Introduction
Breast cancer has been increasing in incidence and
remains the most frequent and the deadliest cancer in women
worldwide (1). Many factors
contribute to the development of breast cancer such as age, life
style and genetic factors (2).
However, in many cases there is no obvious predisposing factor,
supporting the view that a variety of environmental carcinogens may
play a major role in the initiation of breast cancer (3). Polycyclic aromatic hydrocarbons, such
as 7,12-dimethylbenz[a]anthracene (DMBA), are genotoxic
environmental pollutants which have been implicated in the
development of cancer and widely used in experimental models of
breast cancer (4).
While research efforts have mostly focused on the
advanced stages of DMBA-induced mammary gland tumors (5), little is known about the early
preneoplastic changes. In 2008, when the macroscopically normal
looking mammary glands of DMBA-treated rats were analyzed, they
demonstrated a wide range of histopathological and cellular
changes. They varied from increased signs of cell death to
hyperplasia, dysplasia and in situ carcinoma. The stage of
cell death was found to occur earlier than hyperplasia, and was
taken to be the first stage in the multistep process of breast
cancer development in DMBA-treated rats (6). These basic cellular dynamic events
leading to cancer are probably associated with the acquisition of
molecular alterations involving several cancer-driven genes
(7). Identification of these
alterations can thus improve our understanding of carcinogenesis
and help in the development of new diagnostic tools and therapeutic
modalities.
Tumor suppressor genes, such as P53 and BRCA, are
frequently involved in the development of breast cancer (8). While the wild-type p53 protein
suppresses cell growth, its mutated form acts as an oncogene.
Mutations in the P53 gene usually result in stabilization and
accumulation of its translated protein which is a frequent feature
of many malignant tumors (9). It
has been estimated that up to 58% of breast cancer patients have
mutated P53 gene with accumulation of altered p53 protein as
detected by immunological assays (10). Mutant forms of p53 protein lose the
ability to bind DNA and cause abnormal cell growth (11).
BRCA1 protein interacts with many nuclear proteins,
such as Rad51 and BRCA2 (12), and
consequently plays several critical functions in the cell (13). BRCA1 amino terminal ring finger
domain is involved in repression of estrogen receptor-α signaling,
modulation of DNA repair, and apoptosis. The carboxyl terminal
acidic domain of BRCA1 acts as a transcriptional activator when
linked to DNA binding domain. Moreover, BRCA1 plays a role in the
regulation of cell cycle checkpoints and centromeres (13).
Individuals carrying mutations in the BRCA1 gene
have an increased risk of developing breast and ovarian tumors
(14). Mutations in BRCA1 alone
account for ~45% of families with high incidence of breast cancer
and up to 80% of families with both breast and ovarian cancer
(15,16). It has been shown that BRCA1 knockout
mice are hypersensitive to γ-irradiation which induces chromosomal
aberrations (17). Therefore, loss
of transcriptional activation by BRCA1 is an important factor in
oncogenesis (18). BRCA1 is also
involved in the development of sporadic breast cancer by loss of
heterozygosity, downregulation of mRNA expression, and methylation
of the promoter region (19).
In the present study, preneoplastic and neoplastic
mammary gland tissues of DMBA-treated rats were processed for
protein expression and mutation analyses of P53 and BRCA1 genes.
The data provide new information on this commonly used animal model
of breast cancer.
Materials and methods
Animals and study design
Female virgin Wistar rats (43–50 days old) were
supplied by the animal facility of the College of Medicine and
Health Sciences, UAE University. The protocol described below was
approved by the Animal Research Ethics Committee of the College of
Medicine and Health Sciences, UAE University. All rats were kept in
standard conditions with 12:12 light-dark regimen and ad
libitum access to food and water.
Rats were divided into two groups. The first group
included 21 rats and used for breast cancer induction using a
single gavage of DMBA solution containing 80 mg/kg body weight
(5). The second group included 9
rats which received only vehicle (corn oil) to serve as age-matched
control. Thus, each 2 or 3 DMBA-induced rats had a control
littermate.
Mammary glands of all rats were gently palpated
every other day to detect development of any abnormal mass. After
15, 25, 30, 35 or 40 weeks of DMBA or corn oil gavage, rats in each
group were sacrificed by an overdose of anesthetic. For each rat,
the mammary glands of one side were dissected along with their
covering skin and immediately fixed for 12–24 h in Bouin's solution
and processed for immunohistochemistry. The opposite group of
mammary glands were immediately dissected and stored at −80°C for
protein and DNA analysis. In case that a mammary gland had a mass
or tumor, it was weighed and divided for immunohistochemistry and
protein/DNA analyses.
Immunohistochemical studies
Bouin's fixed tissues were dehydrated in graded
ethanol, cleared in xylene, and finally infiltrated and embedded in
paraffin. Tissue blocks were cut at 5 µm thickness. Tissues were
deparaffinized, rehydrated, and washed in phosphate-buffered saline
(PBS). Endogenous peroxidase activity was inhibited by incubating
the tissue sections in methanol containing 1% hydrogen peroxide for
30 min. The slides were placed horizontally in a humid chamber. To
ensure equal conditions for all tissue sections, slides were
drained off, area around sections were wiped dry, and circled with
a thin film using PAP-pen (DakoCytomation, Glostrup, Denmark).
Non-specific binding was blocked by incubating sections in PBS
containing 1% bovine serum albumin for 45 min. Then, sections were
incubated overnight with rabbit polyclonal anti-BRCA1 antibody,
clone I-20 (Santa Cruz Biotechnology Inc., Dallas, TX, USA) at 4°C.
This antibody is specific for the C-terminal region between codons
1823–1842. The catalyzed signal amplification CSA kit
(DakoCytomation) was used according to the manufacturer's
instructions. Tissue sections were counterstained with Harris
hematoxylin.
For quantification, two different approaches were
used. First, semi-quantitatively using the 100× objective of the
light microscope, the overall amount of cells with labeled nuclei
and those with labeled cytoplasm were estimated and scored as low
(±), medium (+), high (++), and very high (+++) expression. The
score of low characterized the moderate staining of widely
scattered cells. Medium was defined by focal moderate staining in
less than half of the cells. High was indicated by focal moderate
staining of more than half of the cells. Very high was
characterized by dark staining of more than half of the cells.
Second, quantitatively, light micrographs prepared using the 100×
objective lens were examined to determine the percentage of labeled
cells in glandular profiles and localization of the BRCA1 protein
(nuclear or cytoplasmic). Only cells with visible nuclei in the
micrographs were considered. The means of labeled cells were
compared in control and DMBA-treated rats using the ANOVA with post
hoc Tukey test and GraphPad Prism Software (GraphPad Inc., San
Diego, CA, USA).
