Introduction
The blood-brain barrier (BBB) is located at the
level of cerebral capillaries in the forefront of the defense line
of the central nervous system (CNS) and restricts the flow of
essential components into and out of the CNS (1,2). The
most important cellular elements of the BBB consist of endothelial
cell, astrocyte, pericyte, and the adjacent neurons (3). The endothelial cells are connected by
junction complex, in which tight junctions (TJs) play a significant
role. Key components of the TJs are the transmembrane protein, and
the best characterized TJs in cerebral endothelial cells are
occluding, claudins and junctional adhesion molecules (JAMs)
(4).
Brain metastases constitute a significant part of
intracranial tumors. The majority of brain metastases originate
from lung cancer, breast cancer and malignant melanoma (5). Metastatic cells invading the CNS
parenchyma, however, have to pass the BBB. Since brain metastases
represent a great therapeutic challenge, it is important to
understand the mechanisms of the interaction of tumor cells with
the BBB to find targets of prevention of brain metastasis
formation.
The attachment of tumor cells to brain endothelial
cells and the transendothelial migration of tumor cells are the key
step in brain metastasis, plenty of molecules and signal pathways
participated in this complicated process. Claudins are small
proteins (20–27 kDa) (6), and the
principal claudin in brain endothelial cells is claudin-5 (CLDN5)
(7). CLDN5 forms the backbone of
the tight junction and was shown to be solely responsible for the
paracellular barrier that exists between epithelial cell, and
absence of CLDN5 leads to a selective opening of the BBB to
molecules smaller than 800 Da (8).
CLDN5 has also recently been shown to directly play a role in the
interaction of metastatic tumor cells and brain endothelial cells
(9), but its mechanism of action
remains largely unknown.
Recent studies have described a complicated
interplay among diverse RNA molecules, including coding and
non-coding RNAs (10,11). Human genome generates plurality of
regulatory RNAs that are either long non-coding RNAs (lncRNAs) or
small non-coding RNA such as microRNAs (miRNAs). LncRNAs are
defined as >200 nucleotides and unable to be translated into
proteins. Accumulating data show that lncRNAs are major regulators
of physiological and disease-related gene expression through
various mechanisms. The dysregulated lncRNA expression has been
documented in various disease states, and their tissue specificity
makes them attractive candidates as diagnostic or prognostic
biomarkers or therapeutic agents. miRNAs are endogenous ~22
nucleotide RNAs, which post-transcriptionally regulate the gene
expression through interaction between their 5′ end and the 3′
untranslated region (UTR) of mRNA. The functions of miRNA have been
elucidated extensively and it participates in regulating a variety
of cellular events (12–14). Recent studies indicated that some
specific lncRNAs can serve as competitive endogenous RNAs (ceRNA)
to control miRNAs available for binding with targets, functionally
sequester miRNAs and as miRNA sponges, thereby alleviating the
inhibitory effect of miRNA on their respective mRNA targets
(15). The function introduces an
extra layer of complexity in the miRNA-target interaction network.
The complexity and diversity of potential ceRNA interactions have
been described with the identification of abundant lncRNAs.
Understanding this novel RNA interaction will lead to significant
insight into gene regulatory networks in a variety of cell process
(16–19).
The involvement of ceRNA regulation as a factor of
CLDN5 modulation in brain vascular endothelial cells has not been
previously investigated. To gain further understanding of how CLDN5
mediates its activities in tumor brain metastasis, herein, in this
study, we examined changes in globe mRNA, miRNA, lncRNA gene
expression when CLDN5 was overexpressed in the human brain vascular
endothelial cell line, hCMEC/D3. We have identified a number of
ceRNAs whose expression levels were altered as a consequence of
high CLDN5 expression. The identified sets of lncRNA, miRNA and
mRNA specific to CLDN-5-overexpressing hCMEC/D3 cells were
subsequently confirmed by quantitative reverse
transcription-polymerase chain reaction (qRT-PCR).
Materials and methods
Cell culture and transduction
Human brain vascular endothelial hCMEC/D3 cells were
obtained from Institut Cochin (Université René Descartes, Paris,
France). Cells were cultured in the EBM-2 medium supplemented with
EGMTM-2 Bullet kit (Lonza, Walkersville, NJ, USA) in a humidified
37°C incubator with an atmosphere of 5% CO2.
In the overexpression experiment, the CLDN5
expression vector was constructed by inserting a human CLDN5 cDNA
into pLL3.7-GFP. CLDN5-pLL3.7 or control pLL3.7 vector were
transfected into 293T cells using Lipofectamine 3000 (Thermo Fisher
Scientific, Inc., Waltham, MA, USA). Supernatant containing
retroviruses was collected and concentrated 48 and 72 h later and
was used for the transduction. The hCMEC/D3 cells were transducted
with the virus-containing medium plus 8 µg/ml polybrene.
Forty-eight hours after beginning the transduction, GFP+
cells were sorted and the expression of CLDN5 was confirmed by
qRT-PCR.
RNA extraction
Total RNA of CLDN5-overexpressing and control
hCMEC/D3 cells were extracted using TRIzol Reagent (Invitrogen Life
Technologies, Carlsbad, CA, USA) according to the manufacturer's
protocol. RNA quantity was evaluated by the 28S/18S ratio and RNA
Integrity Number (RIN) was analyzed on the Agilent 2100 bioanalyzer
using the Eukaryote Total RNA Nano assay (Agilent Technologies,
Waldbronn, Germany). RNA purity was evaluated by the A260/A280
ratio using an RNA 6000 Nano Spectrophotometer (Agilent).
miRNA, mRNA, lncRNA microarray and
computational analysis
For mRNA and lncRNA expression profiling, Affymetrix
Human Transcriptome Array 2.0 (HTA 2.0) (Thermo Fisher Scientific,
Inc.) was used. For miRNA expression profiling, Affymetrix GeneChip
miRNA 4.0 Array (Thermo Fisher Scientific, Inc.) was used. After
hybridization and washing, the arrays were scanned by an Affymetrix
Microarray Scanner (Applied Biosystems, Grand Island, NY, USA). Raw
data of HTA 2.0 were extracted and normalized by
Affymetrix® Transcriptome Analysis Console (TAC)
Software (Thermo Fisher Scientific, Inc.). miRNA QC Tool software
(Thermo Fisher Scientific, Inc.) was used for miRNA 4.0 array data
summarization, normalization, and quality control.
Difference analysis
Two-class differential was used to determine the
differentially expressed miRNA, lncRNA and mRNA between the
pLL3.7-CLDN5-transfected group (n=3) and pLL3.7 control group
(n=3). The random variance model (RVM) t-test was applied to filter
the differentially expressed genes for it can effectively raise the
degrees of freedom in cases of small samples. The false discovery
rate (FDR) was calculated to correct the P-value. P-values <0.05
and FDR <0.05 were considered as significant differences.
The differentially expressed probe sets were
imported into Cluster and Tree View to perform hierarchical cluster
analysis (HCA).
Construction of lncRNA-mRNA
co-expression network
We utilized the expression profile of different
lncRNAs and difference mRNAs to construct the lncRNA-mRNA network.
This network distinctly revealed the relation between the lncRNAs
and mRNAs, found the key regulation and ‘interaction venation’
thoroughly. This network assimilated the scale-free property of the
huge data, to simulate the scale-free relation by the interaction
between lncRNAs and mRNAs and interaction among themselves, and
that is the correlation of pairwise expression. In order to get
reliable and accurate relation of gene interaction and lncRNA
regulation function, we recommend the number of no less than 30 and
the number of genes no less than 200.
For each pair of mRNA-lncRNA, mRNA-mRNA, or
lncRNA-lncRNA, we calculated the Pearson correlation and chose the
significant correlation pairs to construct the network (20). The clustering coefficient represents
the density of each gene with the adjacent gene, and the larger the
clustering coefficient, the greater importance the gene has in
regulating the network.
Gene ontology and pathway
analysis
A gene ontology (GO) analysis was applied to analyze
the main functions of the mRNAs of miRNA-lncRNA-mRNA internets.
Specifically, a two-side Fisher's exact test and a
χ2 test were used to classify the GO category. We
computed P-values of the GO for each differential gene. Enrichment
provides a measure for the significant function: As the enrichment
increases, the corresponding function is more specific. Within the
significant category, the enrichment Re was given as follows:
Re=(nf/n)/(Nf/N), where nf is
the number of flagged genes within the particular category,
n is the total number of genes within the same category,
Nf is the number of flagged genes in the entire microarray,
and N is the total number of genes in the microarray.
