Introduction
Gastric cancer (GC) ranks the third cause of cancer
mortality and the fifth most common cancer worldwide (1), while most patients with GC are
diagnosed at the advanced stage which means a relatively poor
prognosis of overall survival (2).
As is well recognized, cancer results from accumulation of multiple
molecular alterations in the same cells or their descendants
(3). Hence, the identification of
genes with oncogenic potential or tumor-suppressing activity may be
of great use for diagnosis and treatment.
Previously, Zhao et al used an
oligonucleotide microarray containing 38,500 genes in 11 patients
with GC to distinguish malignant lesions from premalignant and
normal ones, and type XI collagen α1 (COL11A1) gene
expression was found upregulated in malignant tissue compared to
premalignant tissue (4). As a
member of minor fibrillar collagens, COL11A1, encoded by
COL11A1 gene located on chromosome 1p21, can be produced by
cartilage and a variety of non-cartilaginous tissues, such as bone,
vitreous, skin, and heart (5).
Alterations of COL11A1, including at least four mRNA
variants, can lead to several diseases such as Stickler syndrome
type 2 (6), Marshall syndrome
(7,8) and lumbar disc disease (9).
Besides, COL11A1 has also been reported to be
upregulated in various cancers, including colorectal, pancreatic,
and ovarian cancer. Fischer et al first reported that
COL11A1 mRNA expression was significantly increased in
colorectal cancer tissue compared with normal colonic tissue
(10), and studies in pancreatic
cancer (11), non-small cell lung
cancer (12) and breast cancer
(13) have also demonstrated
increased levels of COL11A1 in tumor tissue compared with
normal tissue. Recent studies have implicated COL11A1 in
cancer cell growth and tumorigenicity. For example, COL11A1
knockdown in head and neck squamous cell cancer gave rise to a
reduction in cell growth and invasion in vitro (14), and COL11A1 knockdown in
ovarian cancer cells led to a decrease in cell proliferation and
invasion in vitro and the metastasis of an ovarian tumor
xenograft (15). Thus,
COL11A1 may represent a potential therapeutic target in need
of further investigation.
Considering the limited studies regarding the
relationship between COL11A1 and GC, we evaluated the
COL11A1 mRNA expression in GC tissue, investigated the
possible role of COL11A1 in GC proliferation, cell cycle,
apoptosis and invasion and explored the potential molecular
mechanisms of COL11A1 in GC cells in the present study.
Materials and methods
Ethics statement and clinical tissue
samples
This study was approved by the Clinical Research
Ethics Committee of Sir Run Run Shaw Hospital of Zhejiang
University (Hangzhou, Zhejiang, China) (permit number:
20110225-3200). Giving their informed consents to use their gastric
tissue and publish their case details, all 57 consecutive patients
who underwent surgery for GC in Sir Run Run Shaw Hospital were
enrolled in this study. The histologic classification of gastric
carcinoma is based on the 2010 WHO classification system and the
clinicopathologic parameters including age, gender, tumor size,
depth of invasion and lymph node positivity were obtained. The
matched tumor tissues and adjacent non-tumor tissue samples were
frozen immediately in liquid nitrogen and were stored at −80°C
until RNA extraction.
Cell culture
A total of four poorly differentiated (AGS, MKN-45,
BGC-823 and MGC-803), one moderately differentiated (SGC-7901), one
undifferentiated (HGC-27) and one well differentiated (MKN-28)
gastric cancer cell line were included for the purposes of this
study. These cell lines were obtained from the American Type
Culture Collection (Manassas, VA, USA) and Type Culture Collection
of China Academic Science (Shanghai, China). One normal
immortalized gastric epithelial cell line (GES-1) was obtained from
the Beijing Institute for Cancer Research (Beijing, China). The
cells were cultured in RPMI-1640 medium (Gibco, Carlsbad, CA, USA)
supplemented with 10% fetal bovine serum (FBS) (Sijiqing, Huzhou,
Zhejiang, China) and were incubated at 37°C in 5%
CO2.
Reverse transcription quantitative PCR
(RT-qPCR) analysis
Total RNA was extracted using TRIzol reagent (CW
Biotech, Beijing, China), and the reverse transcription reaction
was performed using 1 µg RNA with a reverse transcription kit
(Takara, Otsu, Japan). RT-qPCR reactions were performed using a
SYBR green PCR kit (Takara) in a LightCycler® 480 II
Real-Time PCR System according to the manufacturer's instructions
(Roche Diagnostics, Basel, Switzerland). U6 and
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were used as
endogenous controls for tissues and cells, respectively. The
RT-qPCR conditions are as follows: one cycle of 95°C for 30 sec, 40
cycles of 95°C for 15 sec and 60°C for 60 sec, melting curve
analysis. The mRNA expression levels were determined using the
2−∆Ct method for tissues and the 2−∆∆Ct
method for cells. The forward primer sequence is
5′-AGTGGCATCGGGTAGCAATCA-3′ (located in exon 3–4, nt 806–826) and
the reverse one is 5′-TGTCCCCCTCAAAAACTTCTTCAT-3′ (located in exon
4–5, nt 953–976). The other primer sequences for the RT-qPCR assays
are listed in Table I. All
experiments were performed in triplicate.
 | Table I.The forward and reverse primer
sequences used in experimental procedures. |
Table I.