Sodium dodecyl sulfate polyacrylamide
gel electrophoresis (SDS-PAGE) and western blot analysis
Frozen mammary gland tissues of control and
DMBA-treated rats were homogenized under liquid nitrogen
temperature and then lysed with 1 ml buffer containing: 100 mM
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 10% sucrose, 10
mM 1,4-dithio-DL-threitol, 0.1%
3-[(3-cholamidopropyl)dimethyl-ammoniol]-1-propanesulfonate, 150 mM
NaCl, and protein inhibitors: 100 mM phenylmethylsulfonyl fluoride,
0.1% leupeptin and aprotenin. Tissue lysates were centrifuged at
14,000 rpm for 30 min at 4°C. Protein concentrations in the
supernatants were determined using the Bio-Rad protein assay kit
(Bio-Rad Laboratories, Hercules, CA, USA). Equal amounts of protein
(30 µg) from each sample were mixed with 5X sample buffer
containing Tris-HCl (pH 6.8), 10% glycerol, 2% sodium dodecyl
sulfate, 2-mercaptoethanol, and bromophenol blue. Samples were
boiled for 5 min and electrophoresed on 8% polyacrylamide gel at 80
volts for 2 h. To check the expected size of the protein, Bio-Rad
prestained protein marker was used.
After electrophoresis, proteins in the gels were
transferred onto nitrocellulose membrane. Non-specific binding was
blocked by PBS containing 5% non-fat dry milk and 0.1% Tween-20 for
1 h. Following two washes in PBS-Tween, blots were incubated
overnight with anti-p53 antibody (clone PAb240; dilution 1:1,000;
DakoCytomation) at 4°C. Blots were washed with PBS-Tween, and then
incubated with horseradish peroxidase-conjugated goat anti-mouse
immunoglobulin (Ig) G (Cell Signaling Technology, Danvers, MA, USA)
at dilution 1:1,000 for 2 h at room temperature. Blots were then
washed with PBS-Tween and immunoreactive proteins in the blot were
detected using enhanced chemiluminescence reagents (Pierce
Biotechnology, Rockford, IL, USA) on X-ray film (Fuji Medical,
Japan). To confirm equal loading of proteins, the same blot was
immunoprobed with rabbit polyclonal anti-actin antibody (dilution
1:1,000; clone AC40; Sigma, St. Louis, MI, USA). Films were scanned
using BioDocAnalyze system (Biometra, Göttingen, Germany) to
estimate the intensity of the bands. The percent change in protein
band intensity as compared to control samples was determined.
DNA extraction
Genomic DNA was obtained from mammary glands of
control and DMBA-treated rats using DNA extraction kit (Qiagen,
Toronto, Canada). Tissues were quickly washed in cold PBS, weighed,
and then homogenized in a mortar at liquid nitrogen temperature.
The fine powder was mixed with cold PBS (100 µl PBS per 20 mg
tissue powder). Homogenized tissue was mixed with equal amount of
digestion buffer provided by the kit. QIAamp column containing DNA
was transferred to clean Eppendorf tube and 50 µl elution buffer
was added. The extracted DNA was stored at −20°C until used. DNA
concentration was determined by measuring the optical density at
260 nm using spectrophotometer (WPA; Cambridge, UK).
Polymerase chain reaction
PCR was performed using PuReTaq Ready-to-Go PCR
beads (Amersham Pharmacia Biotech, Uppsala, Sweden). The procedure
was carried out according to the manufacturer's instructions.
Primers of P53 gene were obtained from Pharmacia (Piscataway, NJ,
USA). BRCA1 primers were obtained from Operon Biotechnologies
(Sweden). Sequences of all primers used are listed in Table I.
 | Table I.The primers used in PCR
reactions. |
Table I.
The primers used in PCR
reactions.
| Primers |
| Sequences | Annealing
temperature (°C) |
|---|
| BRCA1/E11 | Forward |
5′-TTTCACCCATACACATTTG-3′ |
|
|
| Reverse |
5′-CCTTTGCCAATATTACCTG-3′ | 48 |
| P53/E5 | Forward |
5′-GACCTTTGATTCTTTCTCCTCTCC-3′ |
|
|
| Reverse |
5′-GGGAGACCCTGGACAACCAG-3′ | 64 |
| P53/E6-7 | Forward |
5′-CTGGTTGTCCAGGGTTCTCC-3′ |
|
|
| Reverse |
5′-CCCAACCTGGCACACAGCTT-3′ | 64 |
| P53/E8-9 | Forward |
5′-CTTACTGCCTTGTGCTGTGC-3′ |
|
|
| Reverse |
5′-CTTAAGGGTGAAATATTCTCC-3′ | 58 |
| P53/E10 | Forward |
5′-GTACTGTGAATATACTTACTTCTCC-3′ |
|
|
| Reverse |
5′-GGGCTGAGGTCACTCACC-3′ | 60 |
The PCR amplifications of P53 exons were performed
using the Techne Genius PCR thermal cycler (Burlington, NJ, USA).
Each sample was initially denatured at 95°C for 5 min, and then
subjected to 40 cycles, each included denaturation at 95°C for 1
min, annealing for 1 min, and then extension at 72°C for 1 min. A
final 5 min extension step was included. PCR products (5 µl) were
loaded into 1.5% agarose gel and run at 100 volts for 1 h at room
temperature. To determine the expected size of PCR products, 100
base pair ladder marker (Amersham Biosciences Corp., Piscataway,
NJ, USA) was used. The bands were visualized by staining the gel
with ethidium bromide (10 mg/ml) and exposing it to the UV
trans-illuminator (Life Technologies, Carlsbad, CA, USA). PCR
amplification of BRCA1 exon 11 was performed with genomic DNA, 2.5
µl of each primer at 10 pmol/µl, and DEPC water was added to a
total volume of 25 µl. The same conditions of P53 exon
amplification were applied, except that the annealing temperature
of BRCA1 exon 11 was adjusted to 48°C.
Single-strand conformation
polymorphism (SSCP)
The p53 PCR-product (5 µl) was denatured by adding 5
µl loading buffer containing formamide, xylene cyanol and
bromophenol blue and incubated at 96°C for 10 min. The mixture was
immediately chilled on ice. Then, 10 µl of the denatured PCR
products were loaded onto a 10% non-denaturating polyacrylamide gel
and run at 60 volts for 4 h at 4°C, using Bio-Rad vertical
electrophoresis. The silver staining kit (Amersham Pharmacia
Biotech) was used to stain DNA bands in the polyacrylamide gels.
The procedure was carried out according to the manufacturer's
instructions. The stained polyacrylamide gels were dried using
cellophane membrane (Promega, Fitchburg, WI, USA) and scanned using
HP Officejet scanner and HP Image Zone software.
Restriction fragment length
polymorphism (RFLP)
BamHI was used to cut the PCR product of
BRCA1. The enzymatic reaction included 3 µl of 10X enzyme buffer E
(Promega), 0.3 µl 1% bovine serum albumin (Promega), 30 U
BamHI enzyme (Promega) and 10 µl BRCA1 PCR product. The
reaction mixture was brought up to 30 µl with DEPC-treated water.