Pathway analysis was used to identify the
significant pathway of the differential mRNAs according to KEGG,
Biocarta, and Reatome. We used Fisher's exact test and the
χ2 test to select the significant pathway, and
the threshold of significance was defined by P-value and FDR. The
enrichment Re was calculated as indicated in the equation
above.
qRT-PCR analysis
Total RNA was reverse-transcribed with a
PrimeScript™ RT reagent kit (Takara Biotechnology Co., Ltd.,
Dalian, China) for cDNA synthesis and genomic DNA removal. For
miRNA detection, total RNA was reverse transcribed using miRNA
specific primers. QPCR was performed according to the instructions
of the SYBR premix Ex Taq™ II kit (Takara Biotechnology Co.) and
carried out in the Takara real-time PCR system. GAPDH was used as
lncRNA and mRNA control and U6 was used as a miRNA control.
Gene-specific primers were designed using primer designing tools
primer 5.0. The primer sequences are listed in Table I. The specificity of amplification
was assessed by dissociation curve analysis, and the relative
abundance of genes was determined with the 2−ΔΔCt
method. All experiments were performed in triplicate.
 | Table I.The primers of miRNA, mRNA, and
lncRNA for qRT-PCR. |
Table I.
The primers of miRNA, mRNA, and
lncRNA for qRT-PCR.
| Primers | Sequence
(5′-3′) |
|---|
| miR-297 | RT:
TCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGACATGCACA |
|
| F:
CTCAACTGGTGTCGTGGAGT |
|
| R:
ACACTCCAGCTGGGATGTATGTGTGCAT |
| miR-610 | RT:
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGATCCCAGCA |
|
| F:
CTCAACTGGTGTCGTGGAGT |
|
| R:
ACACTCCAGCTGGGTGAGCTAAATGTG |
| miR-767-5p | RT:
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGACATGCTCA |
|
| F:
CTCAACTGGTGTCGTGGAGT |
|
| R:
ACACTCCAGCTGGGTGCACCATGGTTGTCT |
| miR-329-3P | RT:
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAAAAGAGGT |
|
| F:
CTCAACTGGTGTCGTGGAGT |
|
| R:
ACACTCCAGCTGGGAACACACCTGGTTAA |
| miR-921 | RT:
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAATCCTG |
|
| F:
CTCAACTGGTGTCGTGGAGT |
|
| R:
ACACTCCAGCTGGGCTAGTGAGGGACAGAACC |
| miR-127-5p | RT:
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAATCAGAGC |
|
| F:
CTCAACTGGTGTCGTGGAGT |
|
| R:
ACACTCCAGCTGGGCTGAAGCTCAGAGGG |
| miR-4786-5p | RT:
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGTGCATC |
|
| F:
CTCAACTGGTGTCGTGGAGT |
|
| R:
ACACTCCAGCTGGGTGAGACCAGGACTGG |
| POTED | F:
GTGGTGCTGTCACTGCTTCCC |
|
| R:
CGCTGTGGTCGTAGTCTCCC |
| TMPRSS15 | F:
GGCAGCTCAAGACATCACCC |
|
| R:
GCCGCCATAATACAGACCCA |
| EFNB2 | F:
GAGCAGGAAGCCGATGTGAC |
|
| R:
GGGAAAACCCAACGCAGAAA |
| IL7R | F:
TGAGTGTCGTCTATCGGGAAGG |
|
| R:
CTGGCGGTAAGCTACATCGTG |
| AMTN | F:
CAGACCCACCCATTGACCCT |
|
| R:
GGATTAGCCCCTGCCTGACT |
| PIK3CG | F:
GCCACTGATCCACTTAACCCTC |
|
| R:
AATTTCTTGCTGTCCCCATTTC |
| CLDN1 | F:
TGAGGATGGCTGTCATTGGG |
|
| R:
ACCTGGCATTGACTGGGGTC |
| OCLN | F:
TCTCCCTCCCTGCTTCCTCT |
|
| R:
GCAATGCCCTTTAGCTTCCA |
|
ENST00000427446 | F:
ATTATCAGCTCGTGAGTACGGACAT |
|
| R:
GAAAACATGGAAGCTCTATTTGGTC |
| n342142 | F:
AGAAAGAGCCAACAACTCCTACAGA |
|
| R:
TGAGTGGTTCAGATTTAGGCACAGA |
|
TCONS_00017956- | F:
GATAATTCCGTTTGACTCCGTTTGA |
| XLOC_008786 | R:
CTAATGGAATCGCATGGTATCTTCA |
|
ENST00000425979 | F:
CAAAGGCAAAGATTGGAGTTGTACT |
|
| R:
ACGAATCCTTGCTTGTTTCTTCTAG |
|
TCONS_00028418-XLOC_013771 n338895 | F:
CTTTCTGGGTCTTCTGTTTCACGAT |
|
| R:
TTTCTGTTCAAGTTTGAGGGTCCTG |
|
| F:
GCACGGTATCATTACTCCCGTTTTA |
|
| R:
AACCTCTGATAAGGACAAGCAACCC |
| n339695 | F:
ATTCTCCTGCCTCAGCCTCCCAAGT |
|
| R:
GTGCGGTGGCTCACGTCTGTAGTCC |
|
TCONS_00022673-XLOC_010971 | F:
AACAGAAGCCCAGGGAGATAAAGAC |
|
| R:
TGGAAATGATCCTGGATTAGGAATG |
Results
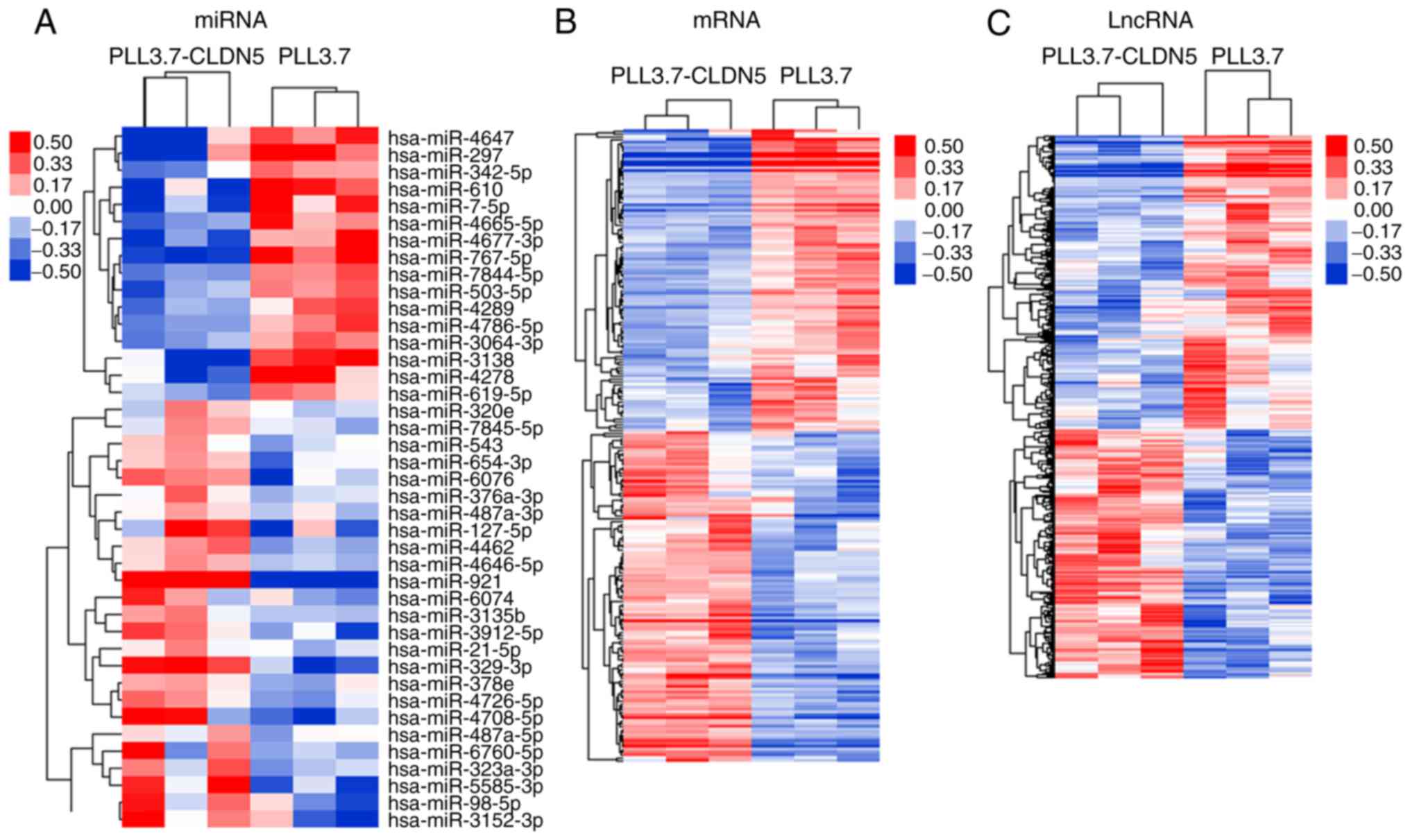
CLDN-5 induces significantly altered
miRNA, lncRNA and mRNA expression patterns in hCMEC/D3 cells
To explore the regulatory mechanism of CLDN5 on
permeability of BBB, we stably transfected the hCMEC/D3 cells with
pLL3.7-CLDN5 or pLL3.7, and then analyzed the changes of miRNA,
lncRNA, and mRNA levels in CLDN-5-overexpressing hCMEC/D3
cells.
