Introduction
Colorectal cancer (CRC) has the fourth-highest
incidence among all cancers and the fifth-highest incidence of
tumor-related mortalities across China (1). Failure of treatments to prevent and
control distant metastasis is the main reason for its high
mortality of 50.7% (2). Therefore,
further investigation of the active proteins or pathways in CRC
progression may contribute to the development of novel therapeutic
targets.
The tripartite motif (TRIM) protein family contains
more than 77 members that are characterized by the presence of an
N-terminal region, which consists of a RING domain, 1 or 2 B-box
motifs, and a coiled-coil region (3). TRIM proteins are implicated in a wide
range of physiologic processes such as immunity, proliferation,
antiviral process, oncogenesis and transcriptional regulation
(4–8). Due to the RING domain, a large part of
the TRIM protein functions as E3 ubiquitin ligases (6). Over the last decade, the role of TRIM
proteins in innate immunity has been extensively studied. In
addition, recent studies have shown that various members of this
family, including TRIM15, TRIM25 and TRIM29, have vital roles in
human tumorigenesis (9–11).
Tripartite motif-containing 59 (TRIM59), a novel
TRIM family member, is related to several cancers. Zhou et
al (2014) reported that TRIM59 expression is markedly increased
in gastric cancer and may promote gastric carcinogenesis by
promoting ubiquitination and degradation of p53 (12). Furthermore, TRIM59 can facilitate
the proliferative and migratory capacity of non-small cell lung
cancer (13). Mouse models of
prostate cancer have shown that TRIM59 may exert its carcinogenic
effects through the RB and Ras signal pathways (14). Collectively, these data suggest that
TRIM59 plays a role in tumor proliferation and migration.
Nevertheless, the exact role of TRIM59 in CRC is still unclear.
Therefore, the present study aimed to examine the
expression and biological function of TRIM59 in CRC.
Materials and methods
CRC patient specimens
A total of 90 human CRC tissues and their
corresponding normal colorectal mucosa were surgically acquired
between June 2009 and June 2011 at The First Affiliated Hospital of
Nanjing Medical University (Jiangsu, China). All tissues were
frozen in liquid nitrogen immediately after surgical exeresis and
stored at −80°C. Written informed consent was obtained from all
patients or their relatives. Tumor-node-metastasis (TNM) stage was
determined on the basis of The National Comprehensive Cancer
Network (2015.2). In the present study, patients who had accepted
any neoadjuvant radiotherapy or chemotherapy were not included.
Cell culture and chemicals
Human colorectal carcinoma cell lines Caco-2, SW480,
HT-29, LoVo, DLD-1, HCT116 and normal human colorectal epithelial
cells (NCM460) were maintained in our laboratory and were cultured
in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10%
fetal bovine serum (FBS) (both from Winsent Inc., St. Bruno,
Quebec, Canada) 100 U/ml penicillin and 100 µg/ml streptomycin at
37°C in an incubator containing 5% CO2. The PI3K
inhibitor LY294002 was purchased from Cell Signaling Technology
(Danvers, MA, USA).
RNA extraction and real-time
quantitative polymerase chain reaction (RT-qPCR)
RNA was extracted from tissues and cells using
TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Complementary DNA
(cDNA) was synthesized using PrimeScript RT reagent kit (Takara,
Dalian, China). RT-qPCR was conducted using a SYBR-Green PCR kit
(Roche Diagnostics, Indianapolis, IN, USA) with a StepOnePlus
Real-Time PCR system (Applied Biosystems, Foster City, CA, USA).
The primer sequences for PCR are presented in Table I. The PCR cycling conditions were as
follows: 95°C for 30 sec, 40 cycles of 95°C for 5 sec and 60°C for
30 sec, and dissociation at 95°C for 15 sec, 60°C for 1 min and
95°C for 15 sec. The date was analyzed using the 2−ΔΔCt
method. All qRT-PCR reactions were performed in triplicate.
 | Table I.Primer sequences used for qRT-PCR. |
Table I.
Primer sequences used for qRT-PCR.
| Primer | Sequence (5′-3′) |
|---|
| GAPDH | F
5′-ACAGTCAGCCGCATCTTCTT-3′ |
|
| R
5′-GACAAGCTTCCCGTTCTCAG-3′ |
| TRIM59 | F
5′-CCTGTGTTTGAGATAGATTTAAGAGC-3′ |
|
| R
5′-GCAACAAGGTGAGACCCAGT-3′ |
Small interfering RNA (siRNA)
interference and plasmid transfection
siRNA targeting human TRIM59 and a scrambled
nucleotide sequence (NC) were designed and synthesized by
GenePharma Corporation (Shanghai, China), and then transfected into
LoVo and DLD-1 cells using Lipofectamine 3000 (Invitrogen)
following the manufacturer's instructions. Assays to assess the
knockdown efficiency were performed 48 h after transfection. The
sequences for the TRIM59 siRNAs were as follows: siRNA1,
5′-CCCUGAACAUUACAGGCAATT-3′; siRNA2, 5′-CCUAUAGAUGACCUUCAAATT-3′;
siRNA3, 5′-CCACUCAAGUGCCCUAAUUTT-3′. The NC sequences were:
5′-UUCUCCGAACGUGUCACGUTT-3′ (sense) and 5′-ACGUGACACGUUCGGAGAATT-3′
(antisense). Expression of the TRIM59 plasmid was purchased from
GeneCopoeia, Inc., Rockville, MD, USA). The primer sequences were
as follows: forward primer, 5′-CAGCCTCGGACTCTAGC-3′ and reverse
primer, 5′-TAATACGACTCACTATAGGG-3′.
Colony-formation assay
The colony-formation assay was performed to measure
cell proliferation. After transient transfection with TRIM59
siRNA3, control siRNA or untreated as a negative control, 500 cells
were planked/well in 6-well plates in triplicate. The culture
medium was changed to DMEM + 10% FBS every second day. When visible
clones were visible, each well was washed with phosphate-buffered
saline (PBS) three times, fixed with methanol for 30 min at room
temperature, and then stained with 0.05% crystal violet for 30 min.