All samples were incubated overnight at 37°C in block heater
(Stuart Scientific, Staffordshire, UK). The digested products were
loaded in 10% polyacrylamide gel for 2 h at 110 V. Gels were
stained in the running TBE buffer containing ethidium bromide.
Bands were finally visualized using the UV trans-illuminator and
photographed.
Results
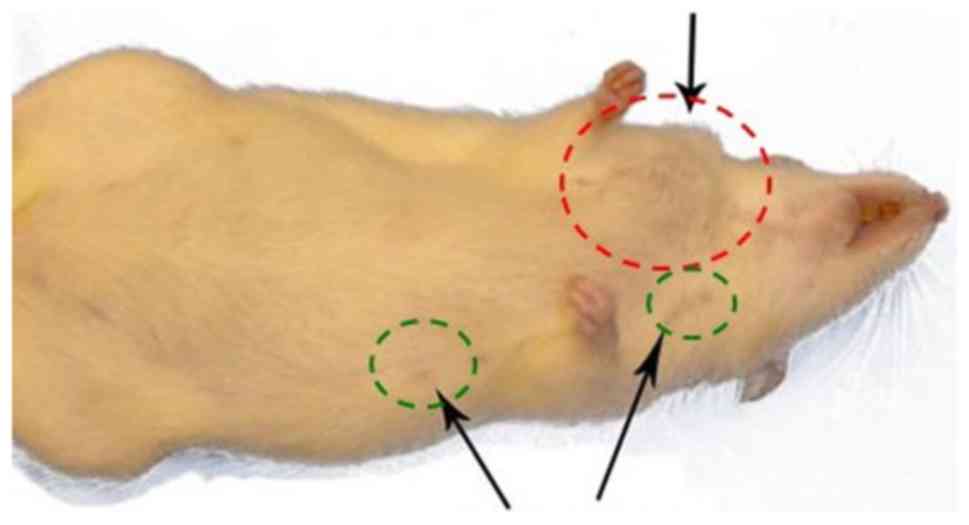
Gross observation of the mammary glands of
DMBA-treated rats revealed tumor formation in the cervical,
thoracic or abdominoinguinal regions (6). Fig. 1
shows an example of these DMBA-treated rats with a tumor in the
cervical mammary gland, but all other mammary glands appeared small
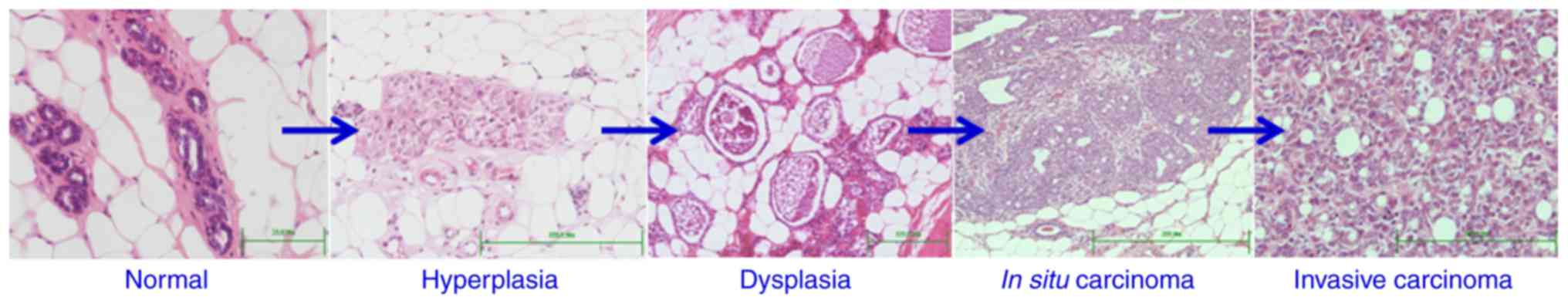
in size as those of the control. However, microscopic examination
of all the mammary glands of DMBA-treated rats sacrificed after 15,
25, 30, 35 and 40 weeks revealed the development of a wide range of
pathological changes. None of the mammary glands appeared
microscopically normal as in the control. The changes included
increased cell death, hyperplasia, dysplasia, adenoma, carcinoma
in situ and invasive carcinoma (Fig. 2). These preneoplastic and neoplastic
lesions were similar to those previously identified and
characterized (6). In addition,
mammary gland tumors were developed in ~29% of the treated rats.
Approximately 14% of all mammary glands examined developed benign
tumors and 19% were invasive or malignant.
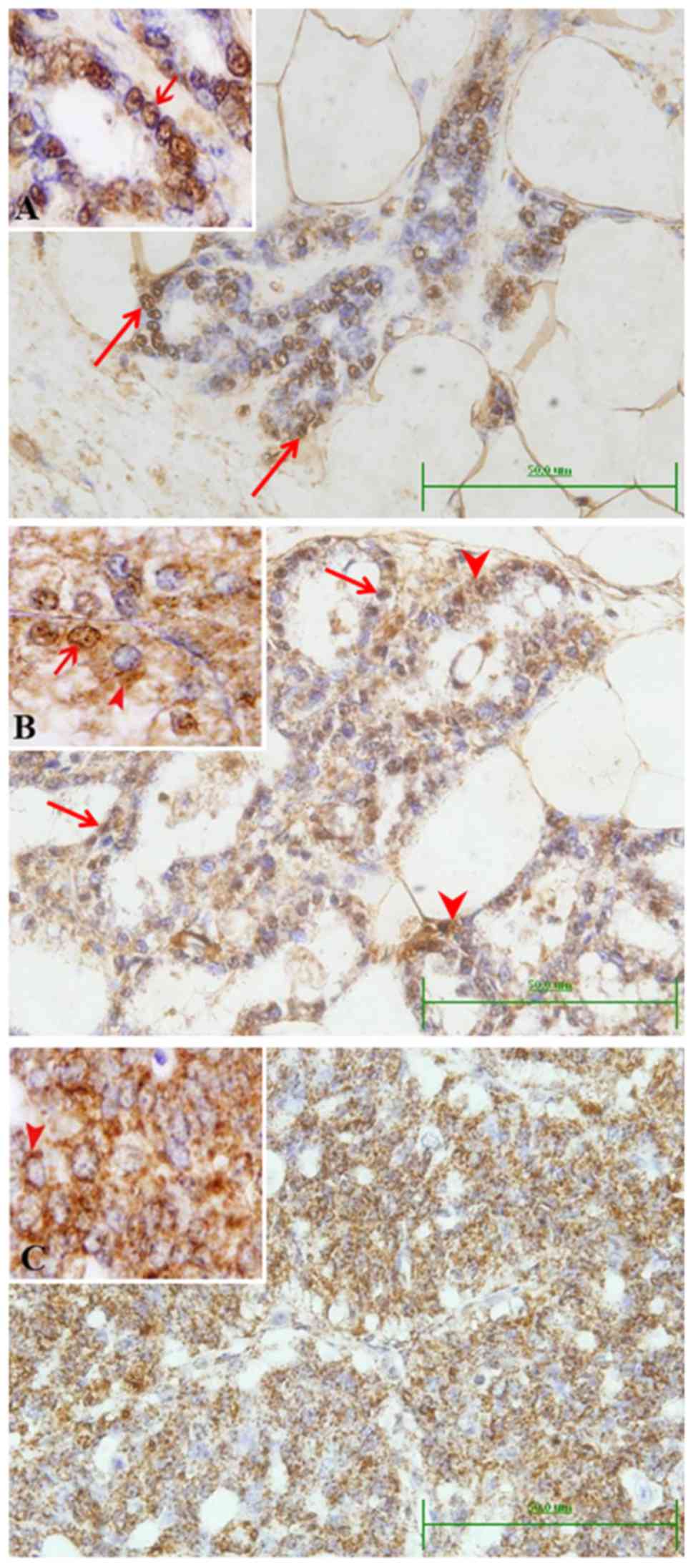
Immunohistochemical localization of
BRCA1
Control rats showed that BRCA1 protein was present
mainly in the nuclei of some luminal epithelial duct cells
(Fig. 3A). Faint cytoplasmic stain
was also observed in cells of some small ducts, which was more
apparent in the large ducts. Early microscopic lesions (hyperplasia
and dysplasia) developed in DMBA-treated rats showed both nuclear
and cytoplasmic localization of BRCA1 (Fig. 3B). This nucleo-cytoplasmic pattern
of BRCA1 expression was also noted in some neoplastic lesions
classified as lactating adenoma and squamous cell papilloma.