In terms of the miRBaseV20 Database, 2578 human
miRNAs were authenticated on the Affymetrix GeneChip microRNA 4.0
Array. Based on the RefSeq, UCSC, GENCODES, MGC, lincRNAs TUCPs,
lincRNAdb and UCSC lincRNA, annotations of lncRNAs and mRNAs, the
probe sets covered 22, 829 lncRNAs and 44, 699 mRNAs on the
Affymetrix GeneChip HTA 2.0 Array.
The miRNA, lncRNA, and mRNA expression patterns were
detected in pLL3.7-CLDN5-transfected group and pLL3.7 control
group. We identified 41 miRNAs, 954 lncRNAs, and 222 mRNAs that had
significant differential expression in the CLDN5-overexpressing
group comparing with the control group (fold change ≥1.2 or ≤0.8,
and P-value <0.05). The hierarchical clustering analysis showed
that with the differentially expression of these miRNAs, lncRNAs
and mRNAs, samples were non-random partitioned, they were divided
into two groups (Fig. 1). Thus, the
miRNA, lncRNA, and mRNA expression signatures identified here were
likely to be representative.
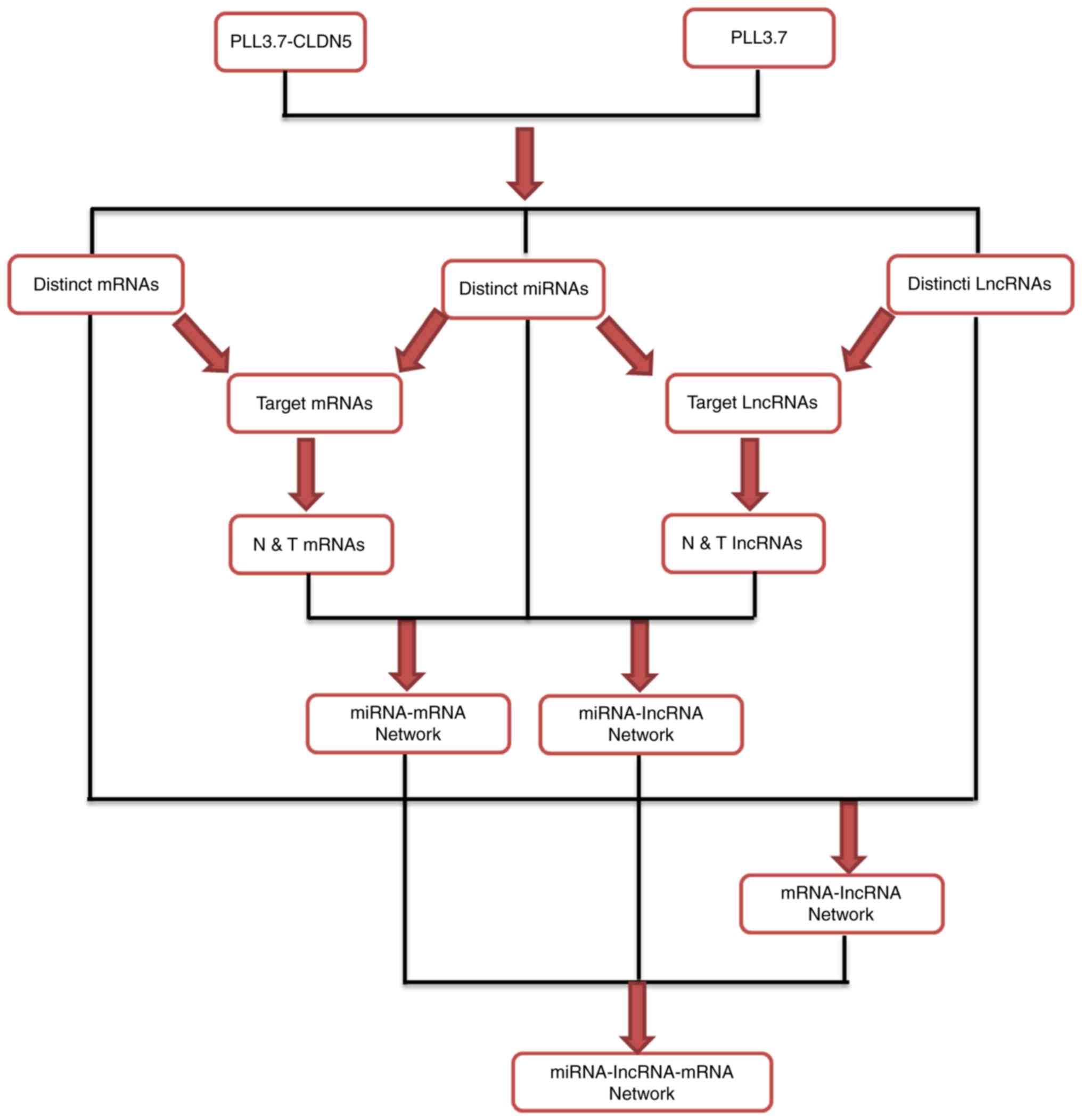
Construction of miRNA-lncRNA-mRNA
interaction of network regulated by reinforced CLDN5 and
identification lncRNAs acting as ceRNAs
The miRNA-lncRNA-mRNA interaction network was
constructed according to the work flow summarized in Fig. 2.
Firstly, the miRanda was applied to analyze the
target mRNAs and lncRNAs of the 41 miRNAs, and then the interaction
of these target mRNAs with distinct mRNAs and target lncRNAs with
distinct lncRNAs, separately termed as target mRNAs (172 mRNAs,
data not shown) and target lncRNAs (681 lncRNAs, data not shown).
Of the target mRNAs and lncRNAs, the mRNAs and lncRNAs were
selected which expression levels were negatively correlated with
miRNA expression, and were termed the N&T mRNAs (152 mRNAs,
data not shown) and N&T lncRNAs (618 lncRNAs, data not shown),
and obtained the miRNA-mRNA, miRNA-lncRNA network.
Secondly, according to the correlation of gene
expression of 222 mRNAs and 954 lncRNAs, we constructed the
lncRNA-mRNA co-expression network (data not shown).
At last, based on the interaction network of
miRNA-mRNA, miRNA-lncRNA, and lncRNA-mRNA, we obtained 1049
feed-forward loop networks and constructed the general
miRNA-lncRNA-mRNA interaction network (data not shown). In this
network, we got 148 miRNAs targeted lncRNAs, and these 148 lncRNAs
were identified as ceRNAs and listed in Table II.
 | Table II.146 lncRNAs as ceRNAs in the
miRNA-lncRNA-mRNA interaction network. |
Table II.
146 lncRNAs as ceRNAs in the
miRNA-lncRNA-mRNA interaction network.