The forward and reverse primer
sequences used in experimental procedures.
| Gene name | Primer
sequences |
|---|
| XIAP | F:
GACAGTATGCAAGATGAGTCAAGTCA |
|
| R:
GCAAAGCTTCTCCTCTTGCAG |
| NEFL | F:
AGCTGGAGGACAAGCAGAAC |
|
| R:
TGCCATTTCACTCTTTGTGG |
| RGS2 | F:
GTTGGGTAGTGAATCAGGAAGC |
|
| R:
GACCACCTATTCCCTTCTTGC |
| ITGB8 | F:
GGCCAAGGTGAAGACAATAGA |
|
| R:
ATCCTCTTGAACACACCATCC |
| WNT5A | F:
ATCAATTCCGACATCGAAGG |
|
| R:
CGTTCACCACCCCTGCT |
| CDK6 | F:
GTGCCCTGTCTCACCCATAC |
|
| R:
GACCCATAAGCCACCAAGG |
| TIAM1 | F:
CAGGTGTTTGGAGAGGGAAC |
|
| R:
AATGTCGCAGTCAGGGTTG |
| U6 | F:
CTCGCTTCGGCAGCACA |
|
| R:
AACGCTTCACGAATTTGCGT |
| GAPDH | F:
GAAGGTGAAGGTCGGAGT |
|
| R:
GAAGATGGTGATGGGATTTC |
Western blotting
The cells were washed in cold PBS and lysed in RIPA
lysis buffer (Beyotime, Jiangsu, China) on ice for 30 min. The cell
lysates were centrifuged at 13,000 × g for 15 min at 4°C, and the
total protein concentration was analyzed with the BCA Protein Assay
kit (Beyotime). Equal amounts of protein samples were separated on
sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels
(SDS-PAGE). The proteins were then transferred to polyvinylidene
difluoride membranes (Millipore, Bedford, MA, USA). The membranes
were blocked with 5% nonfat milk and incubated with primary
antibodies at 4°C overnight. The proteins were detected using
chemiluminescence with a Las-4000 Imaging System (Fujifilm, Tokyo,
Japan). The following primary antibodies were used: COL11A1
(1:1000; Abcam, Cambridge, UK), pro-caspase-3 (1:1000; Cell
Signaling Technology Inc., Danvers, MA, USA), cleaved caspase-3
(1:1000; Cell Signaling Technology Inc.), cyclin D1
(1:1000; Cell Signaling Technology Inc.), p21 (1:1000; Cell
Signaling Technology Inc.), p27 (1:1000; Cell Signaling Technology
Inc.), cyclin-dependent kinase 2 (CDK2) (1:500; Santa Cruz
Biotechnology Inc., Santa Cruz, CA, USA), cyclin-dependent kinase 4
(CDK4) (1:500; Santa Cruz Biotechnology Inc.) and β-tubulin
(1:1000; CW Biotech). All experiments were performed in
triplicate.
Plasmid transfection of stable cell
lines
The COL11A1 short hairpin RNA (shRNA)
plasmids (HSH002603-2-mH1, GeneCopoeia, Rockville, MD, USA) and
COL11A1 vector plasmids (CSHCTR001-mH1, GeneCopoeia) were
purchased and transfected into HGC-27 cells using Lipofectamine
2000 (Invitrogen, Carlsbad, CA, USA) to establish stable cell
lines. After 48 h, the transfectants were selected with 3 µg/ml
puromycin (Amresco, Cleveland, OH, USA) for 2 weeks. A positive
stably transfected clone was isolated and allowed to grow.
Decreased COL11A1 expression was confirmed by RT-qPCR and
western blotting. All experiments were performed in triplicate.
Cell proliferation assay
A total of 3×103 stably transfected cells
in 100 µl medium were seeded into 96-well plates and incubated for
96 h. Then, 20 µl of
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
(MTS) (Promega, Madison, WI, USA) reagent was added to each well
and incubated for 2 h. The absorbance values were measured by a
multi-well plate reader (Molecular Devices, Sunnyvale, CA, USA) at
490 nm. All experiments were performed in triplicate.
Colony formation assay
A total of 6×102 cells were plated into
six-well plates in triplicate. The plates were incubated at 37°C in
5% CO2 for 10 days, and then the colonies were fixed
with methanol and stained with crystal violet before counting.
Values are presented as the mean percentage ± SD from three
individual experiments, and the number for the COL11A1
vector cells was set to 100%. All experiments were performed in
triplicate.
Cell migration and invasion
assays
Cell migration was assessed using Transwell
migration assays (Corning, Inc., Corning, NY, USA). Briefly,
5×104 cells were plated in the upper chamber in 200 µl
medium containing 1% FBS. The lower chamber contained 600 µl medium
containing 10% FBS. After 13 h of incubation, the non-migratory
cells in the upper chamber were carefully removed with a cotton
swab. The migrated cells were fixed with methanol and stained with
DAPI. The cell numbers were counted in five random fields (×400
magnification) using a fluorescence microscope. All experiments
were performed in triplicate.