However, in case of localized and invasive carcinomas (in
situ cribriform, cribriform and papillary carcinoma), BRCA1
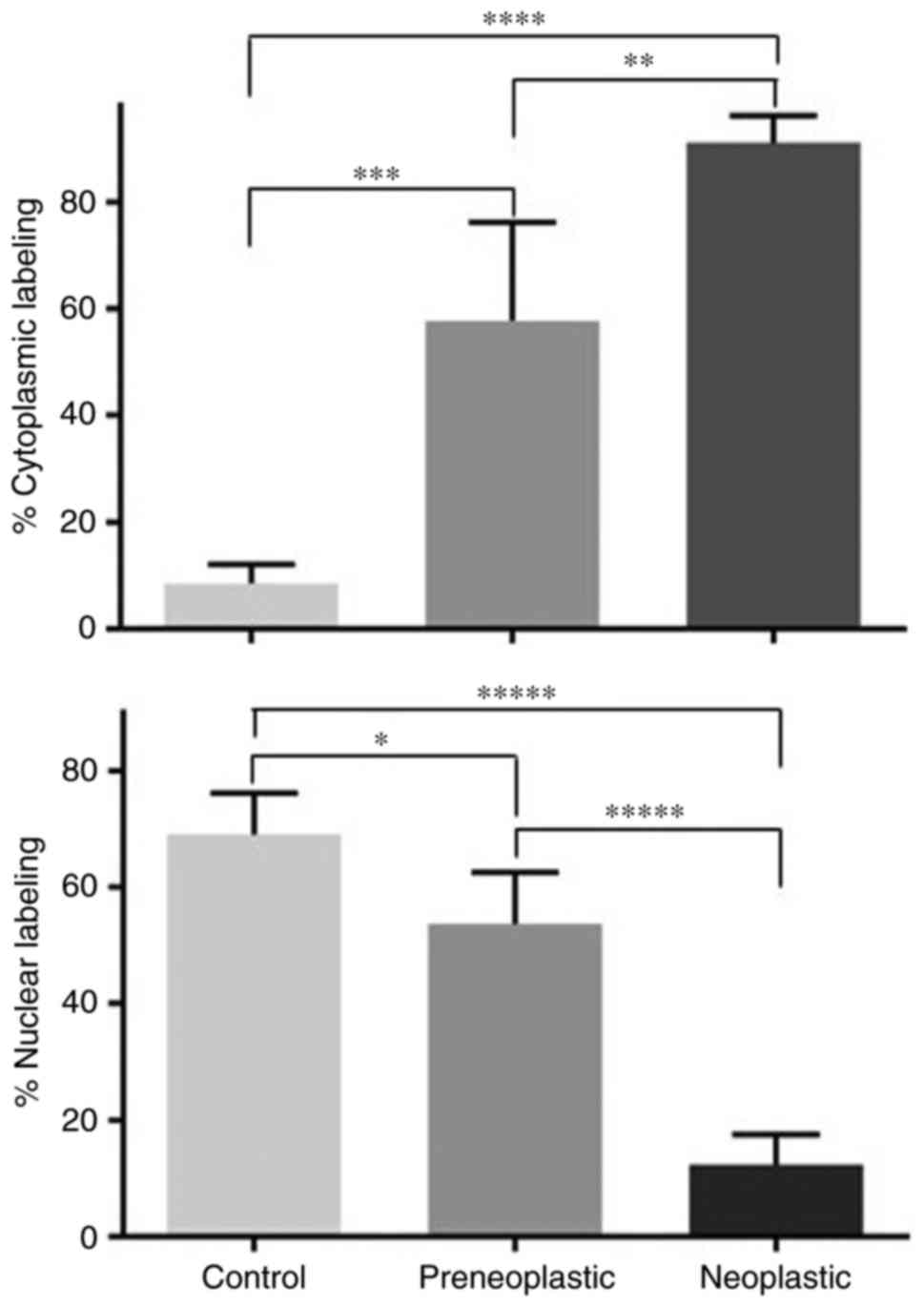
protein became mostly localized in the cytoplasm (Fig. 3C). Scoring of the immunolocalization
of BRCA1 in rat mammary glands with different histopathological
conditions are presented in Table
II. It shows a change in the pattern of the expression of BRCA1
from more nuclear in control tissues to more cytoplasmic in
DMBA-treated tissues. In addition, counts in tissue sections
obtained from different mammary glands of control and DMBA-treated
rats revealed that the percentages of cells with BRCA1 labeled
nuclei gradually dropped from 69 to 54 to 12 in control,
preneoplastic and neoplastic tissues, respectively. The differences
between these percentages of nuclear labeling were statistically
significant (Fig. 4). This change
in BRCA1 nuclear labeling was associated with a significant
increase in the percentages of cells with BRCA1 labeled cytoplasm
from 9 to 58 to 91 in control, preneoplastic and neoplastic
tissues, respectively (Fig. 4).
 | Table II.Protein expression of BRCA1 in
mammary glands of control and DMBA-induced rats. |
Table II.