| No. | lncRNA
accession | No. | lncRNA
accession |
|---|
| 1 |
TCONS_l2_00014900-XLOC_l2_008262 | 75 | n334798 |
| 2 |
TCONS_l2_00020697-XLOC_l2_010802 | 76 | n338832 |
| 3 |
TCONS_00016931-XLOC_007962 | 77 | n340212 |
| 4 |
TCONS_l2_00016969-XLOC_l2_008979 | 78 | n340287 |
| 5 |
TCONS_00008436-XLOC_003882 | 79 | n382215 |
| 6 |
TCONS_l2_00016970-XLOC_l2_008980 | 80 | n405896 |
| 7 |
TCONS_l2_00017319-XLOC_l2_008977 | 81 | n410552 |
| 8 | NR_002813 | 82 | NR_027755 |
| 9 |
TCONS_l2_00012202-XLOC_l2_006419 | 83 |
TCONS_00012035-XLOC_005569 |
| 10 | n409500 | 84 |
TCONS_00014701-XLOC_006799 |
| 11 | n341154 | 85 |
TCONS_00017991-XLOC_008840 |
| 12 |
TCONS_l2_00010766-XLOC_l2_005781 | 86 |
TCONS_00024504-XLOC_011823 |
| 13 | n342063 | 87 |
ENST00000448869 |
| 14 | n386722 | 88 |
ENST00000506895 |
| 15 |
TCONS_00003006-XLOC_001678 | 89 | n326341 |
| 16 | NR_024387 | 90 | n332799 |
| 17 |
TCONS_00007677-XLOC_003770 | 91 | n335107 |
| 18 |
TCONS_00019368-XLOC_009191 | 92 | n335724 |
| 19 | n409372 | 93 | n337920 |
| 20 | n342249 | 94 | n338102 |
| 21 | n408882 | 95 | n338270 |
| 22 |
TCONS_00022673-XLOC_010971 | 96 | n340792 |
| 23 | n334377 | 97 | n378134 |
| 24 | NR_024076 | 98 | n382996 |
| 25 | NR_033360 | 99 | n406201 |
| 26 | NR_038399 | 100 | n407038 |
| 27 |
TCONS_00016279-XLOC_007654 | 101 | n408084 |
| 28 |
TCONS_00026192-XLOC_012669 | 102 | n409198 |
| 29 |
TCONS_00028418-XLOC_013771 | 103 | NR_036676 |
| 30 |
ENST00000440955 | 104 |
OTTHUMT00000318709 |
| 31 | n341106 | 105 |
TCONS_00009631-XLOC_004772 |
| 32 | n409178 | 106 |
TCONS_00012197-XLOC_005754 |
| 33 | NR_027995 | 107 |
TCONS_00026389-XLOC_012738 |
| 34 |
TCONS_00011934-XLOC_005442 | 108 |
TCONS_l2_00020780-XLOC_l2_010854 |
| 35 |
TCONS_00028427-XLOC_013779 | 109 |
TCONS_l2_00025904-XLOC_l2_013423 |
| 36 |
TCONS_00011225-XLOC_005777 | 110 |
ENST00000422082 |
| 37 |
TCONS_00028805-XLOC_013878 | 111 |
ENST00000468202 |
| 38 | n338895 | 112 |
ENST00000511994 |
| 39 | n339264 | 113 |
ENST00000513211 |
| 40 | n381942 | 114 |
ENST00000514265 |
| 41 | NR_002808 | 115 |
ENST00000546710 |
| 42 |
TCONS_00020860-XLOC_010143 | 116 |
ENST00000550035 |
| 43 |
TCONS_00024807-XLOC_011645 | 117 | n324907 |
| 44 |
TCONS_l2_00011136-XLOC_l2_006021 | 118 | n325126 |
| 45 | n338919 | 119 | n325417 |
| 46 | n339042 | 120 | n325964 |
| 47 | n341945 | 121 | n332690 |
| 48 | n342142 | 122 | n333177 |
| 49 | n411744 | 123 | n333293 |
| 50 | NR_038849 | 124 | n335571 |
| 51 |
TCONS_00009713-XLOC_004905 | 125 | n339266 |
| 52 |
ENST00000414772 | 126 | n339695 |
| 53 |
ENST00000427446 | 127 | n340730 |
| 54 |
ENST00000508925 | 128 | n340955 |
| 55 |
ENST00000546560 | 129 | n342367 |
| 56 | n340540 | 130 | n342493 |
| 57 | n342109 | 131 | n345192 |
| 58 | n379390 | 132 | n345550 |
| 59 | n411650 | 133 | n346330 |
| 60 |
TCONS_00028187-XLOC_013543 | 134 | n346394 |
| 61 |
TCONS_l2_00021562-XLOC_l2_010625 | 135 | n383730 |
| 62 | n342193 | 136 | n408256 |
| 63 | n342616 | 137 | n408302 |
| 64 | n342622 | 138 | n410892 |
| 65 | n406194 | 139 | n411520 |
| 66 | n406899 | 140 | NR_033752 |
| 67 | n411603 | 141 | NR_033826 |
| 68 |
OTTHUMT00000363702 | 142 |
OTTHUMT00000370627 |
| 69 |
TCONS_00016056-XLOC_007446 | 143 |
TCONS_00000861-XLOC_000086 |
| 70 |
TCONS_00022099-XLOC_010709 | 144 |
TCONS_00003277-XLOC_002089 |
| 71 |
TCONS_l2_00012572-XLOC_l2_006751 | 145 |
TCONS_00008003-XLOC_003469 |
| 72 |
ENST00000452532 | 146 |
TCONS_00017956-XLOC_008786 |
| 73 |
ENST00000507838 | 147 |
TCONS_00025335-XLOC_012147 |
| 74 | n325350 | 148 |
TCONS_00029068-XLOC_013994 |
Biological role prediction of lncRNAs
function as ceRNAs in CLDN5-overexpressing hCMEC/D3 cells
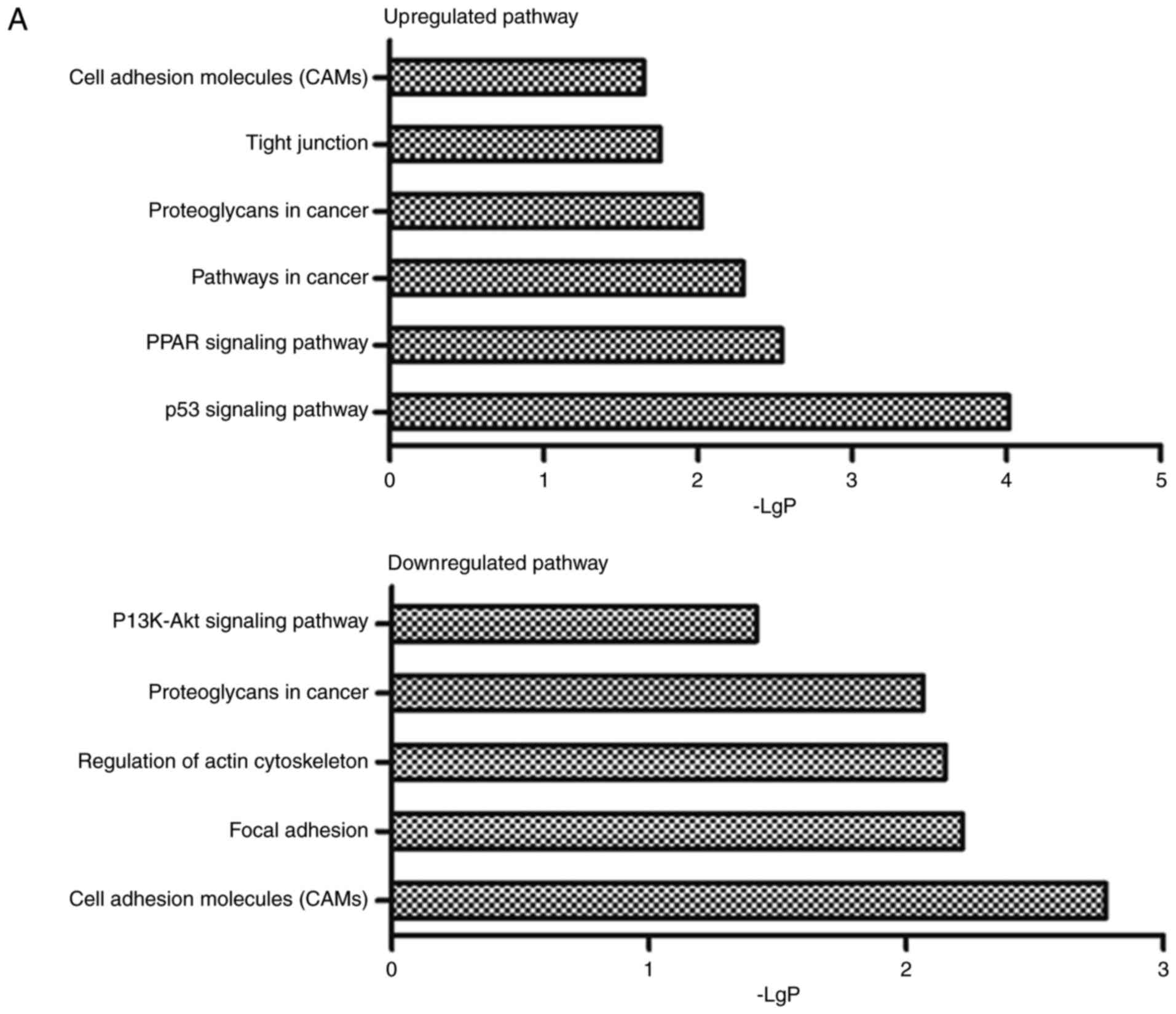
GO and pathway analysis were applied to analyze the
significant function and pathway of the mRNAs that contained in the
1049 feed-forward loop networks. Go analysis results showed that
upregulated and downregulated mRNA respectively, were involved in
122 and 81 items significant functions (P-value <0.05, data not
shown). The pathway analysis revealed that there were 29 and 6
significant pathways corresponding to the up and down regulated
mRNAs separately (P-value <0.05, data not shown).
