Introduction
Worldwide, colorectal carcinoma (CRC) is one of the
most common malignant cancers and is the second most frequent cause
of cancer-related deaths (1–3).
Despite marked advances in diagnostic and therapeutic approaches,
the prognosis for CRC patients remains poor (4). Consequently, there is an urgent need
to develop new CRC treatment methods.
Since homeobox-containing (HOX) genes encode
DNA-binding proteins, they are master transcriptional regulators of
cell differentiation, morphogenesis, and organogenesis during
development (5,6). In humans and other mammals, 39
HOX genes are clustered in four complexes called
HOXA, B, C, and D. These are further
subdivided into 13 paralogous groups based on their sequence
similarities and relative positions along the clusters (7–11).
Mutational analyses demonstrated that HOXD genes play a
pivotal role in determining the regional specificity of cells
(12,13). For example, the HOXD13 gene
is involved in the normal morphogenesis of limbs (12) and of the anal sphincter (13). HOXD3 is a member of the third
paralogous group of the HOXD gene family and is involved in
embryonic development (14,15).
Studies have shown that the human HOXD3 gene
plays a multifunctional role in cancer. For example, HOXD3
overexpression was found to regulate cell adhesion in human
erythroleukemia HEL cells (16),
induce coordinate metastasis-related gene expression, and enhance
the motility and invasiveness of human lung cancer A549 (17–19)
and melanoma cells (20).
Additionally, studies have shown that HOXD3 induces
angiogenesis by increasing pro-angiogenic molecules (21–25).
While these studies clearly demonstrated that HOXD3 is
involved in the development and growth of various types of cancers,
the functional role of HOXD3 in human CRC has not yet been
determined.
In the present study, we demonstrated that
HOXD3 is highly expressed in the human CRC RKO cell line.
Consequently, we used a lentiviral vector to deliver small
interfering RNA (siRNA) to knock down HOXD3 expression in
the RKO cells. Finally, we assessed the effects of HOXD3
knockdown on human CRC cell growth and survival in
vitro.
Materials and methods
Cell lines
DLD-1, HCT-116, SW620, HT-29, and RKO human colon
carcinoma cell lines were purchased from the Shanghai Cell Bank of
the Chinese Academy of Science (Shanghai, China). Cell lines were
maintained in Dulbecco's modified Eagle's medium (DMEM; Corning,
Shanghai, China) supplemented with 10% fetal bovine serum (FBS;
Ausbian, Australia) at 37°C in a 5% CO2 incubator.
Quantitative RT-PCR
Total RNA was extracted from the cells in each group
(DLD-1, HCT-116, SW620, HT-29, and RKO) using TRIzol reagent
(Invitrogen, Carlsbad, CA, USA), according to the manufacturer's
protocol. Two micrograms of total RNA from each sample was reverse
transcribed to single-stranded cDNA. One microgram of cDNA was used
as a template for the quantitative real-time PCR. The primers used
were as follows: HOXD3 forward, 5′-CGG CAA CTT CGT CGA GTC
C-3′ and reverse, 5′-ATG AGG GTC GCA AGG TCC A-3′; and GAPDH
forward, 5′-TGA CTT CAA CAG CGA CAC CCA-3′ and reverse, 5′-CAC CCT
GTT GCT GTA GCC AAA-3′. Cycling conditions for quantitative RT-PCR
were as follows: 95°C for 30 sec, then 45 cycles of 95°C for 5 sec
and 60°C for 30 sec. The PCR products of HOXD3 and
GAPDH were 145 and 121 bp, respectively. The data were
quantified using the 2−ΔΔCt method. All analyses were
performed in triplicate.
Recombinant lentiviral vector production
and cell infection
To create the RNAi target site, the complementary
DNA sequence (CCA AAT CAC AGC CCA ATA T) of HOXD3 was
designed by Shanghai GeneChem Co., Ltd. (Shanghai, China) using the
full-length human sequence (GenBank no. NM_006898). The
HOXD3 hairpin oligonucleotides were synthesized and inserted
into the pGV115-GFP (GeneChem Co. Ltd.) lentiviral vector.
Lentivirus particles were prepared as previously described
(26).
For lentiviral infection, RKO cells were cultured in
6-well plates. The HOXD3-siRNA lentivirus (shHOXD3) or
negative control lentivirus (shCtrl) was added according to a
multiplicity of infection (MOI 1:5). At 3 days post-infection, the
cells were observed for presence of the GFP marker with a
fluorescence microscope (MicroPublisher 3.3RTV; Olympus, Tokyo,
Japan). At 5 days post-infection, the cells were harvested and
knockdown efficiency was analyzed by quantitative RT-PCR and
western blot analysis.
Western blot analysis
While on ice, cell lysates were incubated for 10–15
min in ice-cold lysis buffer (100 mM Tris, pH 6.8, 2%
β-mercaptoethanol, 20% glycerol, 4% SDS). The lysates were
centrifuged at 12,000 × g for 15 min at 4°C, and the supernatants
were collected. Protein concentration was determined using a BCA
protein assay kit (Beyotime, Beijing, China). An equal amount of
total protein from each sample was partially separated in a 10%
SDS-PAGE gel and blotted onto PVDF membranes. Membranes were
incubated with GFP or GAPDH primary antibodies (Santa Cruz
Biotechnology, Santa Cruz, CA, USA) at 4°C overnight, followed by
horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Santa
Cruz Biotechnology) secondary antibody at room temperature.
Enhanced chemiluminescence (ECL) reagent (ECL-Plus/kit; Amersham,
Piscataway, NJ, USA) was used for detection. The amount of GAPDH
detected was used as the protein loading internal control.