Protein expression of BRCA1 in
mammary glands of control and DMBA-induced rats.
|
|
| BRCA1
expression |
|---|
|
|
|
|
|---|
| Weeks post
DMBA | Mammary glands | Nucleus | Cytoplasm |
|---|
| 15 | Control | ++ | + |
|
| Typical
hyperplasia | + | ++ |
|
| Cribriform
carcinoma | ± | +++ |
| 25 | Typical
hyperplasia | + | ++ |
| 30 | Typical
hyperplasia | + | ++ |
|
| In situ
cribriform carcinoma | ± | ++ |
| 35 | Typical
hyperplasia | + | ++ |
|
| In situ
cribriform carcinoma | ± | ++ |
|
| Lactating
adenoma | ++ | ++ |
|
| Cribriform
carcinoma | ± | +++ |
| 40 | Typical
hyperplasia | + | ++ |
|
| Squamous cell
papilloma | + | ++ |
Expression of p53 protein
Homogenized tissue samples of the mammary glands
obtained from control and DMBA-treated rats were analyzed by
western blotting to study the expression of p53 protein during
breast cancer progression. Representative results of 3 different
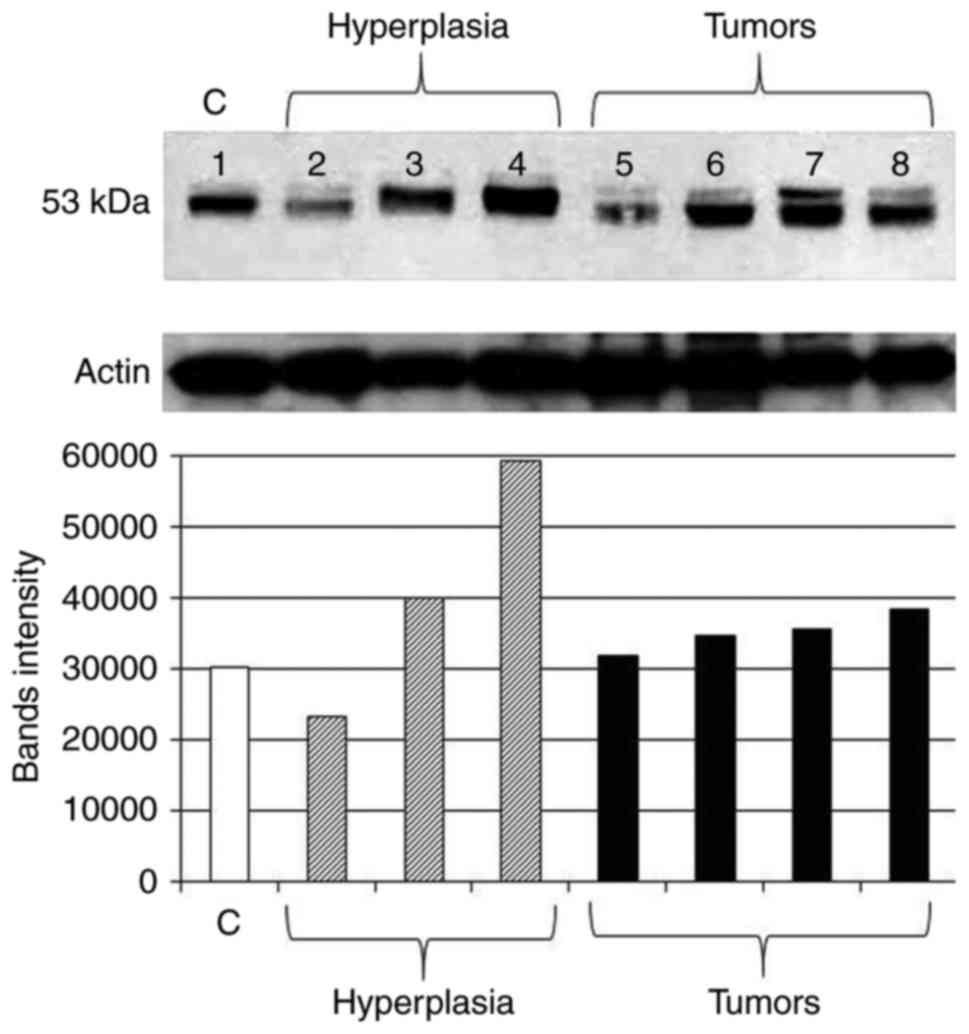
experiments are shown in Fig. 5.
Comparing to control tissue (Fig.
5; lane 1), initial downregulation in some hyperplastic and
benign lesions was observed (Fig.
5; lanes 2 and 5, respectively). These were from mammary glands
of rats treated with DMBA and sacrificed after 35 weeks. In these
rats, some mammary glands progressed toward malignancy, and
therefore, the amount of p53 was increased. This was either the
accumulated wild-type form or the mutant form detected using
anti-p53 antibody clone PAb240 (Fig.
5, lanes 3, 4 and 6–8). Measurement of band densities confirmed
these expression patterns of p53 (Fig.
5). Calculations of percent change in band intensity, as
compared to control, showed an increase in tissues with hyperplasia
by 33 and 96% (except for a 23% decrease in one case) and an
increase in all neoplastic cases by 5, 15, 18 and 27%.