After washing, the colonies (≥50 cells/colony) were counted and
imaged using a digital camera.
Cell-proliferation assay
The cell viability in all samples was measured using
the Cell Counting Kit-8 (CCK-8; Dojindo, Tokyo, Japan) assay
according to the manufacturer's protocol. Cells (2×103)
were seeded in 100 µl complete culture medium in 96-well plates and
cultured overnight. Subsequently, 10 µl CCK-8 was added to each
well at 24, 48, 72 and 96 h. The absorbance was measured using a
microplate reader at a test wavelength of 450 nm and a reference
wavelength of 630 nm. Experiments were performed in triplicate.
Flow cytometric analysis
After a 48-h siRNA transfection, the cells were
collected and resuspended in cold PBS, and then stained with an
Annexin V-FITC Apoptosis Detection Kit I (BD Biosciences, Franklin
Lakes, NJ, USA) following the manufacturer's instructions. Data
were analyzed using flow cytometry (Becton-Dickinson, San Jose, CA,
USA).
Western blotting
Protein lysates from cells were prepared by using a
RIPA kit (Beyotime, Shanghai, China) according to the
manufacturer's protocols. Equivalent amounts of protein were
separated on 10% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis gels and subsequently transferred to polyvinylidene
difluoride membranes (Millipore, Bedford, MA, USA). The membranes
were blocked in 5% non-fat milk at room temperature for 2–4 h, and
then incubated with primary antibodies at 4°C overnight. The
primary antibodies included TRIM59 (1:1,000; Santa Cruz
Biotechnology, Santa Cruz, CA, USA), E-cadherin, vimentin, Snail,
PI3K, p-PI3K, AKT, p-AKT (1:1,000) and glyceraldehyde 3-phosphate
dehydrogenase (GAPDH) (1:10,000) (all from Cell Signaling
Technology). After washing three times with Tris-buffered saline
with Tween-20 (TBST), the membranes were probed with secondary
peroxidase-conjugated antibodies (1:1,000; Beijing Biosynthesis
Biotechnology Co., Ltd., Beijing, China) at room temperature for 2
h. After washing the membranes three times with TBST, the bound
proteins were visualized using ECL Plus (Millipore, Billerica, MA,
USA) with a Bio-Imaging System. Densitometric analysis was
performed to determine the relative expression of the target
protein, normalized to GAPDH.
Wound-healing assay
Cells were seeded into 6-well plates at
4×105 cells/well until they reached 90–100% confluency,
and a linear wound was scratched with a 200-µl pipette tip.
Thereafter, the medium was changed to serum-free DMEM. Cell
migration into the artificial wound area was monitored by
microscopy at two preselected time points (0 and 24 h), and each
experiment was performed in triplicate.
Transwell assay
Cell migration and invasion were assayed using a
24-well Transwell plate with polycarbonate sterile chambers (8 µm
filters; BD Biosciences) with or without Matrigel coating. Cells
(2×104) were incubated with 100 µl serum-free DMEM in
the upper chamber and complete culture medium containing 10% FBS in
the lower chamber. After incubation for 24 h at 37°C, the migrated
or invaded cells were stained with 0.5% crystal violet solution.
Three random fields of migrated cells were selected and counted
using light microscopy.
Statistical analysis
Statistical Program for Social Sciences 20.0 (SPSS,
Inc., Chicago, IL, USA) and GraphPad Prism 5.0 software (GraphPad
Software Inc., La Jolla, CA, USA) were used for statistical
computations. Differences were analyzed using Student's t-test or
one-way analysis of variance. The associations between expression
levels of TRIM59 and clinicopathological features were assessed by
Pearson's correlation test. Kaplan-Meier survival curves and
log-rank test were carried out to analyze the survival data. Data
are expressed as mean ± standard deviation from at least three
experiments. A p-value <0.05 was considered to indicate a
statistical significance.
Results
TRIM59 expression is upregulated in
CRC tissues and cells and is correlated with distant
metastasis
To investigate the functional role of TRIM59 in the
progression of CRC, we first determined the level of TRIM59
expression in 90 pairs of CRC tissues compared with adjacent normal
tissues by using qRT-PCR. We found that TRIM59 was significantly
upregulated in the tumor tissues compared with that noted in the
non-cancerous tissues (Fig. 1A). We
then analyzed the relationship between TRIM59 expression and the
clinicopathological features of the tumor tissue samples (Table II), and classified the 90 patients
into a relative high-TRIM expression group (n=45) and a relative
low-TRIM expression group (n=45) according to the median expression
of TRIM59. High TRIM59 expression levels were significantly
associated with TNM staging (p=0.001; p<0.05), lymph node
metastasis (p=0.011; p<0.05), depth of invasion (p=0.034;
p<0.05), and distant metastasis (p=0.005; p<0.05).
Nevertheless, no significant difference was observed between TRIM59
expression and other clinicopathological characteristics. TRIM59
mRNA expression was detected in six CRC cell lines (LoVo, DLD-1,
SW480, HT-29, HCT-116 and Caco2) and one normal human colon
epithelial cell line (NCM460). Our results showed that the TRIM59
mRNA level was markedly higher in five CRC cell lines than that
noted in the NCM460 cell line (Fig.
1B). We also found that the protein level of TRIM59 was
increased in the CRC cells (Fig.
1C). Kaplan-Meier curves showed that the overall survival for
patients with high-expression of TRIM59 was significantly worse
than that for such patients with low TRIM59 expression (p=0.0056;
p<0.05) (Fig. 1D). Taken
together, these data suggest that TRIM59 was overexpressed in CRC
and may be helpful to evaluate the prognosis of CRC.
 | Table II.Correlation between TRIM59 expression
and clinicopathological features in the colorectal cancer
specimens. |
Table II.