Cell invasion was performed in Transwell chambers
(Corning, Inc.) coated with BD Matrigel. For this assay,
1×105 cells were plated in the upper chamber in 200 µl
of medium containing 1% FBS. The lower chamber was filled with 600
µl of culture medium with 10% FBS. After 24 h, the membranes were
fixed with methanol and stained with gentian violet. The cell
numbers were counted by microscope in five random fields (×200
magnification). All experiments were performed in triplicate.
Cell apoptosis and cell cycle
analysis
For cell apoptosis analysis, 2×105 stable
transfected cells were washed in cold PBS and suspended in 1X
Annexin V Binding Buffer. Then, 5 µl FITC Annexin V (Becton
Dickinson, San Jose, CA, USA) and 5 µl PI (Becton Dickinson)
solutions were added. After incubation for 15 min, the stained
cells were analyzed by flow cytometry on a FACScan analyzer (Becton
Dickinson). All experiments were performed in triplicate.
For cell cycle analysis, 2×105 stably
transfected cells were harvested and washed in PBS. Cellular DNA
was stained in the dark by a Cell Cycle Staining kit (Multisciences
Biotech, Hangzhou, Zhejiang, China) for 30 min at room temperature.
The cells were then analyzed by flow cytometry, and the cell cycle
distribution was determined using ModFit LT software (Verity
Software House, Topsham, ME, USA). All experiments were performed
in triplicate.
cDNA microarray analysis
Total RNA was extracted from stably transfected
HGC-27 cells with COL11A1 shRNA and COL11A1 vector.
Total RNA was amplified, labeled and purified to obtain
biotin-labeled cDNA. The labeled cDNA were hybridized to probes on
Affymetrix U133 plus 2.0 arrays (Shanghai Biotechnology
Corporation, Shanghai, China). The microarray data were analyzed
using Gene Spring Software 11.0. We selected fold change
(shRNA/vector) >3 or <0.333 as the threshold for upregulation
or downregulation. The potential target genes were verified with
RT-qPCR.
Statistical analysis
Wilcoxon matched pairs test was performed to compare
paired data, Mann-Whitney U test was used to analyze and compare
the medians of continuous variables, and Student's t-test was
performed to compare two independent data. A value of p<0.05 was
considered statistically significant. All data were analyzed using
IBM SPSS Statistics 20.0.
Results
We used RT-qPCR analysis to measure the
COL11A1 mRNA expression and found that the COL11A1
mRNA expression level was significantly overexpressed in 57 GC
tissues compared to matched adjacent non-tumor gastric tissue
(p<0.0001, Fig. 1).
In 2/57 patients distant metastases occurred (liver
and peritoneum, respectively), and 56/57 patients were diagnosed
with gastric cancer of the adenocarcinoma subtype, while the
remaining patient was diagnosed with an undifferentiated gastric
carcinoma. The relationship between clinicopathological features
and COL11A1 expression in GC is shown in Table II. COL11A1 mRNA expression
was significantly positively related to age, tumor invasion depth,
tumor size and lymph node positivity (p<0.05, p<0.01,
p<0.05 and p<0.05, respectively), while there were no
significant associations between COL11A1 expression and
patient gender (p>0.05) and degree of differentiation
(p>0.05) in GC.
 | Table II.Clinicopathological characteristics
and COL11A1 mRNA expression in gastric cancer samples. |
Table II.
Clinicopathological characteristics
and COL11A1 mRNA expression in gastric cancer samples.
| Variables | Total | COL11A1
expression (2−∆Ct) (median) | P-value |
|---|
| Age (years) |
|
|
|
|
≤63 | 27 (47%) | 0.00113 | 0.031a |
|
≥64 | 30 (53%) | 0.00548 |
|
| Gender |
|
|
|
|
Male | 39 (68%) | 0.00193 | 0.352 |
|
Female | 18 (32%) | 0.00356 |
|
| Invasion depth |
|
|
|
|
T2–4 | 44 (77%) | 0.00403 | 0.0004b |
|
T1 | 13 (23%) | 0.000409 |
|
| Tumor size
(cm) |
|
|
|
|
<5 | 27 (47%) | 0.000878 | 0.024a |
| ≥5 | 30 (53%) | 0.00630 |
|
| Degree of
differentiation |
|
|
|
|
Poorly | 37 (65%) | 0.00159 | 0.261 |
|
Moderately/Well | 20 (35%) | 0.00630 |
|
| Lymph node
positivity |
|
|
|
|
Yes | 36 (63%) | 0.00455 | 0.019a |
| No | 21 (37%) | 0.000739 |
|
To determine which cell lines to use for further
study, we compared COL11A1 mRNA and protein expression
levels in seven GC cell lines (SGC-7901, MGC-803, BGC-823, HGC-27,
AGS, MKN-28, and MKN-45) with expression levels in normal
immortalized epithelial cells (GES-1) by RT-qPCR and western
blotting. The results showed that the relative COL11A1 mRNA
expression was higher in HGC-27 cells but lower in the other six GC
cells (Fig. 2A), while similar
results were observed by western blotting (Fig. 2B). Hence, HGC-27 was the only chosen
GC cell line for further analysis.