Further analysis indicated there are 10 mRNAs that
contained both in the significant function and pathway (Table III), and they participated in 95
feed-forward loop networks. The miRNAs, lncRNAs, and mRNAs in 95
feed-forward loop are listed in Table
IV.
 | Table III.The 95 feed-forward loops including
miRNAs, lncRNAs and mRNAs. |
Table III.
The 95 feed-forward loops including
miRNAs, lncRNAs and mRNAs.
| No. | miRNA | lncRNA | mRNA |
|---|
| 1 |
hsa-miR-4677-3p | n325350 | IGFBP3 |
| 2 |
hsa-miR-487a-3p |
TCONS_00012035-XLOC_005569 | PIK3CG |
| 3 |
hsa-miR-6760-5p |
TCONS_00012035-XLOC_005569 | PIK3CG |
| 4 | hsa-miR-3135b | NR_038399 | DOCK1 |
| 5 |
hsa-miR-6760-5p | NR_038399 | DOCK1 |
| 6 | hsa-miR-297 | NR_033360 | IGFBP3 |
| 7 |
hsa-miR-4677-3p | NR_033360 | IGFBP3 |
| 8 | hsa-miR-297 |
TCONS_00009713-XLOC_004905 | IGFBP3 |
| 9 | hsa-miR-4289 |
TCONS_00009713-XLOC_004905 | IGFBP3 |
| 10 | hsa-miR-619-5p |
TCONS_00009713-XLOC_004905 | IGFBP3 |
| 11 | hsa-miR-7-5p |
TCONS_00009713-XLOC_004905 | IGFBP3 |
| 12 |
hsa-miR-4726-5p |
TCONS_l2_00011136-XLOC_l2_006021 | DOCK1 |
| 13 |
hsa-miR-6760-5p |
TCONS_l2_00011136-XLOC_l2_006021 | DOCK1 |
| 14 |
hsa-miR-323a-3p |
TCONS_00003006-XLOC_001678 | CDKN1B |
| 15 |
hsa-miR-3912-5p |
TCONS_00003006-XLOC_001678 | CDKN1B |
| 16 | hsa-miR-543 |
TCONS_00003006-XLOC_001678 | CDKN1B |
| 17 |
hsa-miR-6760-5p |
TCONS_00003006-XLOC_001678 | CDKN1B |
| 18 |
hsa-miR-7845-5p |
TCONS_00003006-XLOC_001678 | CDKN1B |
| 19 |
hsa-miR-3912-5p |
TCONS_00003006-XLOC_001678 | PHLPP1 |
| 20 |
hsa-miR-6760-5p |
TCONS_00003006-XLOC_001678 | PHLPP1 |
| 21 |
hsa-miR-7845-5p |
TCONS_00003006-XLOC_001678 | PHLPP1 |
| 22 | hsa-miR-297 |
TCONS_l2_00021562-XLOC_l2_010625 | IGFBP3 |
| 23 | hsa-miR-4289 |
TCONS_l2_00021562-XLOC_l2_010625 | IGFBP3 |
| 24 |
hsa-miR-4677-3p |
TCONS_l2_00021562-XLOC_l2_010625 | IGFBP3 |
| 25 | hsa-miR-619-5p |
TCONS_l2_00021562-XLOC_l2_010625 | IGFBP3 |
| 26 | hsa-miR-7-5p |
TCONS_l2_00021562-XLOC_l2_010625 | IGFBP3 |
| 27 | hsa-miR-297 | n409198 | IGFBP3 |
| 28 |
hsa-miR-4677-3p | n409198 | IGFBP3 |
| 29 | hsa-miR-3135b | n406899 | ARHGEF12 |
| 30 | hsa-miR-4462 | n406899 | ARHGEF12 |
| 31 | hsa-miR-654-3p | n406899 | ARHGEF12 |
| 32 |
hsa-miR-7845-5p | n406899 | ARHGEF12 |
| 33 | hsa-miR-3135b |
TCONS_l2_00014900-XLOC_l2_008262 | DOCK1 |
| 34 |
hsa-miR-323a-3p |
TCONS_l2_00014900-XLOC_l2_008262 | DOCK1 |
| 35 | hsa-miR-329-3p |
TCONS_l2_00014900-XLOC_l2_008262 | DOCK1 |
| 36 | hsa-miR-4462 |
TCONS_l2_00014900-XLOC_l2_008262 | DOCK1 |
| 37 |
hsa-miR-4726-5p |
TCONS_l2_00014900-XLOC_l2_008262 | DOCK1 |
| 38 | hsa-miR-6076 |
TCONS_l2_00014900-XLOC_l2_008262 | DOCK1 |
| 39 |
hsa-miR-6760-5p |
TCONS_l2_00014900-XLOC_l2_008262 | DOCK1 |
| 40 | hsa-miR-921 |
TCONS_l2_00014900-XLOC_l2_008262 | DOCK1 |
| 41 |
hsa-miR-6760-5p | n346394 | CADM1 |
| 42 |
hsa-miR-4665-5p |
ENST00000546560 | PPARD |
| 43 |
hsa-miR-323a-3p |
TCONS_l2_00020697-XLOC_l2_010802 | CDKN1B |
| 44 |
hsa-miR-3912-5p |
TCONS_l2_00020697-XLOC_l2_010802 | CDKN1B |
| 45 | hsa-miR-543 |
TCONS_l2_00020697-XLOC_l2_010802 | CDKN1B |
| 46 |
hsa-miR-6760-5p |
TCONS_l2_00020697-XLOC_l2_010802 | CDKN1B |
| 47 |
hsa-miR-7845-5p |
TCONS_l2_00020697-XLOC_l2_010802 | CDKN1B |
| 48 | hsa-miR-4289 | n335571 | IGFBP3 |
| 49 | hsa-miR-4278 |
ENST00000452532 | PPARD |
| 50 |
hsa-miR-3912-5p |
TCONS_l2_00020697-XLOC_l2_010802 | PHLPP1 |
| 51 |
hsa-miR-4646-5p |
TCONS_l2_00020697-XLOC_l2_010802 | PHLPP1 |
| 52 |
hsa-miR-6760-5p |
TCONS_l2_00020697-XLOC_l2_010802 | PHLPP1 |
| 53 |
hsa-miR-7845-5p |
TCONS_l2_00020697-XLOC_l2_010802 | PHLPP1 |
| 54 | hsa-miR-4289 | n333293 | CDKN1A |
| 55 | hsa-miR-3135b | n409500 | DOCK1 |
| 56 | hsa-miR-4462 | n409500 | DOCK1 |
| 57 |
hsa-miR-4726-5p | n409500 | DOCK1 |
| 58 | hsa-miR-6076 | n409500 | DOCK1 |
| 59 |
hsa-miR-6760-5p | n409500 | DOCK1 |
| 60 | hsa-miR-297 | n408084 | OCLN |
| 61 | hsa-miR-4647 | n408084 | OCLN |
| 62 | hsa-miR-3135b |
TCONS_l2_00020697-XLOC_l2_010802 | DOCK1 |
| 63 |
hsa-miR-323a-3p |
TCONS_l2_00020697-XLOC_l2_010802 | DOCK1 |
| 64 | hsa-miR-4462 |
TCONS_l2_00020697-XLOC_l2_010802 | DOCK1 |
| 65 |
hsa-miR-4726-5p |
TCONS_l2_00020697-XLOC_l2_010802 | DOCK1 |
| 66 |
hsa-miR-6760-5p |
TCONS_l2_00020697-XLOC_l2_010802 | DOCK1 |
| 67 | hsa-miR-921 |
TCONS_l2_00020697-XLOC_l2_010802 | DOCK1 |
| 68 | hsa-miR-619-5p |
TCONS_00029068-XLOC_013994 | CDKN1A |
| 69 | hsa-miR-3135b | NR_024387 | ARHGEF12 |
| 70 | hsa-miR-4462 | NR_024387 | ARHGEF12 |
| 71 | hsa-miR-654-3p | NR_024387 | ARHGEF12 |
| 72 |
hsa-miR-7845-5p | NR_024387 | ARHGEF12 |
| 73 |
hsa-miR-3912-5p |
TCONS_00008436-XLOC_003882 | PHLPP1 |
| 74 |
hsa-miR-4646-5p |
TCONS_00008436-XLOC_003882 | PHLPP1 |
| 75 |
hsa-miR-6760-5p |
TCONS_00008436-XLOC_003882 | PHLPP1 |
| 76 |
hsa-miR-7845-5p |
TCONS_00008436-XLOC_003882 | PHLPP1 |
| 77 | hsa-miR-297 |
ENST00000452532 | OCLN |
| 78 |
hsa-miR-4786-5p | n339695 | CLDN1 |
| 79 |
hsa-miR-4726-5p | n332799 | DOCK1 |
| 80 | hsa-miR-127-5p | n338895 | PIK3CG |
| 81 | hsa-miR-378e | n338895 | PIK3CG |
| 82 |
hsa-miR-5585-3p | n338895 | PIK3CG |
| 83 |
hsa-miR-6760-5p | n338895 | PIK3CG |
| 84 |
hsa-miR-7845-5p | n338895 | PIK3CG |
| 85 | hsa-miR-297 |
TCONS_00022673-XLOC_010971 | OCLN |
| 86 | hsa-miR-3138 |
TCONS_00022673-XLOC_010971 | OCLN |
| 87 | hsa-miR-4647 |
TCONS_00022673-XLOC_010971 | OCLN |
| 88 |
hsa-miR-4786-5p |
TCONS_00022673-XLOC_010971 | OCLN |
| 89 | hsa-miR-619-5p |
TCONS_00022673-XLOC_010971 | OCLN |
| 90 | hsa-miR-7-5p |
TCONS_00022673-XLOC_010971 | OCLN |
| 91 | hsa-miR-767-5p |
TCONS_00022673-XLOC_010971 | OCLN |
| 92 |
hsa-miR-7844-5p |
TCONS_00022673-XLOC_010971 | OCLN |
| 93 | hsa-miR-3138 |
TCONS_00024504-XLOC_011823 | CDKN1A |
| 94 |
hsa-miR-4677-3p |
TCONS_00024504-XLOC_011823 | CDKN1A |
| 95 | hsa-miR-619-5p |
TCONS_00024504-XLOC_011823 | CDKN1A |
 | Table IV.Functional prediction of the lncRNA
ceRNAs based on pathway and GO analysis of mRNAs that locate
together in the miRNA-lncRNA-mRNA feed-forward loop. |
Table IV.