Correlation between TRIM59 expression
and clinicopathological features in the colorectal cancer
specimens.
|
|
| Expression of
TRIM59 |
|
|---|
|
|
|
|
|
|---|
|
Characteristics | No. | Low expression
(n=45) | High expression
(n=45) | P-value |
|---|
| Age (years) |
|
<60 | 31 | 16 | 15 | 0.824 |
|
≥60 | 59 | 29 | 30 |
|
| Sex |
|
Male | 54 | 23 | 31 | 0.085 |
|
Female | 36 | 22 | 14 |
|
| Tumor diameter |
| <5
cm | 54 | 29 | 25 | 0.389 |
| ≥5
cm | 36 | 16 | 20 |
|
| TNM stage |
|
I/II | 35 | 25 | 10 | 0.001a |
|
III/IV | 55 | 20 | 35 |
|
| Lymph node
metastasis |
|
Positive | 42 | 27 | 15 | 0.011a |
|
Negative | 48 | 18 | 30 |
|
| Depth of
invasion |
|
T1+T2 | 25 | 17 | 8 | 0.034a |
|
T3+T4 | 65 | 28 | 37 |
|
| Distant
metastasis |
|
Positive | 14 | 3 | 11 | 0.005a |
|
Negative | 76 | 42 | 34 |
|
| Primary tumor
site |
|
Colon | 42 | 19 | 23 | 0.398 |
|
Rectum | 48 | 26 | 22 |
|
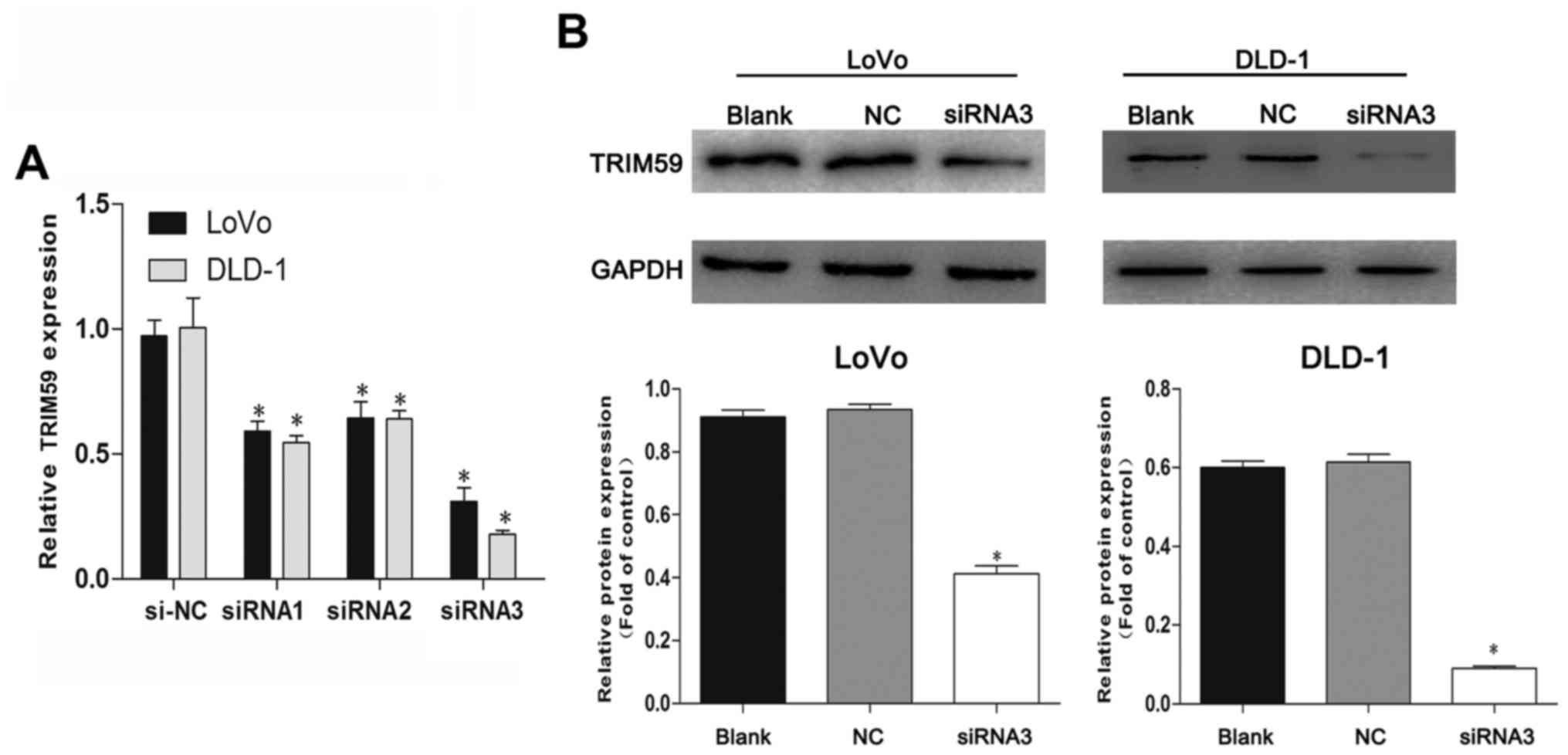
siRNA suppresses TRIM59 expression in
CRC cells
To investigate the functional relevance of TRIM59 in
CRC tumorigenesis, two CRC cell lines (LoVo and DLD-1) which showed
high levels of TRIM59 expression and better growth behavior of all
cells were selected for this assay. RT-PCR showed that TRIM59
siRNAs (siRNA1, siRNA2 and siRNA3) efficiently knocked down TRIM59
compared with cells transfected with control scrambled siRNA (NC)
in the chosen cell lines (Fig. 2A),
and siRNA3 showed the best inhibitory effect on TRIM59 mRNA
expression. Identical results were presented by western blotting,
confirming that TRIM59 protein expression was notably suppressed by
siRNA3 in two cell lines (Fig. 2B).
Accordingly, siRNA3 was selected for the ensuing experiments.
