PPS extract treatment. This result
is mechanistically consistent with the obvious apoptotic phenotype
and increased apoptosis rate of SKOV3 cells measured by flow
cytometry.
In addition, many differentially expressed
transcripts involved in the synthesis of steroid were significantly
upregulated and over-represented, suggesting that cell apoptosis
induced by extract of PPS might be accompanied by the increased
biosynthesis of steroid, probably zymosterol. Therefore, this
steroid could be considered as a novel regulator during ovarian
cancer cell apopotsis after treated with PPS extract.
Acknowledgments
This work was supported by Science and Technology
Foundation of Wenzhou City, China (H20100011), Zhejiang Provincial
Science and Technology Foundation of Chinese Medicine, China
(2012ZB106), Foundation of Science and Technology Innovation Team
of Zhejiang Province, China (2010R50048-13) and Research Initial
Funding in Wenzhou Medical University (QTJ14002).
References
|
1
|
Zhang L, Huang J, Yang N, Greshock J,
Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR,
et al: microRNAs exhibit high frequency genomic alterations in
human cancer. Proc Natl Acad Sci USA. 103:9136–9141. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yap TA, Carden CP and Kaye SB: Beyond
chemotherapy: Targeted therapies in ovarian cancer. Nat Rev Cancer.
9:167–181. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Eisenhauer EL, Tew WP, Levine DA, Lichtman
SM, Brown CL, Aghajanian C, Huh J, Barakat RR and Chi DS: Response
and outcomes in elderly patients with stages IIIC-IV ovarian cancer
receiving platinum-taxane chemotherapy. Gynecol Oncol. 106:381–387.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Boren T, Xiong Y, Hakam A, Wenham R, Apte
S, Chan G, Kamath SG, Chen DT, Dressman H and Lancaster JM:
MicroRNAs and their target messenger RNAs associated with ovarian
cancer response to chemotherapy. Gynecol Oncol. 113:249–255. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li SD, Zhang JR, Wang YQ and Wan XP: The
role of microRNAs in ovarian cancer initiation and progression. J
Cell Mol Med. 14:2240–2249. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li GL, Jiang W, Xia Q, Chen SH, Ge XR, Gui
SQ and Xu CJ: HPV E6 down-regulation and apoptosis induction of
human cervical cancer cells by a novel lipid-soluble extract (PE)
from Pinellia Pedatisecta Schott in vitro. J Ethnopharmacol.
132:56–64. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sun GX, Ding SS and Qian YJ: The
extraction and chemical analysis of proteins from Pinellia
Pedatisecta and their inhibitory effects on the mouse sarcoma-180.
Acta Academiae Medicinae Shanghai. 19:17–20. 1992.
|
|
8
|
Bolger AM, Lohse M and Usadel B:
Trimmomatic: A flexible trimmer for Illumina sequence data.
Bioinformatics. 30:2114–2120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Trapnell C, Pachter L and Salzberg SL:
TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics.
25:1105–1111. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Trapnell C, Roberts A, Goff L, Pertea G,
Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL and Pachter L:
Differential gene and transcript expression analysis of RNA-seq
experiments with Tophat and Cufflinks. Nat Protoc. 7:562–578. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Trapnell C, Hendrickson DG, Sauvageau M,
Goff L, Rinn JL and Pachter L: Differential analysis of gene
regulation at transcript resolution with RNA-seq. Nat Biotechnol.
31:46–53. 2013. View
Article : Google Scholar
|
|
12
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar
|
|
13
|
Ma X, Wehland M, Aleshcheva G, Hauslage J,
Wasser K, Hemmersbach R, Infanger M, Bauer J and Grimm D:
Interleukin-6 expression under gravitational stress due to
vibration and hypergravity in follicular thyroid cancer cells. PLOS
One. 8:e681402013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
15
|
Li GL, Gui SQ and Zhu DH: Effects of
pinellia extraction only or combined with cisplatin on growth of
Hela cells of cervical cancer. Fudan Univ J Med Sci. 34:869–872.