 | Figure 2.COL11A1 expression in seven GC
cell lines compared with normal immortalized epithelial cells
(GES-1). (A) The COL11A1 mRNA expression was higher in
HGC-27 cells but lower in the other six GC cells (SGC-7901,
MGC-803, BGC-823, AGS, MKN-28, and MKN-45) compared with GES-1 by
RT-qPCR analysis. (B) COL11A1 protein expression levels were
higher in HGC-27 cells but lower in the other six GC cells
(SGC-7901, MGC-803, BGC-823, AGS, MKN-28, and MKN-45) compared with
GES-1 by western blotting. |
To validate whether COL11A1 contributed to GC
proliferation, migration and invasion, we transfected HGC-27 cells
with a plasmid encoding COL11A1-silencing shRNA
(COL11A1 shRNA) or COL11A1 vector shRNA
(COL11A1 vector). The decreased COL11A1 mRNA and
protein expression levels were confirmed by RT-qPCR and western
blotting (Fig. 3A).
Then, we examined whether COL11A1 contributed
to GC cell proliferation. The results of MTS assays showed
significant cell growth inhibition in COL11A1 shRNA HGC-27
cells compared with COL11A1 vector cells (p<0.01 at 48,
72 and 96 h) (Fig. 3B).
Consistently, the number of surviving colonies formed on the plates
in COL11A1 shRNA HGC-27 cells was also significantly reduced
compared with COL11A1 vector cells (p<0.01) (Fig. 3C). Thus, COL11A1 contributed
to cell proliferation in GC cells in vitro.
Next, Transwell migration and invasion assays were
performed, and the results demonstrated that COL11A1
knockdown in HGC-27 cells significantly suppressed migration
(p<0.01) and invasion (p<0.01), respectively (Fig. 3D and E).
Cumulatively, these results indicated that
COL11A1 played a role in cell proliferation, migration and
invasion of HGC-27 GC cells in vitro.
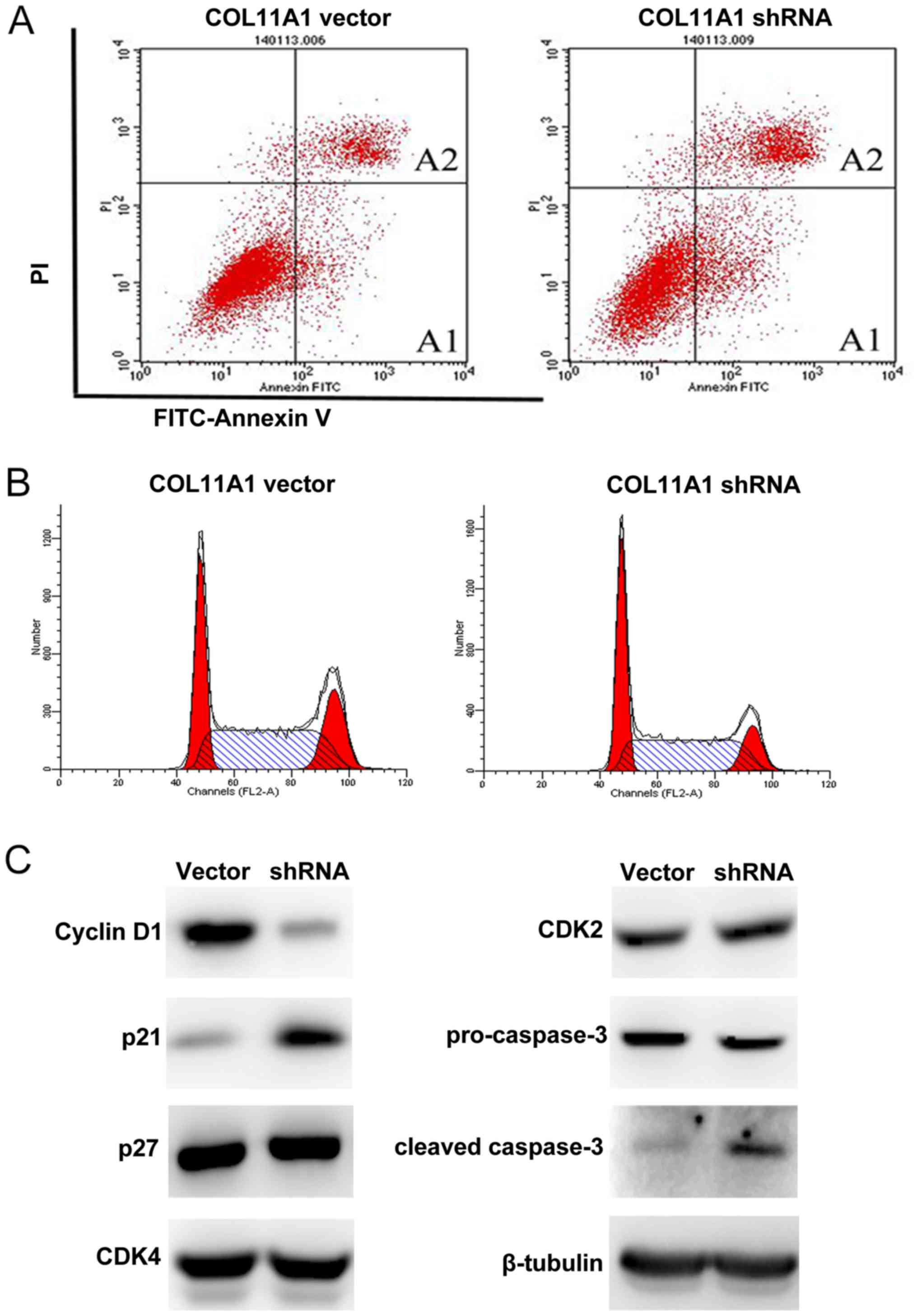
To explore the underlying mechanisms of the growth
inhibition by COL11A1 knockdown, we assessed cell apoptosis
and cell cycle by flow cytometry. COL11A1 suppression
significantly induced apoptosis compared with COL11A1 vector
(Fig. 4A and Table III), and a significant
accumulation of cells in G1 phase was observed in
COL11A1 knockdown cells compared to that in COL11A1
vector cells (Fig. 4B and Table III). Then, we further evaluated
expression levels of several cell cycle and apoptosis-related
proteins, and we found that COL11A1 suppression led to
upregulation of the cell cycle inhibitor p21 but not p27 and
reduced cyclin D1 but not CDK2 and CDK4. We also found
activation of the apoptotic protein caspase-3 (Fig. 4C).