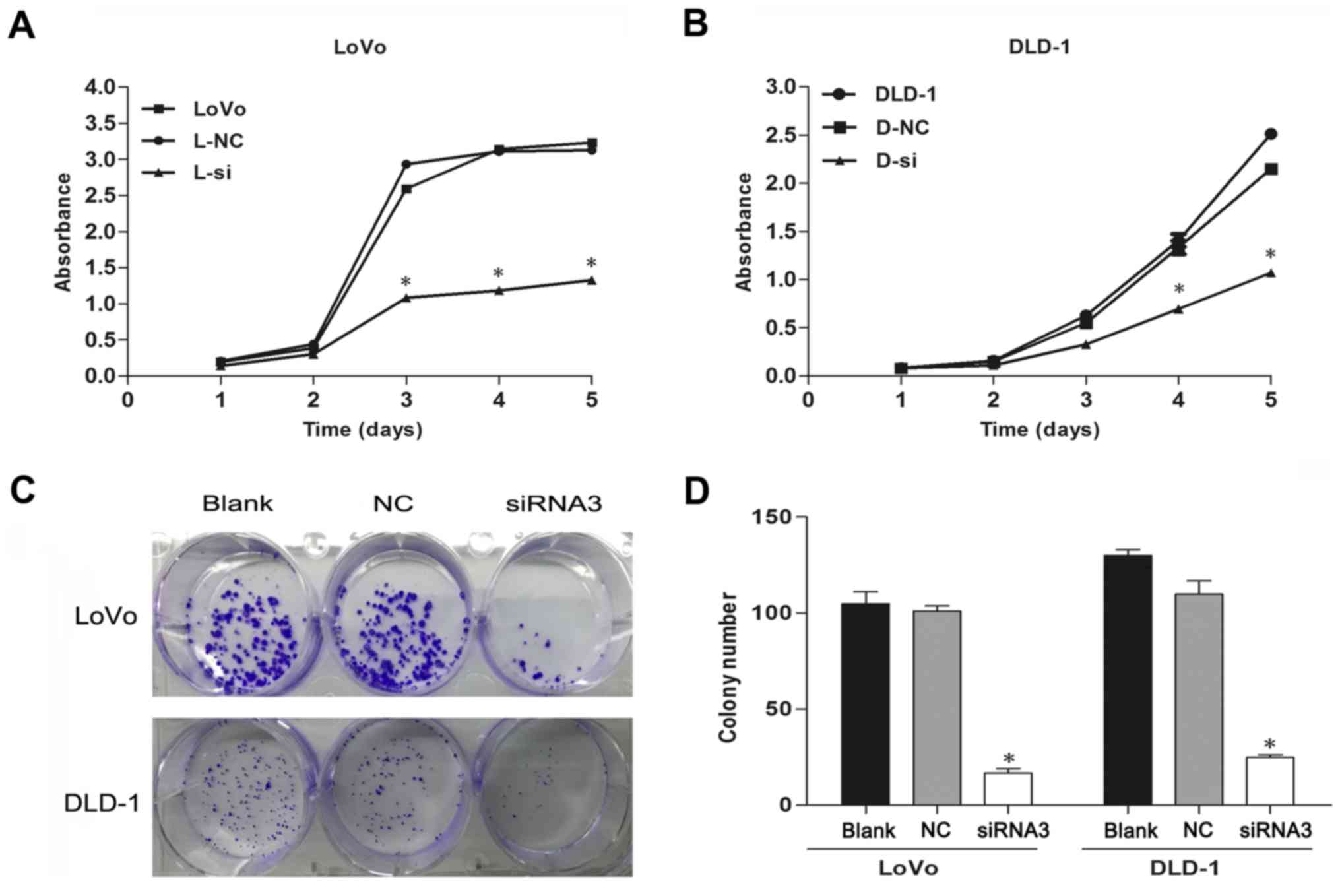
Depletion of TRIM59 negatively
regulates proliferation of CRC cells
As TRIM59 activates proliferation in several
different cell types, we examined whether it facilitates cell
proliferation in human CRC. The CCK-8 assays indicated that
siTRIM59-transfected CRC cells showed evidently decreased
proliferation in comparison to cells transfected with NC (Fig. 3A and B). Moreover, the
colony-formation assay used to further investigate the
anchor-independent growth ability of transfected cells showed that
TRIM59 knockdown attenuated the colony numbers (Fig. 3C and D). Collectively, these results
suggested that interference of TRIM59 reduces the proliferative
capacity of CRC cells.
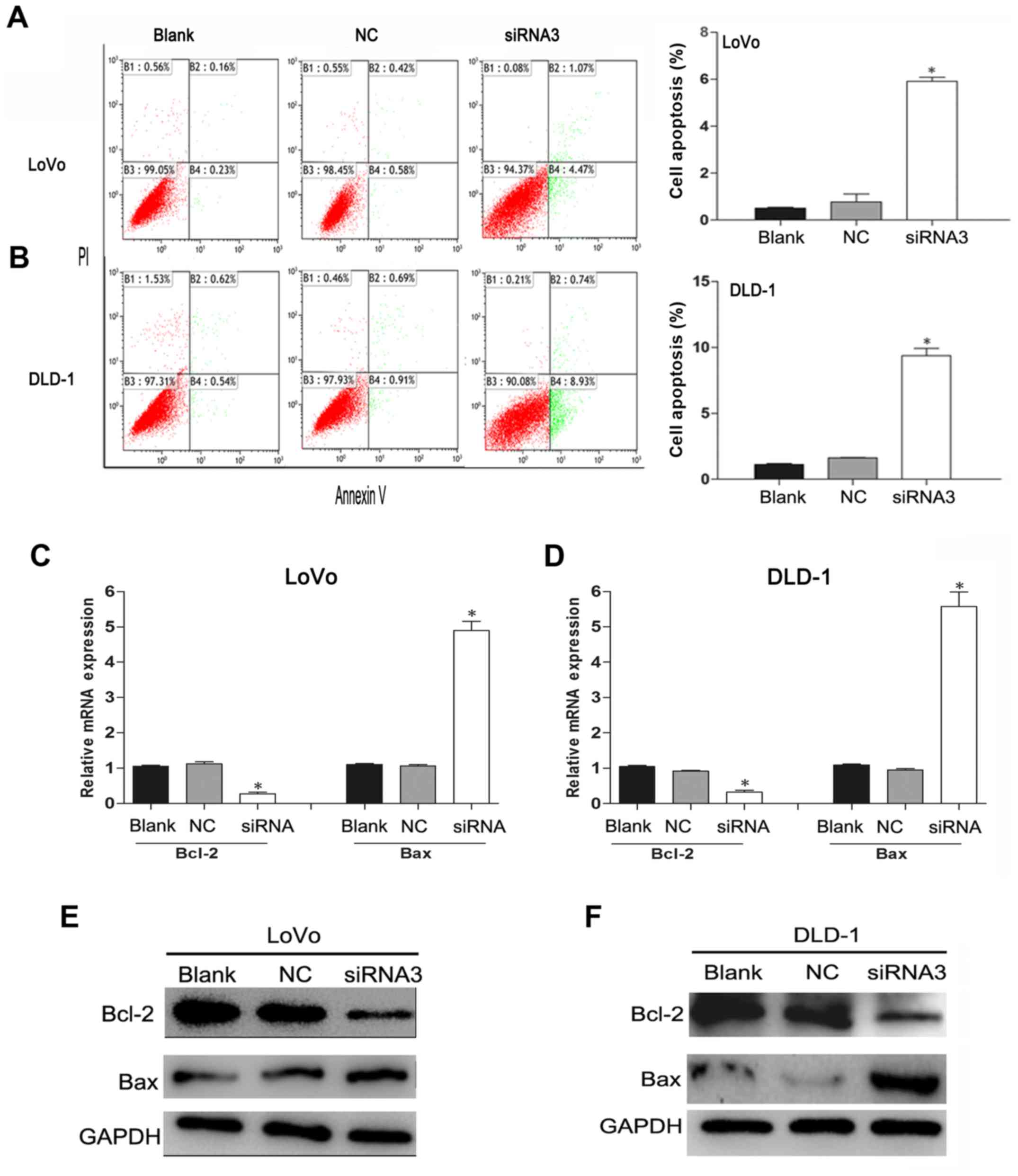
Knockdown of TRIM59 increases
apoptosis of CRC cells through the mitochondrial pathway
To investigate the effect of TRIM59 knockdown
on CRC cell apoptosis, the apoptotic effect after the CRC cells
were transfected with siTRIM59 was analyzed by flow cytometry. The
Annexin V/PI bi-parameter method results implied that the cell
apoptosis rates in the blank group, the NC group, and the siTRIM59
group were as follows: for LoVo cells: 0.39, 1 and 5.54%,
respectively; for DLD-1 cells: 1.16, 1.70 and 9.67%, respectively.
Compared with the blank group and the NC group, markedly elevated