2007.
|
|
16
|
Echevarria F, Norton RA, Nes WD and Lange
Y: Zymosterol is located in the plasma membrane of cultured human
fibroblasts. J Biol Chem. 265:8484–8489. 1990.PubMed/NCBI
|
|
17
|
Lange Y, Echevarria F and Steck TL:
Movement of zymosterol, a precursor of cholesterol, among three
membranes in human fibroblasts. J Biol Chem. 266:21439–21443.
1991.PubMed/NCBI
|
|
18
|
Brown DA and London E: Structure and
function of sphingolipid- and cholesterol-rich membrane rafts. J
Biol Chem. 275:17221–17224. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Edwards PA and Ericsson J: Sterols and
isoprenoids: Signaling molecules derived from the cholesterol
biosynthetic pathway. Annu Rev Biochem. 68:157–185. 1999.
View Article : Google Scholar
|
|
20
|
Strasser A, O'Connor L and Dixit VM:
Apoptosis signaling. Annu Rev Biochem. 69:217–245. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jiang X and Wang X: Cytochrome C-mediated
apoptosis. Annu Rev Biochem. 73:87–106. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ashkenazi A and Dixit VM: Death receptors:
Signaling and modulation. Science. 281:1305–1308. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Deveraux QL and Reed JC: IAP family
proteins - suppressors of apoptosis. Genes Dev. 13:239–252. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bratton SB, Walker G, Srinivasula SM, Sun
XM, Butterworth M, Alnemri ES and Cohen GM: Recruitment, activation
and retention of caspases-9 and -3 by Apaf-1 apoptosome and
associated XIAP complexes. EMBO J. 20:998–1009. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Boocock CA, Charnock-Jones DS, Sharkey AM,
McLaren J, Barker PJ, Wright KA, Twentyman PR and Smith SK:
Expression of vascular endothelial growth factor and its receptors
flt and KDR in ovarian carcinoma. J Natl Cancer Inst. 87:506–516.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Duyndam MC, Hilhorst MC, Schlüper HM,
Verheul HM, van Diest PJ, Kraal G, Pinedo HM and Boven E: Vascular
endothelial growth factor-165 overexpression stimulates
angiogenesis and induces cyst formation and macrophage infiltration
in human ovarian cancer xenografts. Am J Pathol. 160:537–548. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tischer E, Mitchell R, Hartman T, Silva M,
Gospodarowicz D, Fiddes JC and Abraham JA: The human gene for
vascular endothelial growth factor. Multiple protein forms are
encoded through alternative exon splicing. J Biol Chem.
266:11947–11954. 1991.PubMed/NCBI
|
|
28
|
Takahashi Y, Kitadai Y, Bucana CD, Cleary
KR and Ellis LM: Expression of vascular endothelial growth factor
and its receptor, KDR, correlates with vascularity, metastasis, and
proliferation of human colon cancer. Cancer Res. 55:3964–3968.
1995.PubMed/NCBI
|
|
29
|
Leng R, Zha L and Tang L: MiR-718
represses VEGF and inhibits ovarian cancer cell progression. FEBS
Lett. 588:2078–2086. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Iliopoulos O, Levy AP, Jiang C, Kaelin WG
Jr and Goldberg MA: Negative regulation of hypoxia-inducible genes
by the von Hippel-Lindau protein. Proc Natl Acad Sci USA.
93:10595–10599. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Maxwell PH, Wiesener MS, Chang GW,
Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER and
Ratcliffe PJ: The tumour suppressor protein VHL targets
hypoxia-inducible factors for oxygen-dependent proteolysis. Nature.
399:271–275. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nomura S, Yoshitomi H, Takano S, Shida T,
Kobayashi S, Ohtsuka M, Kimura F, Shimizu H, Yoshidome H, Kato A,
et al: FGF10/FGFR2 signal induces cell migration and invasion in
pancreatic cancer. Br J Cancer. 99:305–313. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sugimoto K, Yoshida S, Mashio Y, Toyota N,
Xing Y, Xu H, Fujita Y, Huang Z, Touma M and Wu Q: Role of FGF10 on
tumorigenesis by MS-K. Genes Cells. 19:112–125. 2014. View Article : Google Scholar
|