Cell proliferation assay
After being infected with the shCtrl lentivirus or
shHOXD3 lentivirus, RKO cells were seeded in 96-well plates at a
concentration of 2,000 cells/well and incubated for 5 days at 37°C
with 5% CO2. The cells were counted each day using the
Cellomics ArrayScan™ VT1 HCS automated reader (Cellomics Inc.,
Pittsburgh, PA, USA). At least 800 cells/well were analyzed. At 1,
2, 3, 4, or 5 days post-infection, the cells were incubated with
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT,
5 mg/ml; Promega, Shanghai, China) at a final concentration of 0.5
mg/ml for 4 h. After discarding the supernatants, 150 µl of
dimethyl sulfoxide (DMSO; Sigma-Aldrich Co., LLC, St. Louis, MO,
USA) was added to each well. The plates were read at 490 nm using
an ELISA reader (Tecan Infinite, Männedorf, Switzerland). All
experiments were performed in triplicate.
Cell cycle distribution analysis
Flow cytometry (FCM) was used to determine cell
cycle distribution as previously described (27). Briefly, RKO cells were infected with
shCtrl or shHOXD3 vector and incubated at 37°C for 1, 2, 3, 4, or 5
days. At the indicated time-point, the cells were collected and
washed with ice-cold phosphate-buffered saline (PBS). Cells were
centrifuged at 1,000 × g for 10 min and then fixed in ice-cold 70%
ethanol for 30 min at 4°C. The cells were washed with PBS and then
resuspended and incubated in PBS containing 50 µg/ml
propidium iodide (PI; Sigma-Aldrich) and 100 µg/ml RNase A
(Fermentas, Shanghai, China) in the dark at 4°C for 30 min. Cell
cycle phase was analyzed using a BD FACSCalibur flow cytometer (BD
Biosciences, San Diego, CA, USA). All studies were performed in
triplicate.
Analysis of apoptosis
FCM was used to measure apoptosis and was performed
as previously described (28). At
48 h prior to transfection, the RKO cells were seeded in 6-well
plates. At 72 h post-transfection, the cells were collected and
washed twice with ice-cold PBS. Cell densities were adjusted to
1×106/ml using 1X staining buffer. Cell suspension (100
µl) and 5 µl Annexin V-APC (eBioscience, San Diego,
CA, USA) were thoroughly mixed and incubated in darkness at room
temperature for 15 min. All rates of apoptosis were measured by FCM
within 1 h. Each experiment was performed in triplicate.
Cell colony formation assay
At 72 h post-transfection, the RKO cells were
reseeded at 600 cells/well in 6-well plates and cultured at 37°C
for 10 days. The cells were collected and washed with ice-cold PBS.
When a majority of single colonies contained more than 50 cells,
the samples were fixed using 1 ml/well of 4% paraformaldehyde
(Sinopharm, Shanghai, China) for 30 min at 37°C. According to
instructions, the samples were stained with 500 µl of Giemsa
stain (Dingguo Bio, Shanghai, China) at room temperature for 10 min
and images were acquired. All experiments were performed in
triplicate.
Statistical analyses
Data for each group are presented as the mean ± SD.
Statistical analyses were performed using SPSS for Windows, version
20.0 (IBM SPSS, Chicago, IL, USA). Values of p<0.05 were deemed
statistically significant.
Results
HOXD3 mRNA levels in colorectal cancer
cell lines
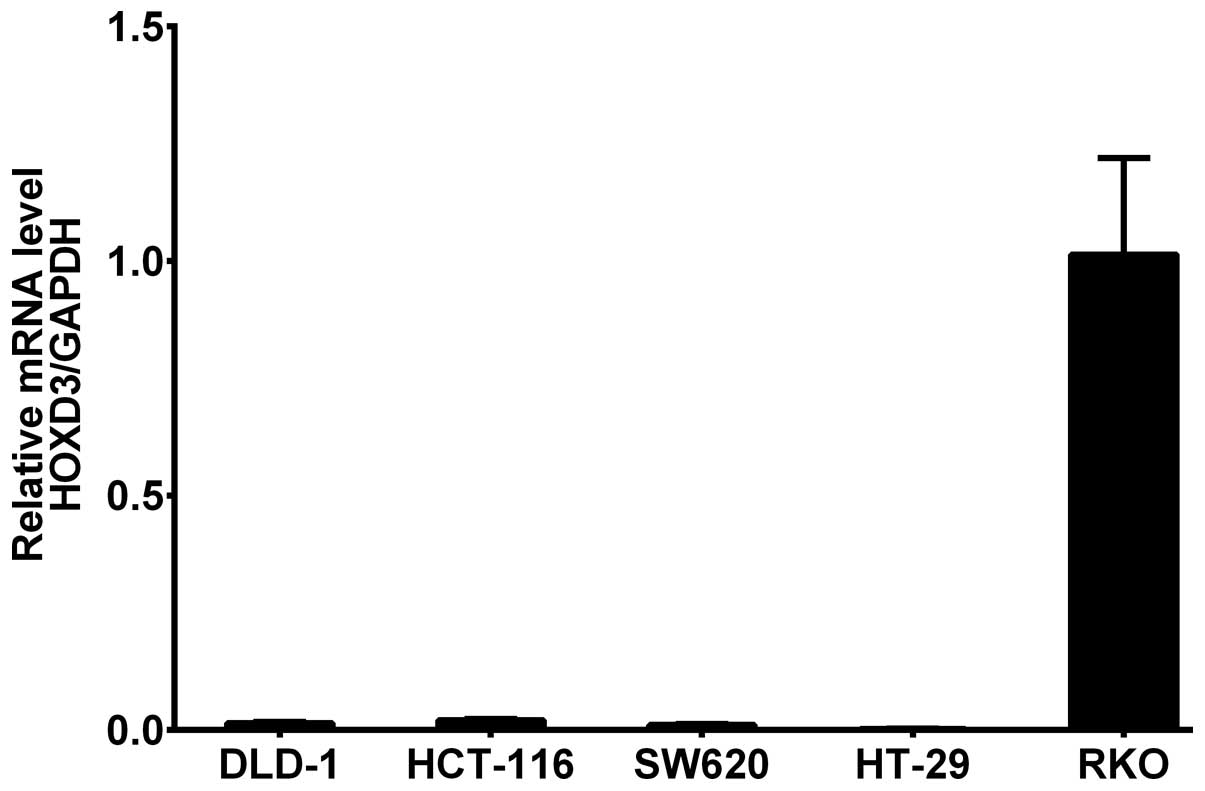
HOXD3 mRNA expression was measured in DLD-1,
HCT-116, SW-620, HT-29, and RKO CRC cell lines by RT-PCR. The
results showed that HOXD3 mRNA was highly expressed in the
RKO cell line (Fig. 1).