cell apoptosis rates were noted in the siTRIM59 group (p<0.05).
However, no significant difference was observed between the blank
and NC groups (p>0.05) (Fig. 4A and
B). In addition, the Bcl-2 family proteins play different roles
in the regulation of apoptosis and mainly affect the mitochondrial
pathway (15). In the Bcl-2 family,
pro-apoptotic member Bax and anti-apoptotic member Bcl-2 are active
effectors and regulators, and the ratio of Bcl-2/Bax is usually
regarded as a criterion in programmed cell death (16). RT-qPCR (Fig. 4C and D) and western blotting
(Fig. 4E and F) analysis indicated
that TRIM59 knockdown suppressed the expression of Bcl-2,
but enhanced the expression of Bax in LoVo and DLD-1 cell lines.
These results indicate that TRIM59 plays an inhibitory role in the
apoptosis of CRC cells, which proved to be linked with the balance
of the Bcl-2/Bax ratio.
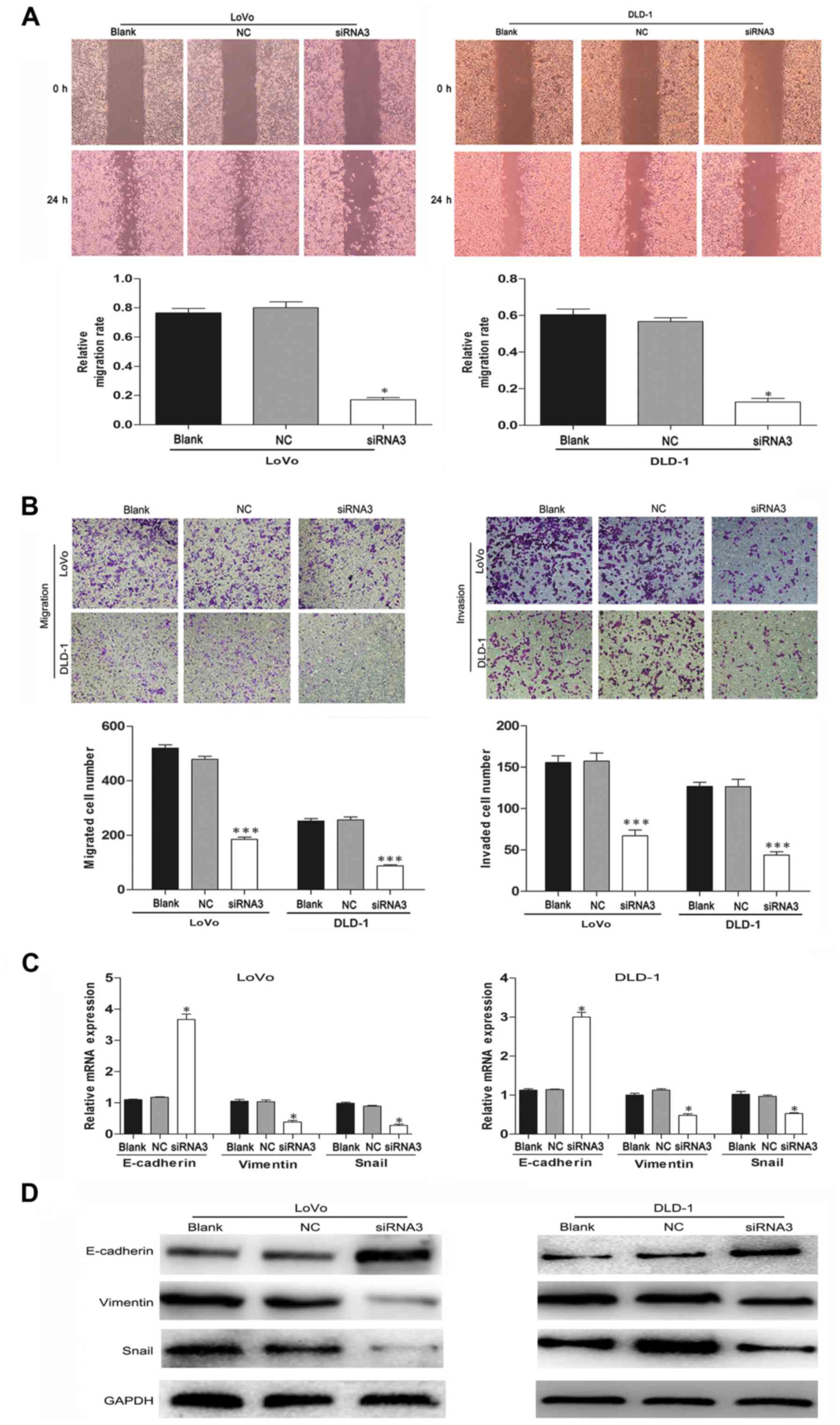
TRIM59 silencing inhibits the motility
and invasiveness of CRC cells
TRIM59 has been reported to promote metastasis in
gastric cancer and other cancer cells. Therefore, wound-healing and
Transwell assays were performed to confirm the prometastatic
properties of TRIM59 on CRC cells. The LoVo and DLD-1 cell
migration effect was strikingly inhibited in the siTRIM59 group
compared with the control group (Fig.