 | Table III.The influences of cell apoptosis and
cell cycle by COL11A1 knockdown. |
Table III.
The influences of cell apoptosis and
cell cycle by COL11A1 knockdown.
| Group | Early apoptotic
cells A1 (%) | Late apoptotic
cells A2 (%) | G1 Phase
(%) | S Phase (%) | G2/M
Phase (%) |
|---|
| COL11A1 vector | 7.42±0.89 | 11.19±0.25 | 28.57±0.24 | 50.50±0.48 | 20.93±0.25 |
| COL11A1 shRNA |
14.94±0.27a |
20.62±0.21a |
36.34±0.38a | 50.38±0.28 |
13.28±0.38a |
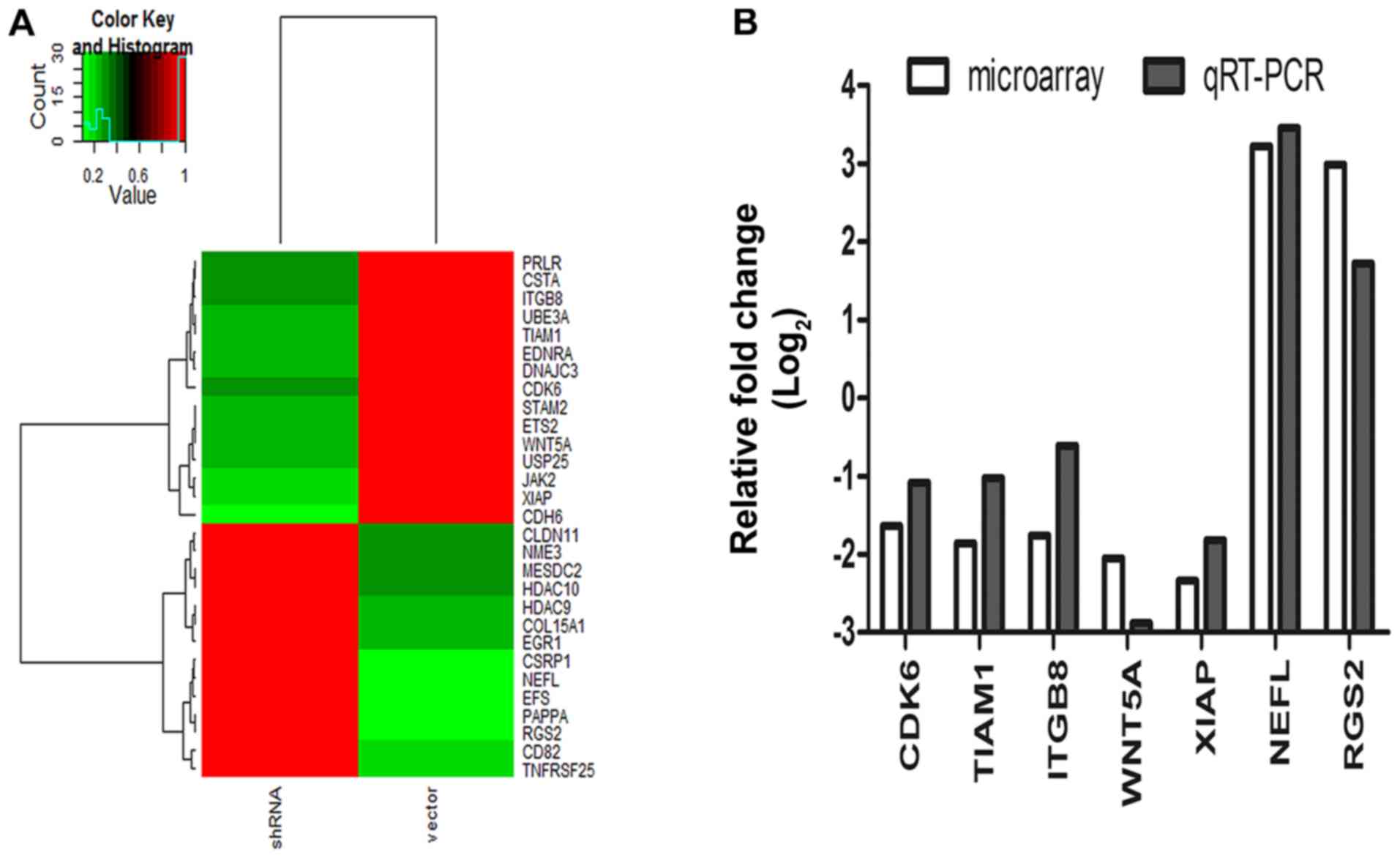
cDNA microarray in HGC-27 cells with and without
COL11A1 knockdown was performed to identify potential
downstream genes of COL11A1 in GC, and the results indicated
that COL11A1 suppression altered the expression of multiple
genes involved in cell proliferation and invasion (Fig. 5A). The results of RT-qPCR further
confirmed that CDK6, TIAM1, XIAP, ITGB8 and WNT5A
were downregulated, and RGS2 and NEFL were
upregulated by COL11A1 suppression in HGC-27 cells (Fig. 5B). These results suggested that
COL11A1 may play a role in tumor development and progression
through regulation of these genes.
Discussion
The gradual accumulation of genetic alternations
contributes to the development and progression of cancers. In the
present study, we observed that overexpression of COL11A1
mRNA was related to tumor age, tumor size, depth of invasion and
lymph node positivity, and that COL11A1 could regulate cell
proliferation, migration and invasion through several potential
downstream genes.
Several lines of evidence indicate that
COL11A1 expression is upregulated in various cancers such as
ovarian (15), colorectal (10), breast (13), pancreatic (16) and head and neck squamous cell
(14) cancers, suggesting an
oncogenic role of COL11A1 in carcinogenesis. However, the
role of COL11A1 in GC has not been elucidated. This study
demonstrated that COL11A1 mRNA was overexpressed in 57 GC
tissues compared with matched non-tumor tissues by RT-qPCR
analysis, which is consistent with previous studies (4). In addition, we analyzed the
relationship between COL11A1 mRNA expression and
clinicopathological characteristics. Consistent with a previous
report in GC by Affymetrix analysis (17), COL11A1 mRNA expression in the
advanced GC was significantly higher than that in the early GC.
Besides, high COL11A1 expression was positively related to
tumor age, tumor size and lymph node positivity, which indicates
COL11A1 may be involved in GC growth and invasion. These
results supports that COL11A1 may play a role in GC