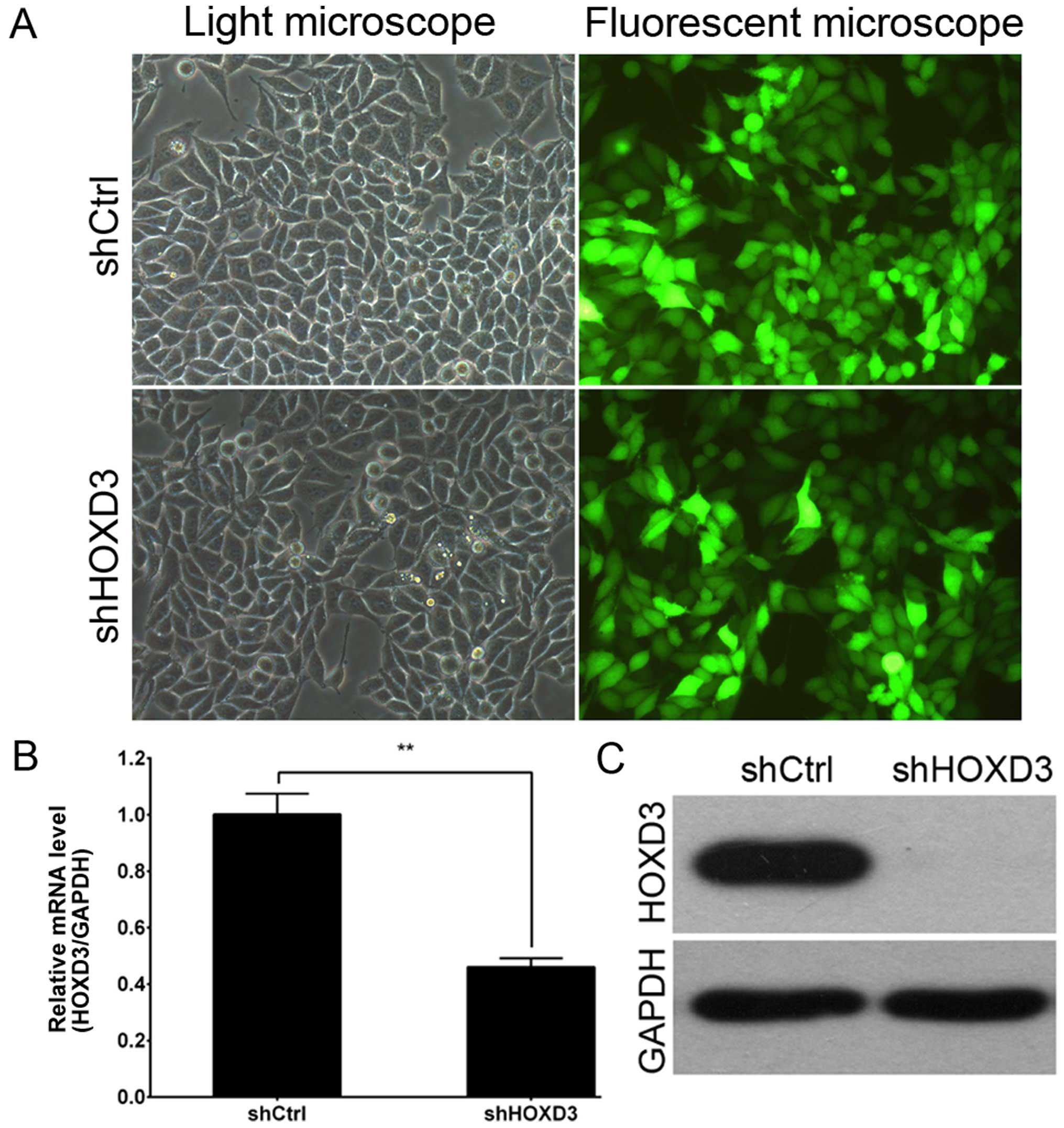
Lentiviral-mediated knockdown of HOXD3 in
RKO cells
To explore the role of HOXD3 in CRC, RKO
cells were infected with the shCtrl lentivirus or shHOXD3
lentivirus. As shown in Fig. 2A, by
3 days post-infection, the proportion of infected RKO cells was
greater than 80% in both the shHOXD3 and shCtrl groups. At 5 days
post-infection, HOXD3 mRNA levels were measured by real-time
PCR. shHOXD3 infected cultures had significantly lower levels of
HOXD3 mRNA when compared to the shCtrl-infected cultures
(Fig. 2B). Fig. 2C shows HOXD3 protein expression as
detected by western blot analysis. HOXD3 levels were greatly
reduced in the shHOXD3 group, indicating effective knockdown of the
target sequence.