5A). Similar results were observed from the Transwell and
Matrigel assays; knockdown of TRIM59 significantly inhibited CRC
cell migration and invasion (Fig.
5B). Thus, TRIM59 functioned as an oncogene accelerator by
suppressing the migration and invasion of CRC cells in
vitro.
TRIM59 facilitates
epithelial-mesenchymal transition (EMT) in CRC cells
EMT is a critical mechanism involved in cell
migration and invasion (17). To
confirm the influence of TRIM59 knockdown on EMT in CRC
cells, we evaluated the expression of EMT-associated genes in LoVo
and DLD-1 cells after transfection. Western blotting and RT-PCR
results showed that the epithelial marker E-cadherin was highly
expressed, whereas the expression of mesenchymal marker vimentin
was significantly decreased when TRIM59 was suppressed in LoVo and
DLD-1 cells (Fig. 5C and D). Snail
is another key regulator of EMT that functions as a transcriptional
repressor of E-cadherin and other epithelial proteins (18,19).
In the present study, the protein levels of Snail were also
markedly reduced in the TRIM59-knockdown cells. These data
suggest that TRIM59 may promote the migration and invasiveness of
CRC cells by regulating the EMT pathway.
Downregulation of TRIM59 restricts
activation of the PI3K/AKT pathway in CRC cells
The PI3K/Akt pathway plays an essential role in cell
proliferation and promotes EMT of various types of cancer (20). Moreover, mouse prostate cancer
models showed that TRIM59 may be linked with the PI3K/Akt pathway
(14). Therefore, we further
examined whether the PI3K/Akt pathway was involved in the
TRIM59-mediated cellular response in CRC. Western blotting
significantly revealed decreased levels of phosphorylated PI3K and
AKT in the cells transfected with siTRIM59. However, there was no
change observed in the expression of total PI3k and AKT (Fig. 6).
To investigate the role of the PI3K/Akt pathway in
TRIM59-induced metastasis in CRC, a specific inhibitor of PI3K
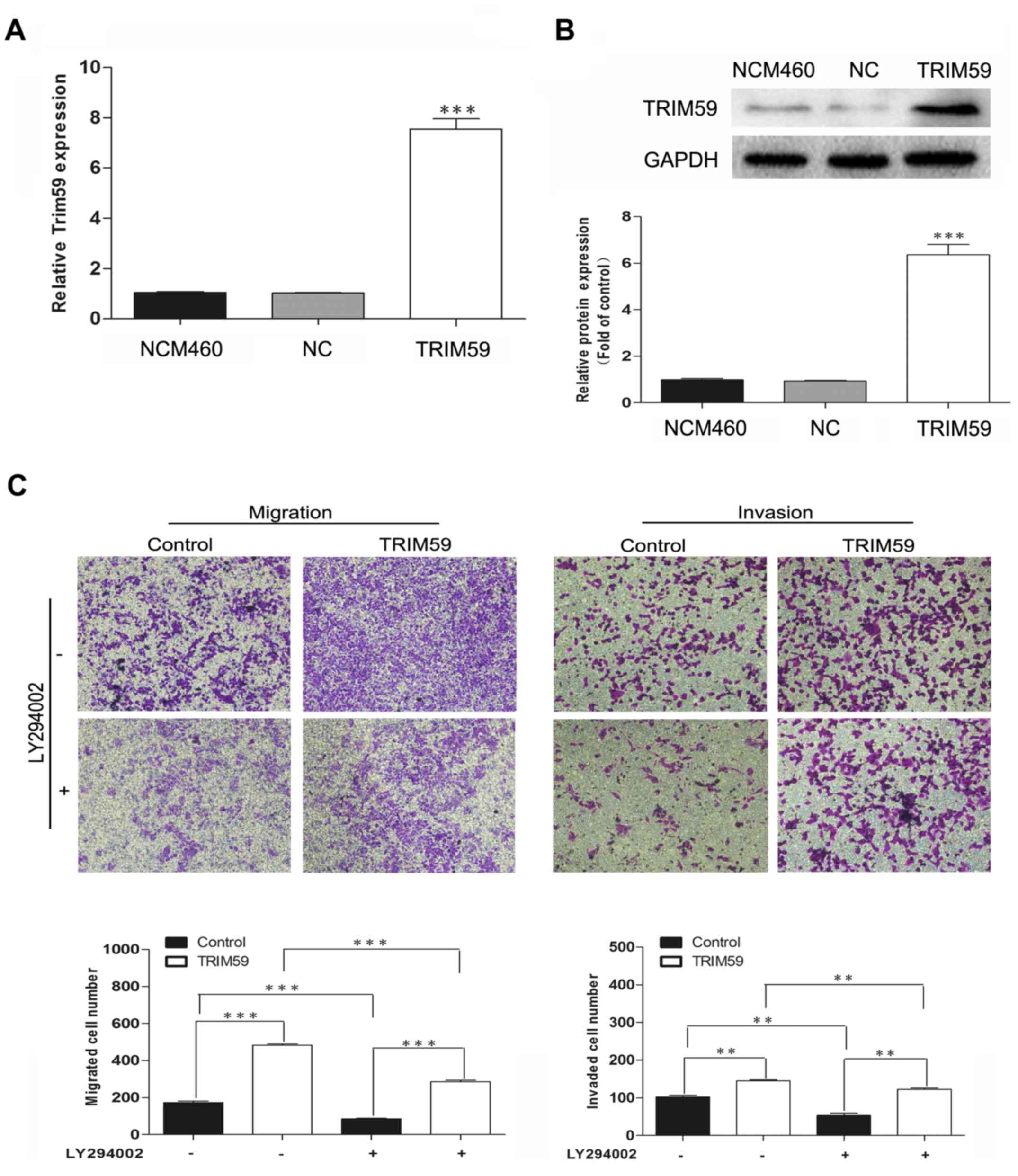
(LY294002) was used. TRIM59 was overexpressed in NCM460, which
contains low expression of TRIM59, and then treated with LY294002.