proliferation and invasion.
To study the biological effects of COL11A1 in
GC, we examined COL11A1 expression in seven GC cell lines
and one GES-1 cell line and found that HGC-27 was the only cell
line showing COL11A1 upregulation compared to GES-1. The
reason why COL11A1 is overexpressed in only one cancer cell
line, out of seven, compared to normal cells needs further
explanation. From our perspective, one explanation may be that
HGC-27, a kind of undifferentiated and relatively less attached
cell, is much more aggressive than the other six cells. Finally, we
chose HGC-27 for the next study.
Based on depletion experiments in vitro, we
found that silencing of COL11A1 significantly decreased the
proliferation, migration and invasion of HGC-27 cells, which is
consistent with two previous reports in ovarian cancer (15) and head and neck squamous cell cancer
(14). Furthermore, we studied the
cell cycle and apoptosis using flow cytometry, and the results
showed that COL11A1 suppression significantly induced cell
cycle arrest at G1 phase and promoted cell apoptosis.
G1/S phase transition is a major checkpoint for cell
cycle progression. Cyclin D1, forming functional kinase
complexes with CDK4 or CDK6, is a periodic regulatory protein that
governs cell cycle transit from G1 phase into S phase
and is abnormally expressed in many human cancers (18,19).
There are also inhibitory proteins preventing the cell cycle. Among
these inhibitors, p21, a potent cyclin-dependent kinase inhibitor
binds and inhibits the activity of cyclin D1-CDK4/6
complexes controlling the transition from G1 to S phase
(20). In the present study,
western blotting demonstrated that suppression of COL11A1
decreased cyclin D1 expression and increased p21
expression. However, there was no significant effect on CDK2, CDK4
or p27. These results suggest that G1/S cell cycle
arrest induced by COL11A1 suppression is mediated through
the p21 and cyclin D1 pathway. In addition, caspase-3 is
a well-recognized indicator of cellular apoptosis (21) and is activated in apoptotic cells by
both the extrinsic and intrinsic pathways (22). Our data indicated that
COL11A1 suppression increased cleaved caspase-3 levels in
HGC-27 cells, and thus promoted apoptosis of HGC-27 cells. Further
studies are required to determine whether the extrinsic or
intrinsic pathway is involved. Despite several attempts, we failed
to establish a successful nude mouse model to investigate the role
of COL11A1 in vivo.