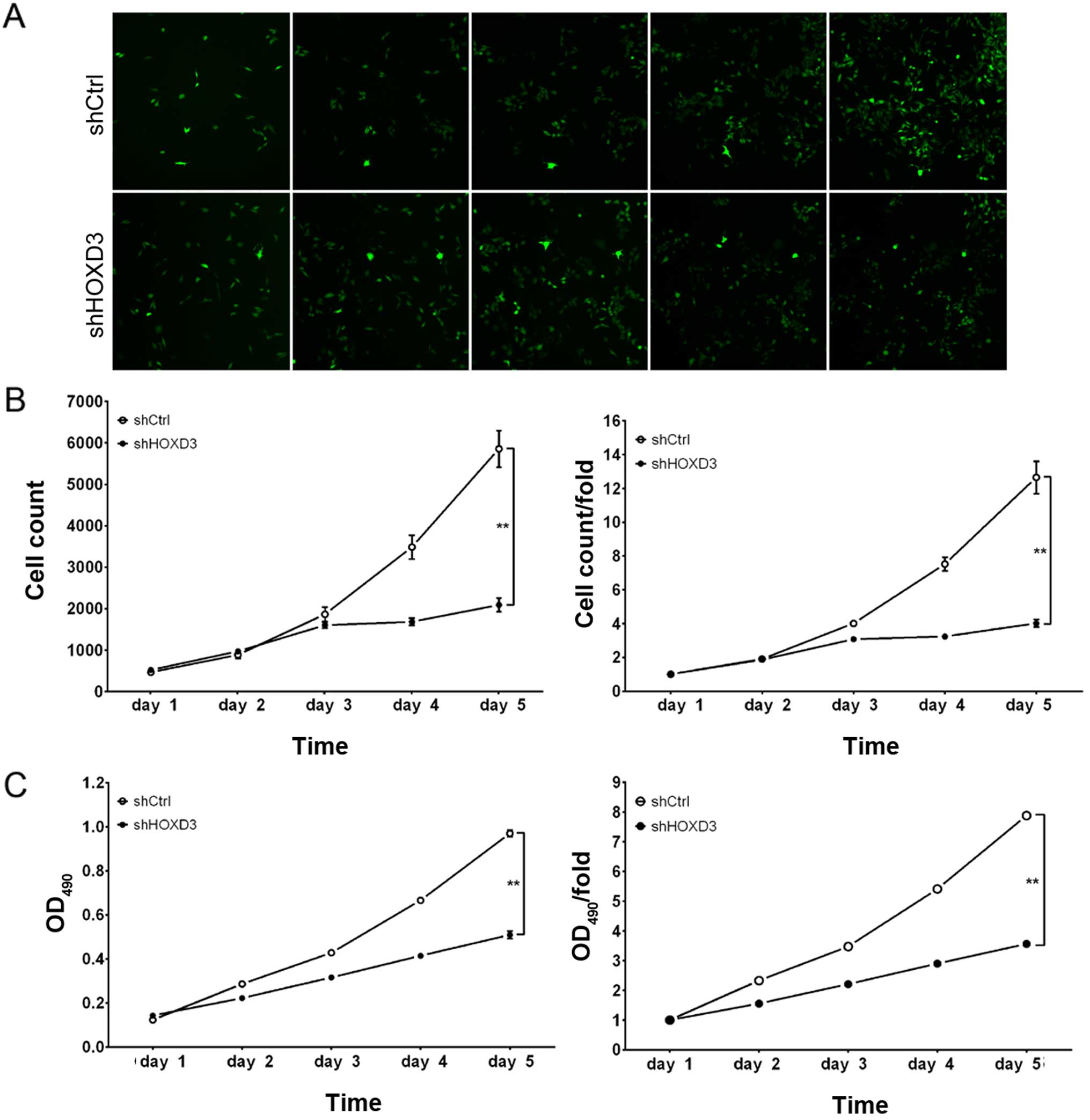
HOXD3 knockdown suppresses RKO cell
proliferation
To examine the effect of HOXD3 knockdown on
cell growth, shCtrl and shHOXD3 infected RKO cells were reseeded in
96-well plates and analyzed at 1, 2, 3, 4, and 5 days
post-infection. As illustrated in Fig.
3A and B, shCtrl cells exhibited extensive proliferation at 5
days post-infection, while the number of shHOXD3 cells increased
slightly. Cell growth rate was defined as: Cell count on day n/cell
count on day 1, where n=2, 3, 4, or 5 (Fig. 3B). These results revealed that
HOXD3 knockdown significantly inhibited the proliferation of
RKO cells.
The effect of HOXD3 protein reduction on RKO cell
proliferation was also determined by MTT assay. Although shCtrl and
shHOXD3 cells had similar in vitro growth on days 1, 2, and
3, the shHOXD3 cells had significantly reduced in vitro
growth on days 4 (shCtrl: 5.41±0.03 vs. shHOXD3: 2.90±0.04,
p<0.01) and 5 (shCtrl: 7.88±0.12 vs. shHOXD3: 3.56±0.12,
p<0.01) (Fig. 3C). Based on
these data, in vitro RKO cell growth was dependent on
HOXD3 expression.