Functional prediction of the lncRNA
ceRNAs based on pathway and GO analysis of mRNAs that locate
together in the miRNA-lncRNA-mRNA feed-forward loop.
| No. | mRNA | Pathway | GO |
|---|
| 1 | IGFBP3 | P53 signaling | Negative regulation
of cell proliferation |
|
|
|
| Regulation of cell
growth |
|
|
|
| Negative regulation
of protein phosphorylation |
|
|
|
| Apoptotic
process |
| 2 | PIK3CG | Focal adhesion | Negative regulation
of apoptotic process |
|
|
| Regulation of actin
cytoskeleton |
|
|
|
| Proteoglycans in
cancer |
|
|
|
| PI3K-Akt
signaling |
|
| 3 | CDKN1A | P53 signaling | Negative regulation
of cell proliferation/cell growth |
|
|
| Pathways in
cancer | Intrinsic apoptotic
signaling |
|
|
| Proteoglycans in
cancer | Negative regulation
of cyclin-dependent protein |
|
|
|
| serine/threonine
kinase activity |
|
|
|
| Negative regulation
of phosphorylation |
|
|
|
| Negative regulation
of apoptotic process |
| 4 | CDKN1B | PI3K-Akt
signaling | Negative regulation
of apoptotic process |
|
|
|
| Cell cycle
arrest |
| 5 | PHLPP1 | PI3K-Akt
signaling | Apoptotic
process |
| 6 | ARHGEF12 | Regulation of actin
cytoskeleton |
|
|
|
| Proteoglycans in
cancer |
|
| 7 | PPARD | PPAR signaling
pathway | Apoptotic
signaling |
|
|
| Pathways in
cancer |
|
| 8 | CADM1 | Cell adhesion
molecules | Homophilic cell
adhesion |
|
|
|
| Apoptotic
process |
|
|
|
| Heterophilic
cell-cell adhesion |
|
|
|
| Unidimensional cell
growth |
|
|
|
| Cell-cell junction
organization |
| 9 | CLDN1 | Tight junction | Cell adhesion |
|
|
| Cell adhesion
molecules | Cell-cell junction
organization |
| 10 | DOCK1 | Focal
adhesion; | Apoptotic
process |
|
|
| Regulation of actin
cytoskeleton |
|
| 11 | OCLN | Apoptotic
process; | Tight junction |
|
|
| Cell-cell junction
organization | Cell adhesion
molecules |
The biological roles of 27 lncRNAs acting as ceRNAs
were predicted through GO and pathway analysis of 10 mRNAs in the
miRNA-lncRNA-mRNA interaction network. In addition, the results
indicated that 10 mRNAs participated in 6 upregulated and 5
downregulated pathways involving in diverse biological processes,
including tight junction, focal adhesion and cell adhesion
molecules, and 10 upregulated and 5 downregulated functions that
containing cell adhesion, heterophilic cell-cell adhesion, cell
growth, cell apoptosis, etc (Fig.
3).
As a consequence, we predicted the important roles
of the 27 ceRNAs in CLDN-5-overexpressing hCMEC/D3 cells.
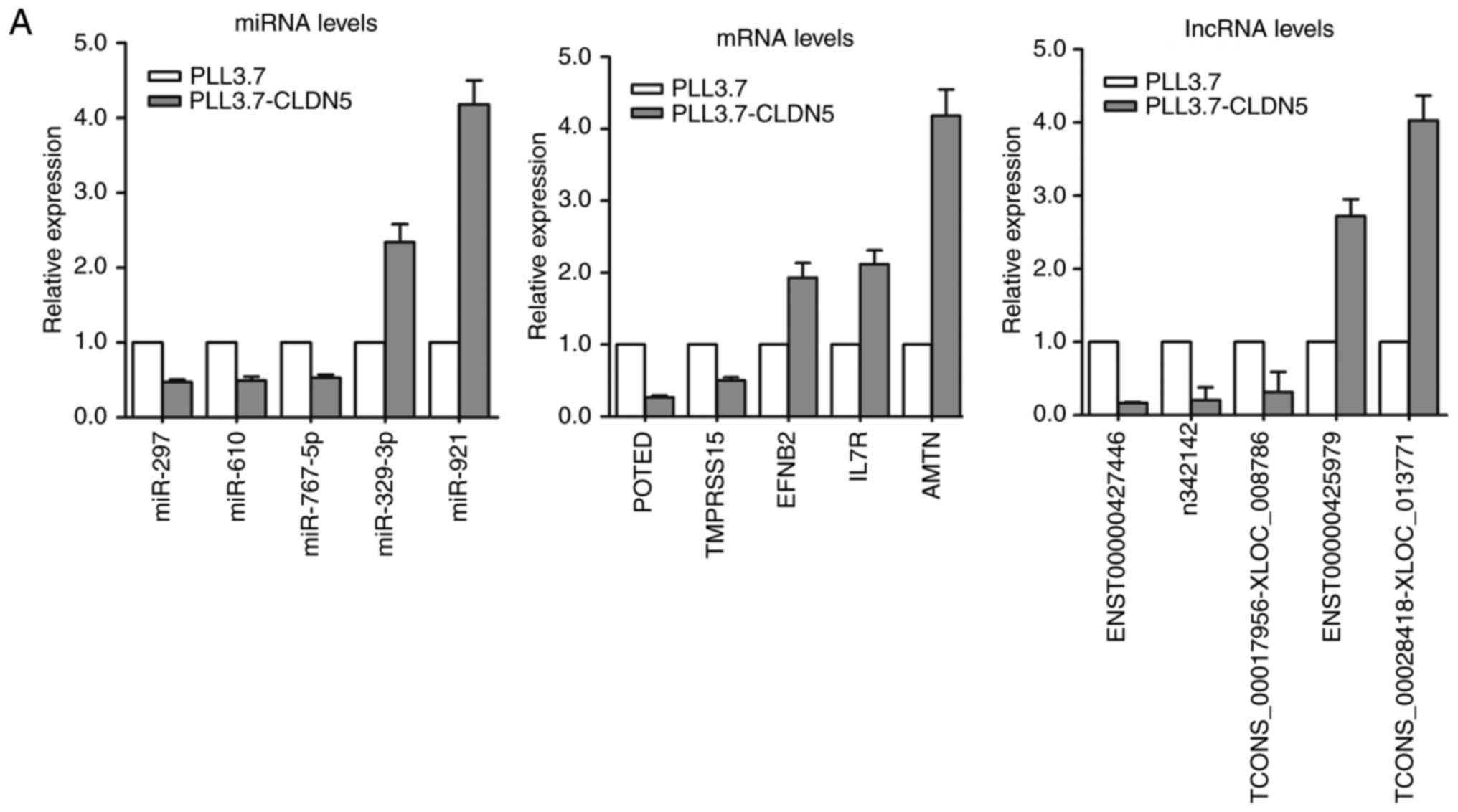
Quantitative real-time RT-PCR
(qRT-PCR) analysis of the distinctive expression of miRNAs,
lncRNAs, and mRNAs in hCMEC/D3 cells with overexpressed CLDN5
To validate the results of microarray analysis, we
selected 5 miRNAs, 5 mRNAs, and 5 lncRNAs with larger fold-change
from the microarray results and analyzed their expression levels by
qRT-PCR in pLL3.7-CLDN5 and pLL3.7 hCMEC/D3 cells. Our results
confirmed the findings of the miRNAs, lncRNAs and mRNAs microarray
dataset (Fig. 4A).