The molecular mechanisms of COL11A1 action in
cancers remain unclear, and the only pathway studied is the
COL11A1-TGF-β1-MMP3 axis through which COL11A1
promotes ovarian cancer aggressiveness. Therefore, we studied the
molecular mechanisms of COL11A1 in GC by cDNA microarray.
Numerous genes were altered in cells with COL11A1 knockdown
compared to cells with COL11A1 vector, and the
representative potential target genes have been previously reported
to participate in cell growth, migration and invasion. As a member
of the inhibitor of apoptosis protein gene family, XIAP can
inhibit caspases and suppress apoptosis, and a previous study
showed that downregulation of XIAP induced apoptosis was
related to activation of caspase-3 in GC (23). CDK6, important for the
G1 phase progression and G1/S transition, was
upregulated in many cancers (24).
Another COL11A1 potential target gene, RGS2, was also
involved in cell growth in breast cancer (25) and prostate cancer (26). In addition to the regulation of cell
proliferation pathways, COL11A1 downregulation decreased
TIAM1 which was involved in cell invasion in retinoblastoma
(27) and gastric cancer (28). Furthermore, NEFL, a putative
tumor suppressor gene, can inhibit cell proliferation and invasion
in head and neck squamous cell carcinoma (29) and neuroblastoma (30). We further validated that CDK6,
TIAM1, XIAP, ITGB8 and WNT5A were downregulated, while
RGS2 and NEFL were upregulated in HGC-27 cells with
COL11A1 suppression using RT-qPCR analysis. Thus, the
identification of potential target genes of COL11A1
supported the hypothesis that COL11A1 may modulate potential
downstream genes to regulate cell proliferation and invasion in
GC.
In conclusion, our study indicates that
COL11A1 may play an oncogenic role in the proliferation,
migration and invasion in gastric cancer and may be a promising
therapeutic target in cancer treatment. Further study is needed to
clarify the potential specific molecular mechanisms of
COL11A1 in gastric cancer.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (81101836, 81372623), Zhejiang Province
Key Science and Technology Innovation Team (2013TD13), and Zhejiang
Province medicine health platform and study plan (YH52013004).
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rohatgi PR, Yao JC, Hess K, Schnirer I,
Rashid A, Mansfield PF, Pisters PW and Ajani JA: Outcome of gastric
cancer patients after successful gastrectomy: Influence of the type
of recurrence and histology on survival. Cancer. 107:2576–2580.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vogelstein B and Kinzler KW: The multistep
nature of cancer. Trends Genet. 9:138–141. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhao Y, Zhou T, Li A, Yao H, He F, Wang L
and Si J: A potential role of collagens expression in
distinguishing between premalignant and malignant lesions in
stomach. Anat Rec (Hoboken). 292:692–700. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yoshioka H, Greenwel P, Inoguchi K, Truter
S, Inagaki Y, Ninomiya Y and Ramirez F: Structural and functional
analysis of the promoter of the human alpha 1(XI) collagen gene. J
Biol Chem. 270:418–424. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Richards AJ, Yates JR, Williams R, Payne
SJ, Pope FM, Scott JD and Snead MP: A family with Stickler syndrome
type 2 has a mutation in the COL11A1 gene resulting in the
substitution of glycine 97 by valine in alpha 1 (XI) collagen. Hum
Mol Genet. 5:1339–1343. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Khalifa O, Imtiaz F, Allam R, Al-Hassnan
Z, Al-Hemidan A, Al-Mane K, Abuharb G, Balobaid A, Sakati N, Hyland
J, et al: A recessive form of Marshall syndrome is caused by a
mutation in the COL11A1 gene. J Med Genet. 49:246–248. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Griffith AJ, Sprunger LK, Sirko-Osadsa DA,
Tiller GE, Meisler MH and Warman ML: Marshall syndrome associated
with a splicing defect at the COL11A1 locus. Am J Hum Genet.
62:816–823. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mio F, Chiba K, Hirose Y, Kawaguchi Y,
Mikami Y, Oya T, Mori M, Kamata M, Matsumoto M, Ozaki K, et al: A
functional polymorphism in COL11A1, which encodes the alpha 1 chain
of type XI collagen, is associated with susceptibility to lumbar
disc herniation. Am J Hum Genet. 81:1271–1277. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fischer H, Stenling R, Rubio C and
Lindblom A: Colorectal carcinogenesis is associated with stromal
expression of COL11A1 and COL5A2. Carcinogenesis. 22:875–878. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Badea L, Herlea V, Dima SO, Dumitrascu T
and Popescu I: Combined gene expression analysis of whole-tissue
and microdissected pancreatic ductal adenocarcinoma identifies
genes specifically overexpressed in tumor epithelia.
Hepatogastroenterology. 55:2016–2027. 2008.PubMed/NCBI
|
|
12
|
Chong IW, Chang MY, Chang HC, Yu YP, Sheu
CC, Tsai JR, Hung JY, Chou SH, Tsai MS, Hwang JJ, et al: Great
potential of a panel of multiple hMTH1, SPD, ITGA11 and COL11A1
markers for diagnosis of patients with non-small cell lung cancer.