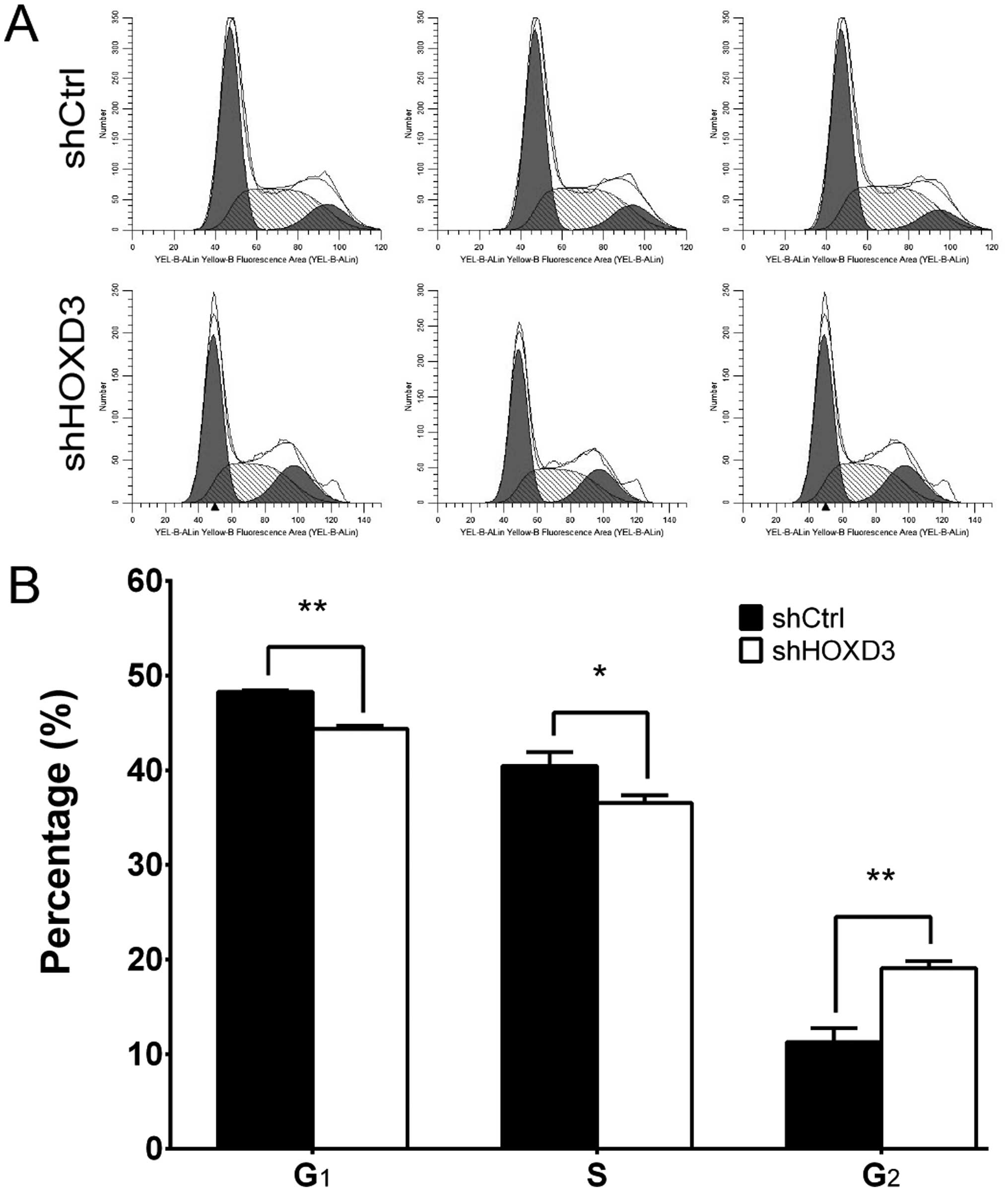
HOXD3 knockdown leads to cell cycle
arrest in the RKO cells
To determine whether HOXD3 is necessary for
cell cycle progression in RKO cells, we measured cell cycle phases
by FCM (Fig. 4A). The shCtrl group
had the following distribution: G1 phase: 48.28±0.16%, S
phase: 40.46±1.46%, G2 phase: 11.26±1.48%. The shHOXD3
group, however, had this distribution: G1 phase:
44.83±0.31%, S phase: 36.56±0.77%, G2 phase:
19.07±0.79%. As shown in Fig. 4B,
shHOXD3 cells had significant decreases in the percentage of cells
in the G1 (p<0.01) and S phases (p=0.015), compared
to the shCtrl cells. Conversely, when compared to the shCtrl cells,
the percentage of shHOXD3 cells in the G2 phase
(p<0.01) was increased. Taken together, these data suggest that
HOXD3 regulates cell growth and can block cell cycle
progression in the G2 phase.