Based on the analysis of 95 miRNA-lncRNA-mRNA
feed-forward loops in Table IV, we
evaluated the expression levels of 3 miRNA, 3 lncRNA, and 3 mRNA
that are respectively located in 3 feed-forward loops. The average
expression level of miR-127-5p was significantly increased, while
miR-4786-5p and miR-297 were reduced in CLDN5-overexpressing
hCMEC/D3 cells compared with control hCMEC/D3 cells. Analysis
showed relatively high expression of miR-127-5p and low expression
of n338895 and PIK3CG, and low expression of miR-4786-5p and
miR-297 and high expression of n339695, TCONS_00022673-XLOC_010971
and CLDN1, OCLN (Fig. 4B). The 3
feed-forward loops detection by qRT-PCR are represented in Fig. 4C.
Discussion
In recent years, studies have confirmed the
dysregulation of lncRNAs by acting as ceRNAs have profound
implications for tumor metastasis. Reports demonstrate that lncRNAs
as ceRNAs play an important role in blood-tumor barrier (BTB), and
their dysfunction leads to the change of permeability of BTB.
Several models have been proposed to explain how ceRNAs regulate
BTB. Examples include lncRNA TUG1, NEAT1, XIST, MALAT1, which
function in ceRNA manner to regulate expression of transcript
factor (TF), such as HSF2, SOX5, FOXC1/ZO-2 or nuclear factor (NF)
NFYA through targeting different miR-144, miR-181d-5p, miR-137, and
miR-140. These TFs and NF regulate the expression of claudin-5,
occludin, and ZO-1, and then impact the permeability of BTB.
Knockdown of lncRNAs TUG1, NEAT1, XIST, and MALAT1 resulted in
increased the permeability of BTBs as well as decreased the
expression of claudin-5, occludin, and ZO-1 (21–26).
Although regulatory networks of ceRNA have now been shown to
contribute to the permeability of BTB, the dysregulation of ceRNAs
as a consequence of alterations in CLDN5 levels in BTB has not been
previously investigated. In this study, we provide a comprehensive
analysis of the roles of CLDN5 mediating the junctional and
adhesion molecules, and signaling pathways through ceRNA
interaction network in brain endothelial cells.
First of all, we used the Affymetrix GeneChip HTA
2.0 Array to analyze the distinct lncRNAs and mRNAs in
CLDN5-overexpressing hCMEC/D3 cells. The sample matched miRNA
expression profiling was determined by the Affymetrix GeneChip
miRNA 4.0 Array. We identified a set of 41 miRNAs, 954 lncRNAs, and
222 mRNAs that differently expressed between
pLL3.7-CLDN5-transfected and pLL3.7 control group. Such
differentiation signified the potential roles of CLDN5 in cerebral
vascular endothelial cells.
Furthermore, we discussed the effect of miRNA
competition on the regulation of both lncRNAs and mRNAs, as well as
the implications of lncRNA function as ceRNA through constructing
the complex miRNA-lncRNA-mRNA interaction network. The possible
biology functions of these regulatory ceRNAs mainly include tight
junction, focal adhesion, cell-cell adhesion, cell growth and
apoptosis. To our knowledge, this is the first study to show the
lncRNA acting as ceRNAs were associated with clautin-5 function in
brain vascular endothelial cells.
Our qRT-PCR expression analysis confirmed there are
a series of miRNAs, lncRNAs, and mRNAs aberrantly expressed in the
CLDN5-overexpressing hCMEC/D3 cells, which indicated that the
differently expressed non-coding and coding RNAs might be
characteristics regulated by CLDN5.
The expression of miR-127 was found to be
upregulated, whereas miR-4786-5p and miR-297 were downregulated by
CLDN5 overexpression. CLDN5 mediated interactions between
metastasis tumor cells and brain endothelial cells. Recent studies
showed miR-127 was aberrantly downregulated and acted as a
functional tumor suppressor in several brain metastatic tumors,
such as breast cancer (27),
osteosarcoma (28), lung cancer
(29), hepatocellular carcinoma
(30), while miR-297 acts as an
oncogene and is upregulated in lung adenocarcinoma and osteosarcoma
(31,32). Based on the findings obtained above,
increased miR-127 and decreased miR-297 expression levels in
claudin-5-overexpressing brain vascular endothelial cells can make
BBB impermeable and reduce brain metastasis from cancer.
The PIK3CG, CLDN1 and OCLN mRNA regulated by ceRNA
play key roles in the BBB. PIK3CG participates in the PI3K-Akt
signaling pathway involved in regulation of numerous important cell
processes including cell growth, differentiation, and metabolism.
PI3K also was found to be among a couple of genes that were
specifically altered in brain metastases of various tumor entities
(33,34), confirming previous reports that
found specific alterations of the PI3K-Akt pathway in melanoma
(35) and non-small lung cancer
(36) brain metastases. The results
of our study suggest that the PI3K-Akt pathway might be a promising
target to prevent brain metastases by overexpression of claudin-5
through ceRNA. Occludin, claudin-1, and claudin-5 are all TJ
proteins, and they interact with other junctional proteins and play
an important role in several pathologies, including tumor brain
metastasis (37,38). Our findings are particularly
interesting in the dysregulated expression of CLDN1 and OCLN mRNA,
in conjunction with the high CLDN5 levels.
Overall, our study identified and analyzed lncRNA
function as ceRNA in CLDN5-overexpressing cerebral vascular
endothelial cells, and in which ceRNA works as a downstream
effector of the CLDN5 to strengthen several signaling pathways and
mediate its role in maintaining BBB permeability. Understanding
these ceRNA regulating networks would advance the development of
prevention and therapy strategies for tumor brain metastasis.
Our findings indicated CLDN5 plays a key role in
regulating BBB permeability through ceRNA network, so the
activation of CLDN5 can inhibit tumor brain metastasis, which would
be applied in medical treatment. Recent reports demonstrated that
some proteases degrade tight junction protein, including CLDN5, and
their inhibitor or antagonist prevents degration of tight junction
protein and attenuates BBB disruption. Examples include telomerase
(39,40), and matrix metalloproteinases (MMPs)
(41). Several models have been
proposed to explain how telomerase regulated tight junction
protein. First, telomerase activity may affect CLDN5; moreover AZT
as an inhibitor of telomerase could modulate CLDN5 expression; at
last, catalytic component of the telomerase holoenzyme may affect
wnt signaling pathways (40).
Earlier studies have shown that MMPs and their inhibitors-TIMPs
play an essential role in the permission of drugs to cross the BBB
(42,43).
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (no. 81471229) and the Natural Science
Foundation of Beijing City (nos. 7152098 and 7142054).
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cardoso FL, Brites D and Brito MA: Looking
at the blood-brain barrier: Molecular anatomy and possible
investigation approaches. Brain Res Rev. 64:328–363. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tam SJ and Watts RJ: Connecting vascular
and nervous system development: Angiogenesis and the blood-brain
barrier. Annu Rev Neurosci. 33:379–408. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hawkins BT and Davis TP: The blood-brain
barrier/neurovascular unit in health and disease. Pharmacol Rev.
57:173–185. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bauer HC, Traweger A, Zweimueller-Mayer J,
Lehner C, Tempfer H, Krizbai I, Wilhelm I and Bauer H: New aspects
of the molecular constituents of tissue barriers. J Neural Transm
(Vienna). 118:7–21. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Eichler AF, Chung E, Kodack DP, Loeffler
JS, Fukumura D and Jain RK: The biology of brain
metastases-translation to new therapies. Nat Rev Clin Oncol.
8:344–356. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Furuse M, Fujita K, Hiiragi T, Fujimoto K
and Tsukita S: Claudin-1 and −2: Novel integral membrane proteins
localizing at tight junctions with no sequence similarity to
occludin. J Cell Biol. 141:1539–1550. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ohtsuki S, Yamaguchi H, Katsukura Y,
Asashima T and Terasaki T: mRNA expression Levels of tight junction
protein genes in mouse brain capillary endothelial cells highly
purified by magnetic cell sorting. J Neurochem. 104:147–154.
2008.PubMed/NCBI
|
|
8
|
Nitta T, Hata M, Gotoh S, Seo Y, Sasaki H,
Hashimoto N, Furuse M and Tsukita S: Size-selective loosening of
the blood-brain barrier in claudin-5-deficient mice. J Cell Biol.