Transwell assays showed that overexpression of TRIM59 facilitated
cell migration and invasion; however, this enhancement of
metastasis was inhibited by LY294002 exposure (Fig. 7). These results confirmed the
involvement of the PI3K/Akt pathway in the promotion of CRC cell
migration and invasion by TRIM59.
Discussion
Recently, TRIM proteins have been identified as
important regulators of cellular processes in a variety of human
cancers (21), and various TRIM
proteins have been found to be highly expressed in CRC (9,22).
Although the oncogenic effect of TRIM59 on progression has been
confirmed in various cancers, including gastric cancer (12), osteosarcoma (23), lung (13) and cervical cancer (24), the expression and function of TRIM59
in CRC remain unclear.
In the present study, we detected the expression
level of TRIM59 in CRC to assess its potential role as a clinically
relevant prognostic or predictive marker. We first detected whether
TRIM59 was expressed in CRC tissues and cell lines. RT-qPCR
analysis of samples from 90 patients revealed that TRIM59 was
overexpressed in CRC tissues as compared to normal colorectal
tissues. Further analysis demonstrated that TRIM59 expression was
significantly associated with TNM stage, status of lymphatic node,
and presence of distant metastasis. Kaplan-Meier and Cox regression
analysis revealed that the TRIM59 expression pattern was associated
with poor prognosis of patients with CRC. These data indicate that
TRIM59 expression could be a potential therapeutic target for
diagnosis and prognosis.
We also evaluated the functions of TRIM59 in CRC
cell lines. Knockdown of TRIM59 significantly suppressed
cell growth in the LoVo and DLD-1 cell lines and promoted CRC cell
apoptosis and decreased the ratio of Bcl-2/Bax. However, no
significant changes were observed in the distribution of CRC cells
in the G0/G1, S and G2/M phases, indicating that TRIM59 was not
involved in cell cycle control. These data suggest that TRIM59
could suppress proliferation of CRC cells by inducing cell
apoptosis.
Our clinical analysis showed that high expression of
TRIM59 was associated with distant metastasis and TNM stage; we
further found that TRIM59 enhanced the infiltrative capability of
the CRC cells. Results of in vitro experiments indicated
that silencing of the TRIM59 gene inhibited the migration
and invasion of CRC cells. Metastasis of CRC remains the leading
cause of the low 5-year overall survival rate in CRC (25). EMT has been reported to play
critical and intricate roles in promoting tumor metastasis. This
process is defined as the loss of cell-cell adhesion molecules,
downregulation of epithelial differentiation markers, and
transcriptional induction of mesenchymal markers (26). Thus, we determined the potential
target protein levels associated with EMT-induced markers after the
knockdown of TRIM59. Our results showed that depletion of
TRIM59 significantly weakened vimentin and Snail expression and
promoted E-cadherin expression. These data support the hypothesis
that downregulation of TRIM59 may inhibit the progression of EMT by
modulating EMT-related gene expression.
EMT is mediated by several different signaling
pathways (27). Valiyeva et
al reported TRIM59 as an early signal transducer in two
oncogene routes: the Ras/ERK/PI3K/AKT pathway and the SV40
Tag/p53/pRB route (14). We
hypothesized that oncogenic TRIM59 functions through the PI3k/AKT
pathway in CRC. In the present study, we found that suppression of
TRIM59 expression notably decreased the protein levels of
phosphorylated PI3K and AKT, while their total protein levels
remained unchanged. After treatment with LY294002, a specific
inhibitor of PI3K, the facilitating roles of TRIM59 overexpression
on cell migration and invasion were evidently decreased. These data
suggest that TRIM59 may play an important role in CRC cell
metastasis via activation of the PI3k/Akt signaling pathway.
Further in-depth clinical research should focus on clarifying the
role of TRIM59 in CRC.
In summary, we reported that TRIM59 expression is
strikingly upregulated in human CRC, indicating that TRIM59 plays a
key oncogenic role as a negative prognostic factor in patients with
CRC. Moreover, TRIM59 facilitates the proliferation, migration and
invasion of CRC cells and regulates cell metastasis through the
PI3K/AKT pathway. Therefore, TRIM59 may serve as a potential
prognostic and therapeutic target of CRC. Future studies should
further investigate the mechanism by which TRIM59 is involved in
the development of CRC.