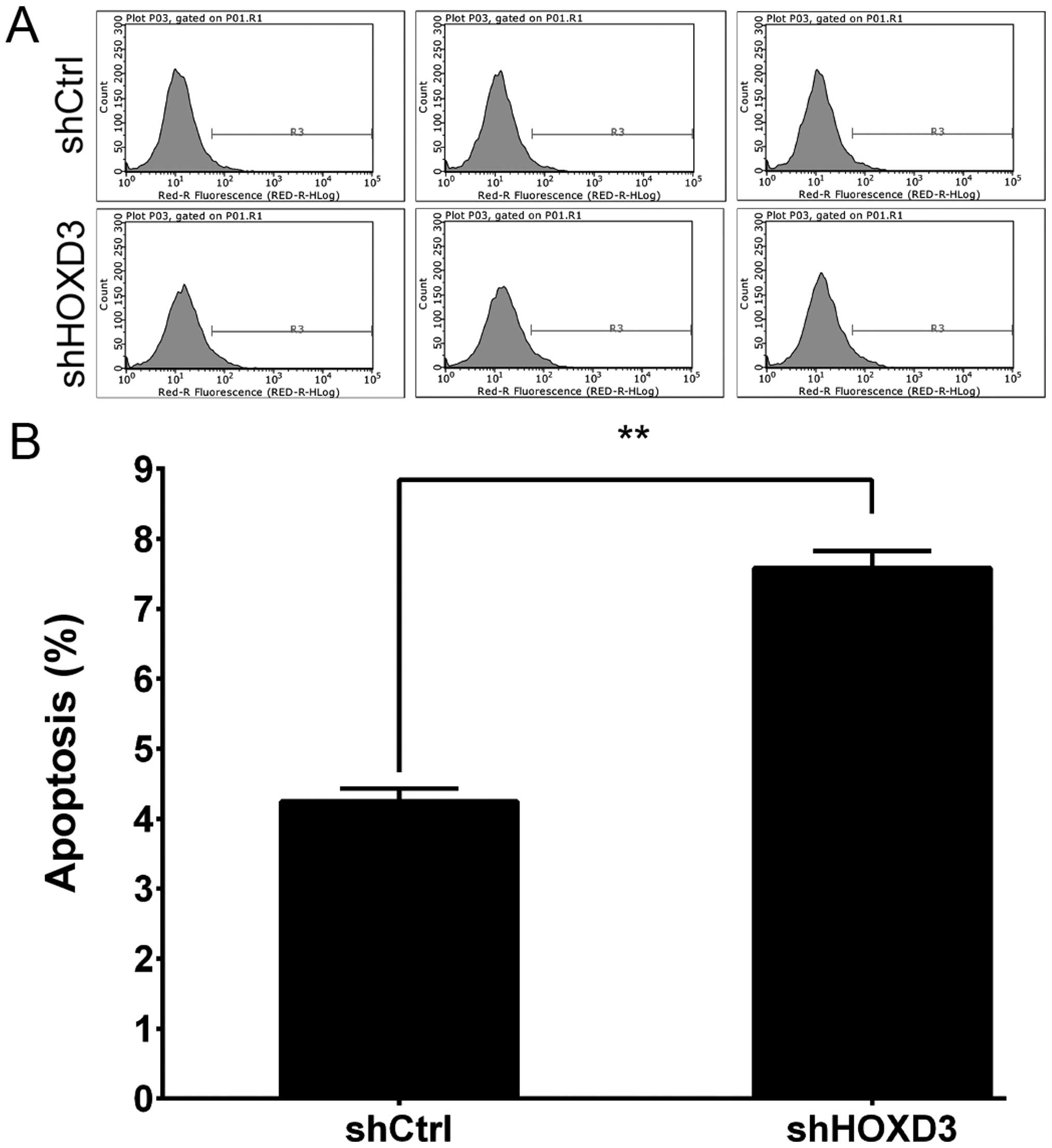
HOXD3 knockdown increases RKO cell
apoptosis
To test whether HOXD3 expression affects RKO
cell death, we knocked down HOXD3 and measured apoptosis.
Annexin V staining followed by FCM was used to determine cell
apoptosis (Fig. 5A). As shown in
Fig. 5B, apoptosis was
significantly increased in the shHOXD3 group compared to the shCtrl
group (shCtrl: 4.24±0.19% vs. shHOXD3: 7.58±0.25%, p<0.01).
These results indicate that HOXD3 expression is an important
determinant of apoptosis in RKO cells.
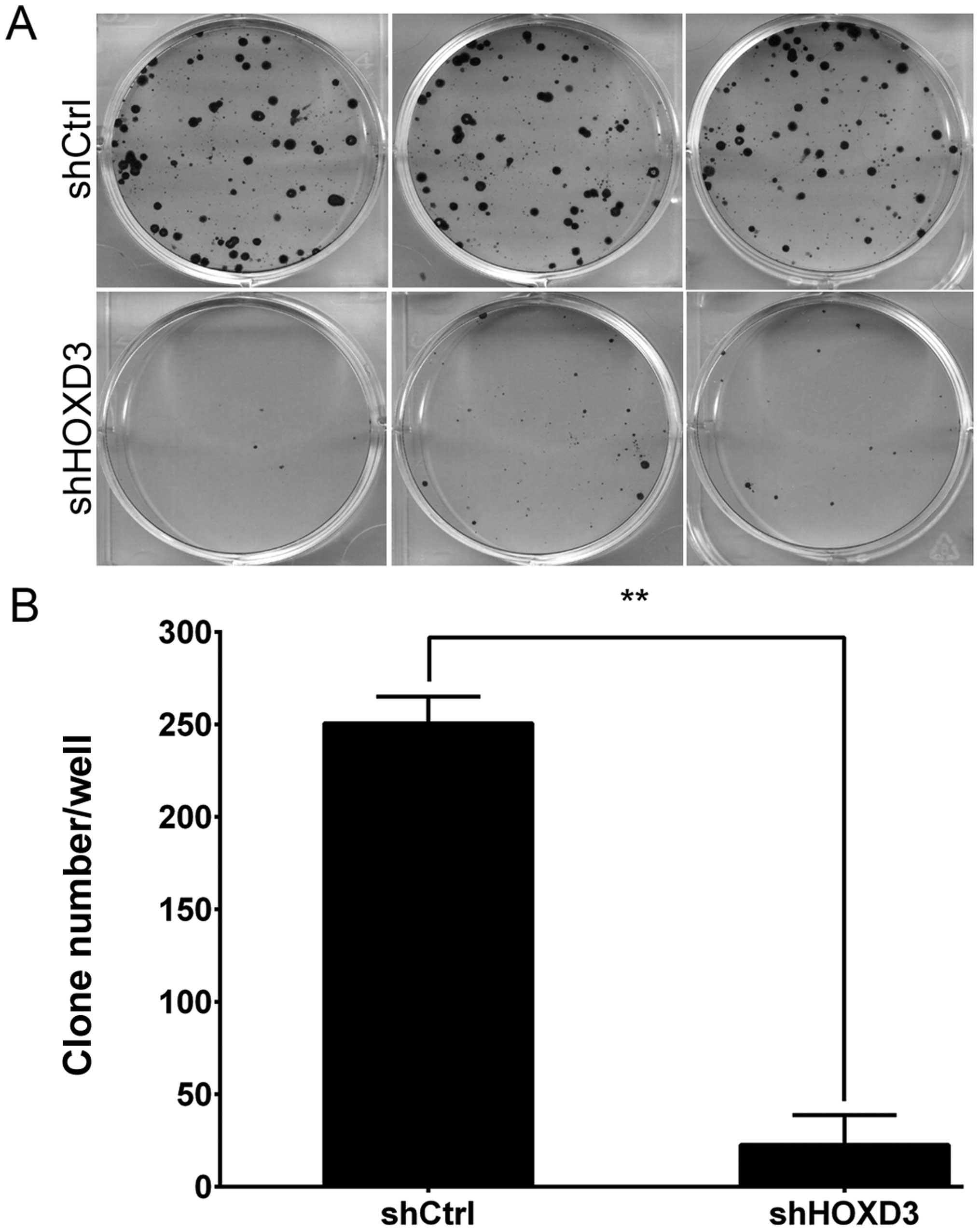
HOXD3 knockdown inhibits colony formation
in RKO cells
Finally, we used Giemsa staining to measure the
effects of HOXD3 knockdown on colony formation in RKO cells
(Fig. 6A). As shown in Fig. 6B, the cell number in a single colony
was significantly reduced in the shHOXD3 group compared to the
shCtrl group (shCtrl: 251±14 vs. shHOXD3: 23±16, p<0.01). This
results indicate that reduction of endogenous HOXD3
expression levels significantly inhibits colorectal carcinoma
growth.
Discussion
Colorectal cancer is the third most common global
cancer, and half of CRC patients die within 5 years of diagnosis
(1). Importantly, gene therapy is
being investigated as a potential cancer treatment method (29). Despite recent advances, however, the
prognosis of patients with CRC remains poor. In order to develop
new therapeutic treatments, it is particularly important to unravel
the underlying mechanisms of CRC development and progression.
To our knowledge, HOXD3 expression in CRC has
been largely unstudied. The present study is the first to measure
HOXD3 expression in five CRC cell lines and find high levels
of expression in RKO cells. Related to our findings, previous
studies have shown that abnormal HOXD3 expression is
associated with oncogenesis and tumor suppression (14,15,17).
One study showed that samples from patients with invasive breast
cancer had high HOXD3 expression levels, and those patients
had poor 5-year survival rates (30). Additionally, HOXD3
overexpression in lung cancer A549 cells led to increased
expression of the adhesion molecule E-cadherin (17). Another study determined that
HOXD3 overexpression altered the adhesive properties of
erythroleukemia HEL cells (16).
In order to assess HOXD3 function in CRC cell