161:653–660. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ouban A and Ahmed AA: Claudins in human
cancer: A review. Histol Histopathol. 25:83–90. 2010.PubMed/NCBI
|
|
10
|
Zamore PD and Haley B: Ribo-gnome: The big
world of small RNAs. Science. 309:1519–1524. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: The ceRNA hypothesis: The rosetta stone of a hidden
RNA language. Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Karreth FA, Tay Y, Perna D, Ala U, Tan SM,
Rust AG, DeNicola G, Webster KA, Weiss D, Perez-Mancera PA, et al:
In vivo identification of tumor-suppressive PTEN ceRNAs in an
oncogenic BRAF-induced mouse model of melanoma. Cell. 147:382–395.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tay Y, Kats L, Salmena L, Weiss D, Tan SM,
Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, et al:
Coding-independent regulation of the tumor suppressor PTEN by
competing endogenous mRNAs. Cell. 147:344–357. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y,
Chen N, Sun F and Fan Q: CREB up-regulates long non-coding RNA,
HULC expression through interaction with microRNA-372 in liver
cancer. Nucleic Acids Res. 38:5366–5383. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fan M, Li X, Jiang W, Huang Y, Li J and
Wang Z: A long non-coding RNA, PTCSC3, as a tumor suppressor and a
target of miRNAs in thyroid cancer cells. Exp Ther Med.
5:1143–1146. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Panzitt K, Tschernatsch MM, Guelly C,
Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder
R, Trauner M and Zatloukal K: Characterization of HULC, a novel
gene with striking up-regulation in hepatocellular carcinoma, as
noncoding RNA. Gastroenterology. 132:330–342. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jendrzejewski J, He H, Radomska HS, Li W,
Tomsic J, Liyanarachchi S, Davuluri RV, Nagy R and de la Chapelle
A: The polymorphism rs944289 predisposes to papillary thyroid
carcinoma through a large intergenic noncoding RNA gene of tumor
suppressor type. Proc Natl Acad Sci. 109:pp. 8646–8651. 2012;
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Johnsson P, Ackley A, Vidarsdottir L, Lui
WO, Corcoran M, Grandér D and Morris KV: A pseudogene
long-noncoding-RNA network regulates PTEN transcription and
translation in human cells. Nat Struct Mol Biol. 20:440–446. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Prieto C, Risueño A, Fontanillo C and De
las Rivas J: Human gene coexpression landscape: Confident network
derived from tissue transcriptomic profiles. PLoS One. 3:e39112008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cai H, Xue Y, Wang P, Wang Z, Li Z, Hu Y,
Li Z, Shang X and Liu Y: The long noncoding RNA TUG1 regulates
blood-tumor barrier permeability by targeting miR-144. Oncotarget.
6:19759–19779. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhao L, Wang P, Liu Y, Ma J and Xue Y:
miR-34c Regulates the permeability of blood-tumor barrier via
MAZ-mediated expression changes of ZO-1, Occludin, and Claudin-5. J
Cell Physiol. 230:716–731. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Miao YS, Zhao YY, Zhao LN, Wang P, Liu YH,
Ma J and Xue YX: MiR-18a increased the permeability of BTB via
RUNX1 mediated down-regulation of ZO-1, occludin and claudin-5.
Cell Signal. 27:156–167. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Guo J, Cai H, Zheng J, Liu X, Liu Y, Ma J,
Que Z, Gong W, Gao Y, Tao W and Xue Y: Long non-coding RNA NEAT1
regulates permeability of the blood-tumor barrier via
miR-181d-5p-mediated expression changes in ZO-1, occludin, and
claudin-5. Biochim Biophys Acta. 1863:2240–2254. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yu H, Xue Y, Wang P, Liu X, Ma J, Zheng J,
Li Z, Li Z, Cai H and Liu Y: Knockdown of long non-coding RNA XIST
increases blood-tumor barrier permeability and inhibits glioma
angiogenesis by targeting miR-137. Oncogenesis. 6:e3032017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ma J, Wang P, Yao Y, Liu Y, Li Z, Liu X,
Li Z, Zhao X, Xi Z, Teng H, et al: Knockdown of long non-coding RNA
MALAT1 increases the blood-tumor barrier permeability by
up-regulating miR-140. Biochim Biophys Acta. 1859:324–338. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang S, Li HJ, Wang J, Wang D, Yao A and
Li Q: Prognostic and biological significance of microRNA-127
expression in human breast cancer. Dis Markers. 2014:4019862014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang J, Hou W, Chai M, Zhao H, Jia J, Sun
X, Zhao B and Wang R: MicroRNA-127-3p inhibits proliferation and
invasion by targeting SETD8 in human osteosarcoma cells. Biochem
Biophys Res Commun. 469:1006–1011. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shi L, Wang Y, Lu Z, Zhang H, Zhuang N,
Wang B, Song Z, Chen G, Huang C, Xu D, et al: miR-127 promotes EMT
and stem-like traits in lung cancer through a feed-forward
regulatory loop. Oncogene. 36:1631–1643. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huan L, Bao C, Chen D, Li Y, Lian J, Ding
J, Huang S, Liang L and He X: MicroRNA-127-5p targets the
biliverdin reductase B/nuclear factor-κB pathway to suppress cell
growth in hepatocellular carcinoma cells. Cancer Sci. 107:258–266.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sun Y, Zhao J, Yin X, Yuan X, Guo J and Bi
J: miR-297 acts as an oncogene by targeting GPC5 in lung
adenocarcinoma. Cell Prolif. 49:636–643. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang Y and Kong D: Knockdown of lncRNA
MEG3 inhibits viability, migration, and invasion and promotes
apoptosis by sponging miR-127 in osteosarcoma cell. J Cell Biochem.
119:669–679. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bai Y, Zhang Y, Hua J, Yang X, Zhang X,
Duan M, Zhu X, Huang W, Chao J, Zhou R, et al: Silencing
microRNA-143 protects the integrity of the blood-brain barrier:
Implications for methamphetamine abuse. Sci Rep. 6:356422016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Brastianos PK, Carter SL, Santagata S,
Cahill DP, Taylor-Weiner A, Jones RT, Van Allen EM, Lawrence MS,
Horowitz PM, Cibulskis K, et al: Genomic characterization of brain
metastases reveals branched evolution and potential therapeutic
targets. Cancer Discov. 5:1164–1177. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen G, Chakravarti N, Aardalen K, Lazar
AJ, Tetzlaff MT, Wubbenhorst B, Kim SB, Kopetz S, Ledoux AA, Gopal
YN, et al: Molecular profiling of patient-matched brain and
extracranial melanoma metastases implicates the PI3K pathway as a
therapeutic target. Clin Cancer Res. 20:5537–5546. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li Q, Yang J, Yu Q, Wu H, Liu B, Xiong H,
Hu G, Zhao J, Yuan X and Liao Z: Associations between
single-nucleotide polymorphisms in the PI3K-PTEN-AKT-mTOR pathway
and increased risk of brain metastasis in patients with non-small
cell lung cancer. Clin Cancer Res. 19:6252–6260. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Piontek J, Winkler L, Wolburg H, Müller
SL, Zuleger N, Piehl C, Wiesner B, Krause G and Blasig IE:
Formation of tight junction: Determinants of homophilic interaction
between classic claudins. FASEB J. 22:146–158. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wilhelm I, Molnár J, Fazakas C, Haskó J
and Krizbai IA: Role of the blood-brain barrier in the formation of
brain metastases. Int J Mol Sci. 14:1383–1411. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Huang W, Rha GB, Chen L, Seelbach MJ,
Zhang B, András IE, Bruemmer D, Hennig B and Toborek M: Inhibition
of telomerase activity alters tight junction protein expression and
induces transendothelial migration of HIV-1-infected cells. Am J
Physiol Heart Circ Physiol. 298:H1136–1145. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Armando RG, Gomez DM and Gomez DE: AZT
exerts its antitumoral effect by telomeric and non-telomeric
effects in a mammary adenocarcinoma model. Oncol Rep. 36:2731–2736.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yang Y and Rosenberg GA: MMP-mediated
disruption of claudin-5 in the blood-brain barrier of rat brain
after cerebral ischemia. Methods Mol Biol. 762:333–345. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
De Lorenzo MS, Alonso DF and Gomez DE:
Nafoxidine modulates the expression of matrix-metalloproteinase-2
(MMP-2) and tissue inhibitor of metalloproteinases-1 (TIMP-1) in
endothelial cells. Anticancer Res. 20(1A): 1–400. 2000.PubMed/NCBI
|
|
43
|
Cyr M, Calon F, Morissette M and Di Paolo
T: Estrogenic modulation of brain activity: implications for
schizophrenia and Parkinson's disease. J Psychiatry Neurosci.
27:12–27. 2002.PubMed/NCBI
|