SSCP analysis of P53 gene in rat
mammary glands
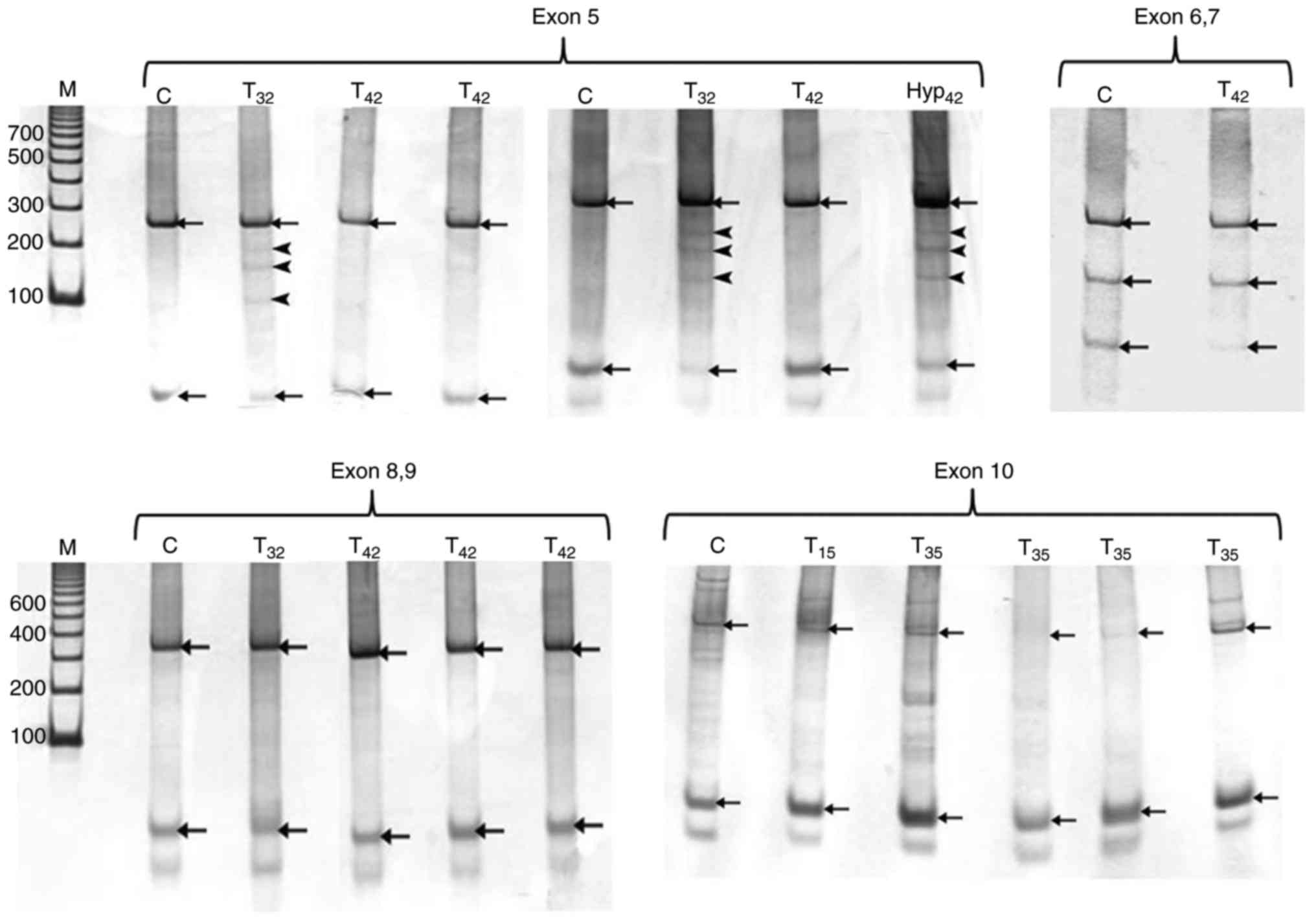
PCR products of exon 5 revealed 270 bp amplicon for
all samples when compared to the standard DNA molecular marker.
SSCP analysis of this 270 bp product of control mammary glands
showed two bands as indicated in Fig.
6. Out of 9 tumors, two of them (22%) exhibited band shift (3
extra bands) with electrophoresis in addition to the two bands
appeared in control sample. These two tumors were developed in rats
treated with DMBA and sacrificed after 25 and 32 weeks. In
addition, some hyperplastic mammary glands showed band shifting
similar to that of tumor samples as shown in Fig. 6. PCR amplification of exon 6–7
produced 300 bp band for all samples in agarose gel. SSCP analysis
displayed 3 bands representing the normal pattern of P53 exon 6–7
(Fig. 6). Analysis of DMBA-treated
samples revealed bands similar to those of control with no shift
detected in the polyacrylamide gels (Fig. 6). All PCR products of P53 exon 8–9
from control and treated mammary glands showed single amplicon
indicated by agarose gel electrophoresis at 350 bp. The SSCP
analysis of P53 exon 8–9 showed two bands similar to the pattern of
exon 5. SSCP analysis of the 410 bp fragment of exon 10 of P53 gene
showed two main bands in the control and DMBA-treated mammary gland
(Fig. 6).
RFLP analysis of BRCA1 gene in rat
mammary glands
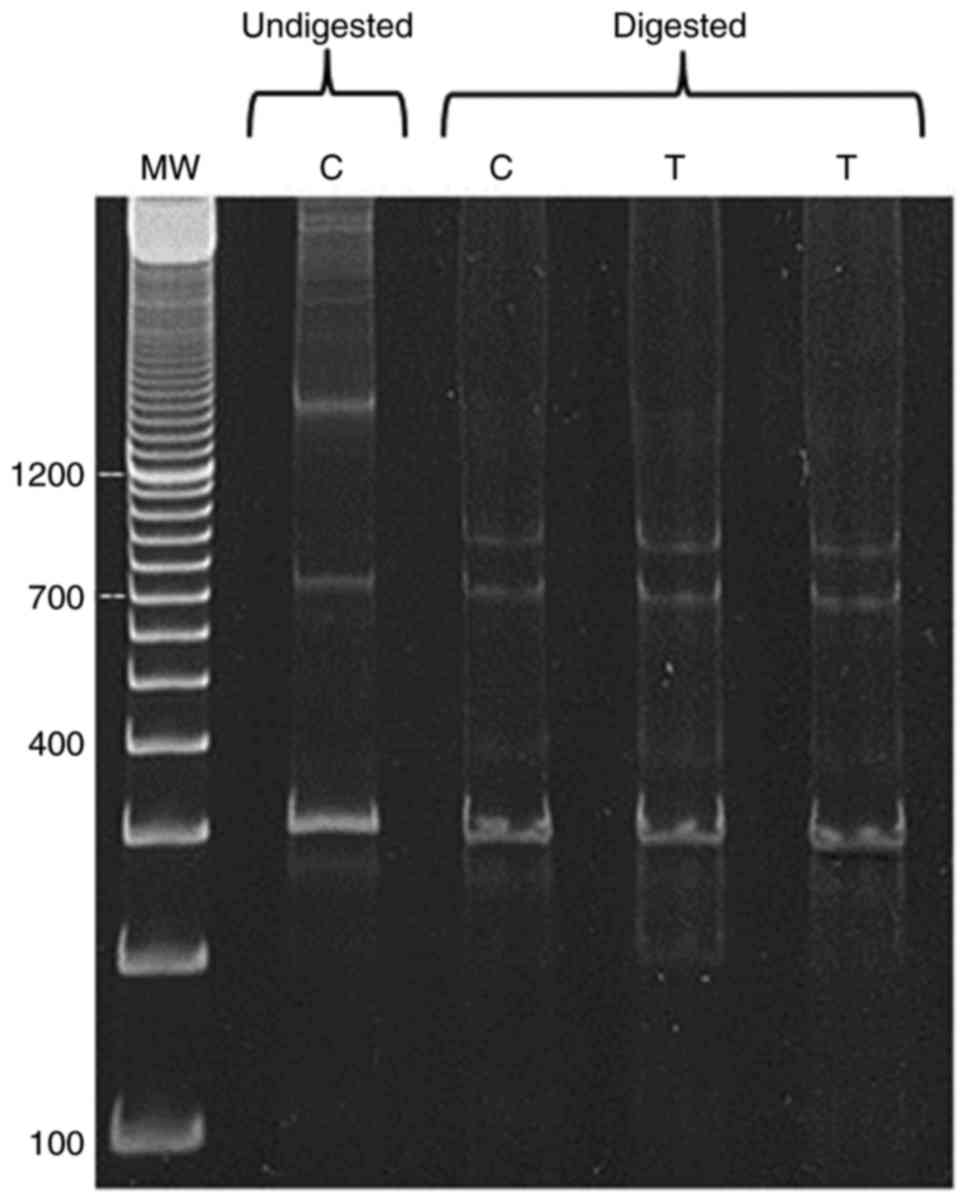
The PCR products of exon 11 of BRCA1 gene revealed
more than one band on the gel (Fig.