lines, we constructed the shHOXD3 lentiviral vector, which
efficiently silenced HOXD3 in the RKO cell line. Compared to
shCtrl cells, the shHOXD3-infected cells had decreased
proliferation and significantly decreased proportions of cells in
the G1 and S cell cycle phases. Significant increases in
the G2 phase population were also detected by FCM in the
shHOXD3-infected cells. Additionally, HOXD3 knockdown
increased apoptosis and decreased colony formation in the RKO
cells. Taken together, these results suggest that HOXD3
promotes RKO cell growth. Further study is ongoing to validate the
anti-apoptotic role of HOXD3 in other CRC cell lines.
In conclusion, we demonstrated in RKO cells that
downregulation of HOXD3 expression by RNA interference
inhibited cell proliferation and induced cell apoptosis. Therefore,
in CRC cases where HOXD3 is overexpressed, HOXD3
knockdown by lentivirus-siRNA may be a valuable candidate
treatment.
Acknowledgments
This study was supported by the Anhui Natural
Science Research Project (no. KJ2014A147).
References
|
1
|
Cunningham D, Atkin W, Lenz HJ, Lynch HT,
Minsky B, Nordlinger B and Starling N: Colorectal cancer. Lancet.
375:1030–1047. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lima JP, de Souza FH, de Andrade DA,
Carvalheira JB and dos Santos LV: Independent radiologic review in
metastatic colorectal cancer: Systematic review and meta-analysis.
Radiology. 263:86–95. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jemal A, Murray T, Samuels A, Ghafoor A,
Ward E and Thun MJ: Cancer statistics, 2003. CA Cancer J Clin.
53:5–26. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Qian WF, Guan WX, Gao Y, Tan JF, Qiao ZM,
Huang H and Xia CL: Inhibition of STAT3 by RNA interference
suppresses angiogenesis in colorectal carcinoma. Braz J Med Biol
Res. 44:1222–1230. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Taniguchi Y, Sato M, Tanaka O, Sekiguchi
M, Inoko H and Kimura M: HOXD3 regulates expression of JAGGED1, a
ligand for Notch receptors. Nucleic Acids Res. (Suppl 1): 43–44.
2001. View Article : Google Scholar
|
|
6
|
Hutlet B, Theys N, Coste C, Ahn MT,
Doshishti-Agolli K, Lizen B and Gofflot F: Systematic expression
analysis of Hox genes at adulthood reveals novel patterns in the
central nervous system. Brain Struct Funct. 221:1223–1243. 2016.