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal cancer
|
|
TRIM
|
tripartite motif
|
|
DMEM
|
Dulbecco's modified Eagle's medium
|
|
FBS
|
fetal bovine serum
|
|
RT-qPCR
|
real-time quantitative polymerase
chain reaction
|
|
cDNA
|
complementary DNA
|
|
NC
|
nucleotide sequence
|
|
siRNA
|
small interfering RNA
|
|
TBST
|
Tris-buffered saline with Tween-20
|
|
EMT
|
epithelial-mesenchymal transition
|
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Glockzin G, Schlitt HJ and Piso P:
Therapeutic options for peritoneal metastasis arising from
colorectal cancer. World J Gastrointest Pharmacol Ther. 7:343–352.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yamada Y, Takayama KI, Fujimura T,
Ashikari D, Obinata D, Takahashi S, Ikeda K, Kakutani S, Urano T,
Fukuhara H, et al: A novel prognostic factor TRIM44 promotes cell
proliferation and migration, and inhibits apoptosis in testicular
germ cell tumor. Cancer Sci. 108:32–41. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ozato K, Shin DM, Chang TH and Morse HC
III: TRIM family proteins and their emerging roles in innate
immunity. Nat Rev Immunol. 8:849–860. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
James LC, Keeble AH, Khan Z, Rhodes DA and
Trowsdale J: Structural basis for PRYSPRY-mediated tripartite motif
(TRIM) protein function. Proc Natl Acad Sci USA. 104:6200–6205.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gack MU, Shin YC, Joo CH, Urano T, Liang
C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, et al: TRIM25
RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated
antiviral activity. Nature. 446:916–920. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Khatamianfar V, Valiyeva F, Rennie PS, Lu
WY, Yang BB, Bauman GS, Moussa M and Xuan JW: TRIM59, a novel
multiple cancer biomarker for immunohistochemical detection of
tumorigenesis. BMJ Open. 2:22012. View Article : Google Scholar
|
|
8
|
Nisole S, Stoye JP and Saïb A: TRIM family
proteins: Retroviral restriction and antiviral defence. Nat Rev
Microbiol. 3:799–808. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee OH, Lee J, Lee KH, Woo YM, Kang JH,
Yoon HG, Bae SK, Songyang Z, Oh SH and Choi Y: Role of the focal
adhesion protein TRIM15 in colon cancer development. Biochim
Biophys Acta. 1853:409–421. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhu Z, Wang Y, Zhang C, Yu S, Zhu Q, Hou K
and Yan B: TRIM25 blockade by RNA interference inhibited migration
and invasion of gastric cancer cells through TGF-β signaling. Sci
Rep. 6:190702016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tan ST, Liu SY and Wu B: TRIM29
overexpression promotes proliferation and survival of bladder
cancer cells through NF-κB signaling. Cancer Res Treat.
48:1302–1312. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhou Z, Ji Z, Wang Y, Li J, Cao H, Zhu HH
and Gao WQ: TRIM59 is up-regulated in gastric tumors, promoting
ubiquitination and degradation of p53. Gastroenterology.
147:1043–1054. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhan W, Han T, Zhang C, Xie C, Gan M, Deng
K, Fu M and Wang JB: TRIM59 promotes the proliferation and
migration of non-small cell lung cancer cells by upregulating cell
cycle related proteins. PLoS One. 10:e01425962015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Valiyeva F, Jiang F, Elmaadawi A, Moussa
M, Yee SP, Raptis L, Izawa JI, Yang BB, Greenberg NM, Wang F, et
al: Characterization of the oncogenic activity of the novel TRIM59
gene in mouse cancer models. Mol Cancer Ther. 10:1229–1240. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ma L and Li W: Emodin inhibits LOVO
colorectal cancer cell proliferation via the regulation of the
Bcl-2/Bax ratio and cytochrome c. Exp Ther Med. 8:1225–1228.
2014.PubMed/NCBI
|
|
16
|
Zhu W, Ye L, Zhang J, Yu P, Wang H, Ye Z
and Tian J: PFK15, a small molecule inhibitor of PFKFB3, induces
cell cycle arrest, apoptosis and inhibits invasion in gastric
cancer. PLoS One. 11:e01637682016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ye X and Weinberg RA:
Epithelial-mesenchymal plasticity: A central regulator of cancer
progression. Trends Cell Biol. 25:675–686. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kim JH, Park S, Chung H and Oh S: Wnt5a
attenuates the pathogenic effects of the Wnt/β-catenin pathway in
human retinal pigment epithelial cells via down-regulating
β-catenin and Snail. BMB Rep. 48:525–530. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yook JI, Li XY, Ota I, Fearon ER and Weiss
SJ: Wnt-dependent regulation of the E-cadherin repressor snail. J
Biol Chem. 280:11740–11748. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Vivanco I and Sawyers CL: The
phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev
Cancer. 2:489–501. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen Y, Guo Y, Yang H, Shi G, Xu G, Shi J,
Yin N and Chen D: TRIM66 overexpresssion contributes to
osteosarcoma carcinogenesis and indicates poor survival outcome.
Oncotarget. 6:23708–23719. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang J, Zhu J, Dong M, Yu H, Dai X and Li
K: Knockdown of tripartite motif containing 24 by lentivirus
suppresses cell growth and induces apoptosis in human colorectal
cancer cells. Oncol Res. 22:39–45. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liang J, Xing D, Li Z, Shen J, Zhao H and
Li S: TRIM59 is upregulated and promotes cell proliferation and
migration in human osteosarcoma. Mol Med Rep. 13:5200–5206.
2016.PubMed/NCBI
|
|
24
|
Aierken G, Seyiti A, Alifu M and Kuerban
G: Knockdown of tripartrtite-59 (TRIM59) inhibits cellular
proliferation and migration in human cervical cancer cells. Oncol
Res. 25:381–388. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Townsend AR, Chong LC, Karapetis C and
Price TJ: Selective internal radiation therapy for liver metastases
from colorectal cancer. Cancer Treat Rev. 50:148–154. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen B, Zeng X, He Y, Wang X, Liang Z, Liu
J, Zhang P, Zhu H, Xu N and Liang S: STC2 promotes the
epithelial-mesenchymal transition of colorectal cancer cells
through AKT-ERK signaling pathways. Oncotarget. 7:71400–71416.
2016.PubMed/NCBI
|