7, lane undigested C). Therefore, the RFLP method was used to
detect polymorphism. BamHI was found to be the appropriate
cutter for BRCA1 fragment. PCR product of BRCA1 exon 11 was at
1,600 bp, which was the biggest size obtained using specific primer
sequence mentioned in the Material and methods section. In Fig. 7, the digested products using
BamHI showed no remarkable difference between control and
tumor samples.
Discussion
The present study demonstrates that DMBA-induced
preneoplastic and neoplastic lesions develop a block in the nuclear
translocation of BRCA1 protein and upregulation and polymorphisms
of P53 gene.
Various studies previously indicated that BRCA1 acts
as a nuclear-cytoplasmic shuttle protein, and its transport is
altered by DNA damage (20,21). Thus, it seems that breast cancer
progression is associated with the production of an inactive form
of BRCA1 which cannot be targeted to the nucleus and therefore,
remains in the cytoplasm of cancer cells. This could be due to
impairment of nuclear localization signals and/or the nuclear
export signal of BRCA1 protein (20). The data presented in the present
study indicate that alterations in the localization of BRCA1
protein occur in preneoplastic lesions of the DMBA-rat model of
mammary gland carcinogenesis.
P53 is a tumor-suppressor gene which has an apparent
role in the development of breast cancer in humans and rodents
(22,23). In the present study, the expression
of p53 protein was studied in mammary glands of DMBA-treated rats
using an antibody that detects both wild-type and mutant forms of
p53 (24). Using SDS-PAGE and
western blot analysis, the p53 protein is found to be
differentially expressed during mammary gland carcinogenesis. It
tends to be downregulated in preneoplastic lesions, but becomes
upregulated in malignant tumors. These findings correlate with
previous studies in human breast cancer (25). A significant correlation between p53
mutations and p53 overexpression was reported in both canines and
humans (26,27). In addition, p53 protein expression
in benign tumors is much less compared to that in malignant lesions
(26).
Mutations in p53 gene are very common in human
cancers; they occur in 20–40% of breast cancer cases (28). In addition, 50% of cancers of colon,
stomach, and liver are characterized by p53 mutations (29). Most of these P53 mutations are
clustered within exons 4–8, which encode a highly conserved region,
containing the DNA binding domain of the protein (29,30).
In the present study, exons 5, 6–7, 8–9 and 10 of p53 gene were
studied for possible mutations that may occur during breast cancer
development. Using SSCP, it was possible to detect P53 polymorphism
in some rats with hyperplastic mammary glands (Fig. 6). In humans, previous studies
reported P53 mutations in individuals with preinvasive mammary
gland lesions: Atypical ductal hyperplasia and ductal carcinoma
in situ (31,32). In mice, overexpression of p53 and
its mutation in hyperplastic mammary glands was also reported
(33).
It has been estimated that 33.3% of the mammary
gland tumors developed in the present study acquire polymorphism in
P53 exon 5. In the DMBA-treated rats, base substitutions at A or G
in the sense strand of the cDNA accounted for 95% of all the point
mutations of P53 gene. The predominance of purine (A or G)
mutations is consistent with the fact that DMBA-adduct formation
preferentially occurs on dA and dG (34,35),
leading to depurination (36).
There may be a concern that some models of breast
cancer may not reflect the disease in humans. This is not the case
for the DMBA model. It has been demonstrated that the different
forms of preneoplastic and neoplastic histopathological changes of
DMBA animal model are very similar to those in humans (6). Therefore, the in vivo model of
DMBA-induced breast cancer is useful for understanding of disease
progression and early detection in humans. In addition, the
observations that BRCA1 and P53 genes are altered in DMBA breast
cancer model are not surprising. They were also described in human
breast cancer tissues (8,10,19).
However, the present study has probed some questions regarding p53
and BRCA1 proteins. In humans, it is known that hyperplastic
mammary gland lesions precede tumor formation. However, the
associated biochemical changes are not well characterized, at least
for mammary gland lesions due to DMBA. The present study has
demonstrated that alteration of BRCA1 subcellular localization and
upregulation and mutations of P53 expression are common changes.
Therefore, due to the similarities of the multistep process of
breast cancer development in humans and rats, the limitations of
DMBA model are minimal.
In conclusion, the present study demonstrates that
the morphological changes in mammary gland induced by a single
gavage of DMBA are associated with alterations in the expression of
p53 and BRCA1 proteins. An initial downregulation followed by
upregulation in the expression of p53 follows the sequence of the
morphological changes. This is associated with polymorphism of exon
5 of P53 gene which is detected as early as during hyperplasia.
While no change in the level of BRCA1 protein was detected, its
translocation from the cytoplasm to nucleus is blocked during
breast cancer progression. Therefore, it seems that P53 mutations
in exon 5 and blocking the nuclear translocation of BRCA1 are
important events for mammary gland carcinogenesis in DMBA-treated
rats.
Acknowledgements
The present study was supported by funds from the
UAEU and Terry Fox Foundation for Cancer Research. The kind
assistance of Ms. Salma Awad with some biochemical procedures is
highly appreciated.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dieterich M, Stubert J, Reimer T, Erickson
N and Berling A: Influence of lifestyle factors on breast cancer
risk. Breast Care. 9:407–414. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Seltenrich N: Institutes in the Lead:
Identifying environmental factors in breast cancer. Environ Health
Perspect. 124:A199–A205. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rennó AL, Alves-Júnior MJ, Rocha RM, De
Souza PC, de Souza VB, Jampietro J, Vassallo J, Hyslop S, Anhê GF,
de Moraes Schenka NG, et al: Decreased expression of stem cell
markers by simvastatin in 7,12-dimethylbenz(a)anthracene
(DMBA)-induced breast cancer. Toxicol Pathol. 43:400–410. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Russo J: Significance of rat mammary
tumors for human risk assessment. Toxicol Pathol. 43:145–170. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Al-Dhaheri WS, Hassouna I, Al-Salam S and
Karam SM: Characterization of breast cancer progression in the rat.