View Article : Google Scholar
|
|
7
|
Duboule D and Dollé P: The structural and
functional organization of the murine HOX gene family resembles
that of Drosophila homeotic genes. EMBO J. 8:1497–1505.
1989.PubMed/NCBI
|
|
8
|
Taniguchi Y, Tanaka O, Sekiguchi M,
Takekoshi S, Tsukamoto H, Kimura M, Imai K and Inoko H: Enforced
expression of the transcription factor HOXD3 under the control of
the Wnt1 regulatory element modulates cell adhesion properties in
the developing mouse neural tube. J Anat. 219:589–600. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
McGinnis W and Krumlauf R: Homeobox genes
and axial patterning. Cell. 68:283–302. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Deschamps J: Ancestral and recently
recruited global control of the Hox genes in development. Curr Opin
Genet Dev. 17:422–427. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Toshner M, Dunmore BJ, McKinney EF,
Southwood M, Caruso P, Upton PD, Waters JP, Ormiston ML, Skepper
JN, Nash G, et al: Transcript analysis reveals a specific HOX
signature associated with positional identity of human endothelial
cells. PLoS One. 9:e913342014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dollé P, Dierich A, LeMeur M, Schimmang T,
Schuhbaur B, Chambon P and Duboule D: Disruption of the Hoxd-13
gene induces localized heterochrony leading to mice with neotenic
limbs. Cell. 75:431–441. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kondo T, Dollé P, Zákány J and Duboule D:
Function of posterior HoxD genes in the morphogenesis of the anal
sphincter. Development. 122:2651–2659. 1996.PubMed/NCBI
|
|
14
|
Condie BG and Capecchi MR: Mice homozygous
for a targeted disruption of Hoxd-3 (Hox-4.1) exhibit anterior
transformations of the first and second cervical vertebrae, the
atlas and the axis. Development. 119:579–595. 1993.PubMed/NCBI
|
|
15
|
Manley NR and Capecchi MR: Hox group 3
paralogs regulate the development and migration of the thymus,
thyroid, and parathyroid glands. Dev Biol. 195:1–15. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Taniguchi Y, Komatsu N and Moriuchi T:
Overexpression of the HOX4A (HOXD3) homeobox gene in human
erythroleukemia HEL cells results in altered adhesive properties.
Blood. 85:2786–2794. 1995.PubMed/NCBI
|
|
17
|
Hamada Ji, Omatsu T, Okada F, Furuuchi K,
Okubo Y, Takahashi Y, Tada M, Miyazaki YJ, Taniguchi Y, Shirato H,
et al: Overexpression of homeobox gene HOXD3 induces coordinate
expression of metastasis-related genes in human lung cancer cells.
Int J Cancer. 93:516–525. 2001. View
Article : Google Scholar
|
|
18
|
Miyazaki YJ, Hamada J, Tada M, Furuuchi K,
Takahashi Y, Kondo S, Katoh H and Moriuchi T: HOXD3 enhances
motility and invasiveness through the TGF-beta-dependent and
-independent pathways in A549 cells. Oncogene. 21:798–808. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ohta H, Hamada J, Tada M, Aoyama T,
Furuuchi K, Takahashi Y, Totsuka Y and Moriuchi T:
HOXD3-overexpression increases integrin alpha v beta 3 expression
and deprives E-cadherin while it enhances cell motility in A549
cells. Clin Exp Metastasis. 23:381–390. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Okubo Y: Overexpression of the human
HOXD3-antisense in melanoma cells results in decreased invasive
activity. Hokkaido Igaku Zasshi. 76:239–250. 2001.In Japanese.
PubMed/NCBI
|
|
21
|
Boudreau N, Andrews C, Srebrow A, Ravanpay
A and Cheresh DA: Induction of the angiogenic phenotype by Hox D3.
J Cell Biol. 139:257–264. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hansen SL, Myers CA, Charboneau A, Young
DM and Boudreau N: HoxD3 accelerates wound healing in diabetic
mice. Am J Pathol. 163:2421–2431. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hansen SL, Young DM and Boudreau NJ: HoxD3
expression and collagen synthesis in diabetic fibroblasts. Wound
Repair Regen. 11:474–480. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhong J, Eliceiri B, Stupack D, Penta K,
Sakamoto G, Quertermous T, Coleman M, Boudreau N and Varner JA:
Neovascularization of ischemic tissues by gene delivery of the
extracellular matrix protein Del-1. J Clin Invest. 112:30–41. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen Y, Xu B, Arderiu G, Hashimoto T,
Young WL, Boudreau N and Yang GY: Retroviral delivery of homeobox
D3 gene induces cerebral angiogenesis in mice. J Cereb Blood Flow
Metab. 24:1280–1287. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lois C, Hong EJ, Pease S, Brown EJ and
Baltimore D: Germline transmission and tissue-specific expression
of transgenes delivered by lentiviral vectors. Science.
295:868–872. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Milner AE, Levens JM and Gregory CD: Flow
cytometric methods of analyzing apoptotic cells. Methods Mol Biol.
80:347–354. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Koopman G, Reutelingsperger CP, Kuijten
GA, Keehnen RM, Pals ST and van Oers MH: Annexin V for flow
cytometric detection of phosphatidylserine expression on B cells
undergoing apoptosis. Blood. 84:1415–1420. 1994.PubMed/NCBI
|
|
29
|
Guinn BA and Mulherkar R: International
progress in cancer gene therapy. Cancer Gene Ther. 15:765–775.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shaoqiang C, Yue Z, Yang L, Hong Z, Lina
Z, Da P and Qingyuan Z: Expression of HOXD3 correlates with shorter
survival in patients with invasive breast cancer. Clin Exp
Metastasis. 30:155–163. 2013. View Article : Google Scholar
|