Oncol Rep. 16:981–988. 2006.PubMed/NCBI
|
|
13
|
Feng Y, Sun B, Li X, Zhang L, Niu Y, Xiao
C, Ning L, Fang Z, Wang Y, Zhang L, et al: Differentially expressed
genes between primary cancer and paired lymph node metastases
predict clinical outcome of node-positive breast cancer patients.
Breast Cancer Res Treat. 103:319–329. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sok JC, Lee JA, Dasari S, Joyce S,
Contrucci SC, Egloff AM, Trevelline BK, Joshi R, Kumari N, Grandis
JR, et al: Collagen type XI α1 facilitates head and neck squamous
cell cancer growth and invasion. Br J Cancer. 109:3049–3056. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu YH, Chang TH, Huang YF, Huang HD and
Chou CY: COL11A1 promotes tumor progression and predicts poor
clinical outcome in ovarian cancer. Oncogene. 33:3432–3440. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
García-Pravia C, Galván JA,
Gutiérrez-Corral N, Solar-García L, García-Pérez E, García-Ocaña M,
Del Amo-Iribarren J, Menéndez-Rodríguez P, García-García J, de Los
Toyos JR, et al: Overexpression of COL11A1 by cancer-associated
fibroblasts: Clinical relevance of a stromal marker in pancreatic
cancer. PLoS One. 8:e783272013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Vecchi M, Nuciforo P, Romagnoli S,
Confalonieri S, Pellegrini C, Serio G, Quarto M, Capra M, Roviaro
GC, Avesani E Contessini, et al: Gene expression analysis of early
and advanced gastric cancers. Oncogene. 26:4284–4294. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hunter T and Pines J: Cyclins and cancer.
II: Cyclin D and CDK inhibitors come of age. Cell. 79:573–582.
1994.
|
|
19
|
Kozar K and Sicinski P: Cell cycle
progression without cyclin D-CDK4 and cyclin D-CDK6 complexes. Cell
Cycle. 4:388–391. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yoon MK, Mitrea DM, Ou L and Kriwacki RW:
Cell cycle regulation by the intrinsically disordered proteins p21
and p27. Biochem Soc Trans. 40:981–988. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Salvesen GS: Caspases: Opening the boxes
and interpreting the arrows. Cell Death Differ. 9:3–5. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ghavami S, Hashemi M, Ande SR, Yeganeh B,
Xiao W, Eshraghi M, Bus CJ, Kadkhoda K, Wiechec E, Halayko AJ, et
al: Apoptosis and cancer: Mutations within caspase genes. J Med
Genet. 46:497–510. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tong QS, Zheng LD, Wang L, Zeng FQ, Chen
FM, Dong JH and Lu GC: Downregulation of XIAP expression induces
apoptosis and enhances chemotherapeutic sensitivity in human
gastric cancer cells. Cancer Gene Ther. 12:509–514. 2005.PubMed/NCBI
|
|
24
|
Wiedemeyer WR, Dunn IF, Quayle SN, Zhang
J, Chheda MG, Dunn GP, Zhuang L, Rosenbluh J, Chen S, Xiao Y, et
al: Pattern of retinoblastoma pathway inactivation dictates
response to CDK4/6 inhibition in GBM. Proc Natl Acad Sci USA.
107:11501–11506. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lyu JH, Park DW, Huang B, Kang SH, Lee SJ,
Lee C, Bae YS, Lee JG and Baek SH: RGS2 suppresses breast cancer
cell growth via a MCPIP1-dependent pathway. J Cell Biochem.
116:260–267. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wolff DW, Xie Y, Deng C, Gatalica Z, Yang
M, Wang B, Wang J, Lin MF, Abel PW and Tu Y: Epigenetic repression
of regulator of G-protein signaling 2 promotes androgen-independent
prostate cancer cell growth. Int J Cancer. 130:1521–1531. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Subramanian N, Navaneethakrishnan S,
Biswas J, Kanwar RK, Kanwar JR and Krishnakumar S: RNAi mediated
Tiam1 gene knockdown inhibits invasion of retinoblastoma. PLoS One.
8:e704222013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhu JM and Yu PW: Downregulation of T-cell
lymphoma invasion and metastasis-inducing factor 1 induces
cytoskeletal rearrangement and inhibits the invasive capacity of
gastric cancer cells. Mol Med Rep. 8:425–433. 2013.PubMed/NCBI
|
|
29
|
Huang Z, Zhuo Y, Shen Z, Wang Y, Wang L,
Li H, Chen J and Chen W: The role of NEFL in cell growth and
invasion in head and neck squamous cell carcinoma cell lines. J
Oral Pathol Med. 43:191–198. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Capasso M, Diskin S, Cimmino F, Acierno G,
Totaro F, Petrosino G, Pezone L, Diamond M, McDaniel L, Hakonarson
H, et al: Common genetic variants in NEFL influence gene expression
and neuroblastoma risk. Cancer Res. 74:6913–6924. 2014. View Article : Google Scholar : PubMed/NCBI
|