Ann NY Acad Sci. 1138:121–131. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ciriello G, Miller ML, Aksoy BA,
Senbabaoglu Y, Schultz N and Sander C: Emerging landscape of
oncogenic signatures across human cancers. Nat Genet. 45:1127–1133.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Peng L, Xu T, Long T and Zuo H:
Association between BRCA status and P53 status in breast cancer: A
meta-analysis. Med Sci Monit. 22:1939–1945. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rivlin N, Brosh R, Oren M and Rotter V:
Mutations in the p53 tumor suppressor gene: Important milestones at
the various steps of tumorigenesis. Genes Cancer. 2:466–474. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Elledge RM and Allred DC: The P53 tumor
suppressor gene in breast cancer. Breast Cancer Res Treat.
32:39–47. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Maslon MM and Hupp TR: Drug discovery and
mutant p53. Trends Cell Biol. 20:542–555. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jiang Q and Greenberg RA: Deciphering the
BRCA1 tumor suppressor network. J Biol Chem. 290:17724–17732. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rosen EM, Fan S, Pestell RG and Goldberg
ID: BRCA1 gene in breast cancer. J Cell Physiol. 196:19–41.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Miki Y, Swensen J, Shattuck-Eidens D,
Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM,
Ding W, et al: A strong candidate for the breast and ovarian cancer
susceptibility gene BRCA1. Science. 266:66–71. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Easton DF, Bishop DT, Ford D and Crockford
GP: Genetic linkage analysis in familial breast and ovarian cancer:
Results from 214 families. The breast cancer linkage consortium. Am
J Hum Genet. 52:678–701. 1993.PubMed/NCBI
|
|
16
|
Keshavarzi F, Javadi GR and Zeinali S:
BRCA1 and BRCA2 germline mutations in 85 Iranian
breast cancer patients. Fam Cancer. 11:57–67. 2011. View Article : Google Scholar
|
|
17
|
Shen SX, Weaver Z, Xu X, Li C, Weinstein
M, Chen L, Guan XY, Ried T and Deng CX: A targeted disruption of
the murine Brca1 gene causes gamma-irradiation
hypersensitivity and genetic instability. Oncogene. 17:3115–3124.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ottini L, D'Amico C, Noviello C, Lauro S,
Lalle M, Fornarini G, Colantuoni OA, Pizzi C, Cortesi E, Carlini S,
et al: BRCA1 and BRCA2 mutations in central and
southern Italian patients. Breast Cancer Res. 2:307–310. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fraser JA, Reeves JR, Stanton PD, Black
DM, Going JJ, Cooke TG and Bartlett JM: A role for BRCA1 in
sporadic breast cancer. Br J Cancer. 88:1263–1270. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Feng Z, Kachnic L, Zhang J, Powell SN and
Xia F: DNA damage induces p53-dependent BRCA1 nuclear export. J
Biol Chem. 279:28574–28584. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang ES and Xia F: BRCA1 16 years later:
DNA damage-induced BRCA1 shuttling. FEBS J. 277:3079–3085. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hollstein M, Sidransky D, Vogelstein B and
Harris CC: p53 mutation in human cancers. Science. 253:49–53. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zurer I, Hofseth LJ, Cohen Y, Xu-Welliver
M, Hussain SP, Harris CC and Rotter V: The role of p53 in base
excision repair following genotoxic stress. Carcinogenesis.
25:11–19. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gannon JV, Greaves R, Iggo R and Lane DP:
Activating mutation in p53 produce a common conformational effect.
A monoclonal antibody specific for the mutant form. EMBO J.
9:1595–1602. 1990.PubMed/NCBI
|
|
25
|
Andersen TI, Holm R, Nesland JM, Heimdal
KR, Ottestad L and Børresen AL: Prognostic significance of TP53
alterations in breast carcinoma. Br J Cancer. 68:540–548. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lee CH, Kim WH, Lim JH, Kang MS, Kim DY
and Kweon OK: Mutation and overexpression of p53 as a prognostic
factor in canine mammary tumors. J Vet Sci. 5:63–69. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rossner P Jr, Gammon MD, Zhang YJ, Terry
MB, Hibshoosh H, Memeo L, Mansukhani M, Long CM, Garbowski G,
Agrawal M, et al: Mutations in p53, p53 protein
overexpression and breast cancer survival. J Cell Mol Med.
13:3847–3857. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kalemi TG, Lambropoulos AF, Gueorguiev M,
Chrisafi S, Papazisis KT and Kotsis A: The association of p53
mutations and p53 codon 72, Her 2 codon 655 and MTHFR C677T
polymorphisms with breast cancer in Northern Greece. Cancer Lett.
222:57–65. 2004. View Article : Google Scholar
|
|
29
|
Jerry DJ, Kittrell FS, Kuperwasser C,
Laucirica R, Dickinson ES, Bonilla PJ, Butel JS and Medina DA:
Mammary-specific model demonstrates the role of the p53 tumor
suppressor gene in tumor development. Oncogene. 19:1052–1058. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Malisic E, Jankovic R, Slavkovic D,
Milovic-Kovacevic M and Radulovic S: P53 gene mutations and codon
72 polymorphism in ovarian carcinoma patients from Serbia. J BUON.
15:101–106. 2010.PubMed/NCBI
|
|
31
|
Keohavong P, Gao WM, Mady HH,
Kanbour-Shakir A and Melhem MF: Analysis of p53 mutations in cells
taken from paraffin-embedded tissue sections of ductal carcinoma in
situ and atypical ductal hyperplasia of the breast. Cancer Lett.
212:121–130. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhou W, Muggerud AA, Vu P, Due EU, Sørlie
T, Børresen-Dale AL, Wärnberg F and Langerød A: Full sequencing of
TP53 identifies identical mutations within in situ and invasive
components in breast cancer suggesting clonal evolution. Mol Oncol.
3:214–219. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jerry DJ, Ozbun MA, Kittrell FS, Lane DP,
Medina D and Butel JS: Mutations in p53 are frequent in the
preneolpastic stage of mouse mammary tumor development. Cancer Res.
53:3374–3381. 1993.PubMed/NCBI
|
|
34
|
Manjanatha MG, Chen JB, Shaddock JG Jr,
Harris AJ, Shelton SD and Casciano DA: Molecular analysis of
lacI mutations in Rat2™ cells exposed to
7,12-dimethylbenz[a]anthracene: Evidence for DNA sequence
and DNA strand biases for mutation. Mutat Res. 372:53–64. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chatterjee M, Janarthan M, Manivannan R,
Rana A and Chatterjee M: Combinatorial effect of fish oil (Maxepa)
and 1α,25-dihydroxyvitamin D3 in the chemoprevention of
DMBA-induced mammary carcinogenesis in rats. Chem Biol Interact.
188:102–110. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chakravarti D, Pelling JC, Cavalieri EL
and Rogan EG: Relating aromatic hydrocarbon-induced DNA adducts and
c-H-ras mutations in mouse skin papillomas: The role of
apurinic sites. Proc Natl Acad Sci USA. 92:10422–10426. 1995.
View Article : Google Scholar : PubMed/NCBI
|