Introduction
Overactive glucose metabolism termed as the ‘Warburg
effect’ plays a vital role in cancer growth. Tumor cells tend to
have a higher level of glucose uptake and glycolytic metabolism to
satisfy themselves with a high energy need (1). Therefore, targeting glucose metabolism
is considered as a therapeutic approach for cancer treatment
(2). Inhibition of glucose uptake
and metabolism that aim to generate an energy deprivation state can
facilitate the effect of other anticancer therapies.
Berberine is a botanical alkaloid from the
Ranunculaceae and Papaveraceae plant families. It is the active
component of Chinese medicine Rhizoma coptidis, and has been widely
used for treating metabolic diseases, including obesity and
diabetes (3,4). One of the rationale for this treatment
is that berberine increased cellular glucose uptake and metabolism
(5–7). On the other hand, the effect of
berberine against cancer has also been widely explored (8). Numerous studies have demonstrated that
berberine inhibits the growth of broad types of tumor cells, and is
thus recognized as a potential multispectrum anticancer therapeutic
agent (8–10). Given that cancer growth needs
overspeed glucose metabolism, it raised a problem that berberine's
promoting effect on glucose metabolism appears to be contradictory
to its anticancer effect. However, as most of the studies on
berberine's glucose metabolism function were carried out in
non-tumor metabolic tissues and cells, we hypothesized that
berberine may have a distinct role on cellular glucose metabolism
in cancer cells.
In the present study, we investigated the
berberine's effects on colon cancer cell lines. We revealed that
berberine inhibits glucose uptake and reduces the transcription of
glucose metabolism relative genes in colon cancer cells. These
effects may be mediated by the inhibition of HIF-1α protein
synthesis through suppression of mTOR pathway.
Materials and methods
Materials
Berberine, anti-rabbit IgG, anti-mouse IgG and
anti-β-actin IgG were purchased from Sigma-Aldrich (Shanghai,
China). Antibodies for HIF-1α (cat. no. 14179), mTOR (cat. no.
2983) and phospho-mTOR (Ser2448) (cat. no. 2971) were purchased
from Cell Signaling Technology Inc. (Beverly, MA, USA).
2-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-2-deoxyglucose
(2-NBDG) was purchased from Thermo Fisher Scientific (Eugene, OR,
USA). All other reagents including DFX and MG132 were obtained from
Sigma-Aldrich unless stated otherwise.
Cell lines and cell culture
Human colon cancer cell lines HCT116 and KM12C were
obtained from the Cell Bank of the Chinese Academy of Sciences
(Shanghai, China). HCT116 cells were cultured in McCoy'5A medium
(Thermo Fisher Scientific, Hudson, NH, USA). KM12C cells were
cultured in Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher
Scientific). All the medium was supplemented with 10% fetal bovine
serum (FBS; GE Healthcare Life Sciences, HyClone Laboratories,
Logan, UT, USA), 100 µg/ml of streptomycin (Life Technologies;
Thermo Fisher Scientific) and 100 units of penicillin.
MTT assay
Cell viability was detected using the
3-(4,5-dimethyl thiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay. Cells were seeded at a density of 1×104
cells/well in 96-well plates and then treated with 0–100 µM
berberine as indicated. At the time-points (24 h for HCT116 cells
and 15 h for KM12C cells), 50 µl of 5 mg/ml MTT solution was added
to each well and incubated at 37°C for 4 h. The formazan crystals
formed were then dissolved in 150 µl of dimethyl sulfoxide (DMSO),
and the absorbance of the solution was then obtained on a
microplate reader at λ570 nm. Results are presented as percentage
loss of cell viability compared with the control.
Colony-forming assay
Cells were seeded in a 6-well plate one day before
the experiment. Cells were treated with berberine for 24 h (HCT116)
or 15 h (KM12C), and then washed with phosphate-buffered saline
(PBS), harvested by trypsinization, counted, and were seeded into
6-well dishes at 600 cells/well. The cells were incubated for
another 10 days, fixed and stained with 1% crystal violet in
ethanol. Then, the colonies in 6-well plates were photographed
using a scanner (Epson Perfection V330; Epson Corp., Nagano,
Japan).
Glucose uptake assay
Glucose uptake of cells was measured by 2-NBDG
uptake as previously described (11). Briefly, the cells were seeded in a
12-well plate at a density of 70–80%, and treated with 0–100 µM of
berberine for 24 h (HCT116) or 15 h (KM12C). After treatment, the
cells were harvested and resuspended in Krebs-Ringer's HEPES Buffer
(KRB) solution, and incubated with 100 nmol/l 2-NBDG at 37°C in 5%
CO2 for 30 min. The 2-NBDG uptake reaction was
terminated by removing the incubation medium and washing the cells
twice with pre-cold PBS. Cells were resuspended in 1 µg/ml
propidium iodide (PI) solution to exclude dead cells. Glucose
uptake was determined by measuring the fluorescence intensity of
2-NBDG in PI-negative cells.
Real-time quantitative PCR
Cells were seeded in a 6-well plate at a density of
70–80% and treated with 0–100 µM berberine for 24 h (HCT116) or 15
h (KM12C). Total RNA was isolated using TRIzol reagent (Takara
Biotechnology Co., Ltd., Dalian, China). RNA was
reverse-transcribed into cDNA using PrimeScript™ RT
reagent kit (Takara Biotechnology Co., Ltd.) according to the
manufacturer's protocol. Real-time quantitative polymerase chain
reaction (PCR) was carried out with the SYBR-Green I fluorescent
dye method (SYBR® Premix Ex Taq™ II; Takara
Biotechnology Co., Ltd.) and the StepOnePlus Real-Time PCR
apparatus (Applied Biosystems; Thermo Fisher Scientific). The
sequences of primers used were as follows: Forward,
5′-TATTGCACTGCACAGGCCACATTC-3′ and reverse,
5′-TGATGGGTGAGGAATGGGTTCACA-3′ for HIF-1α; forward,
5′-GGCATTGATGACTCCAGTGTT-3′ and reverse, 5′-ATGGAGCCCAGCAGCAA-3′
for GLUT1; forward, 5′-TCACGGAGCTCAACCATGAC-3′ and reverse,
5′-CTGCAGTAGGGTGAGTGGTG-3′ for HK2; forward,
5′-GCCCGACGTGCATTCCCGATTCCTT-3′ and reverse,
5′-GACGGCTTTCTCCCTCTTGCTGACG-3′ for LDHA; forward,
5′-CGTGTACTACAATGAGGCTGC-3′ and reverse, 5′-CTGGTCTGAAGATCTGGCCG-3′
for β-tubulin. The amplification specificity was checked by melting
curve analysis. The relative expression of miRNA was calculated by
the 2−ΔΔCt method as described by Livak and Schmittgen
(12).
Western blotting
Cells at 60–80% confluence were washed with PBS and
lysed directly into SDS-PAGE loading buffer. A total of 20 µg of
protein was analyzed by SDS-PAGE and transferred to PVDF membranes.
All primary antibodies were used at 1:1,000 in 5% milk in
Tris-buffered saline with 0.05% Tween. Immunopositive bands were
visualized by Amersham ECL™ Plus Western Blotting Detection kit (GE
Healthcare, Chicago, IL, USA).
Statistical analysis
The SigmaPlot version 11.0 software package (Systat
Software, Inc., San Jose, CA, USA) was used for statistical
analysis. The results are presented as mean ± standard error (SEM).
Data were analyzed by one way analysis of variance (ANOVA) or the
Student's t-test. P<0.05 was considered to indicate a
statistically significant result.
Results
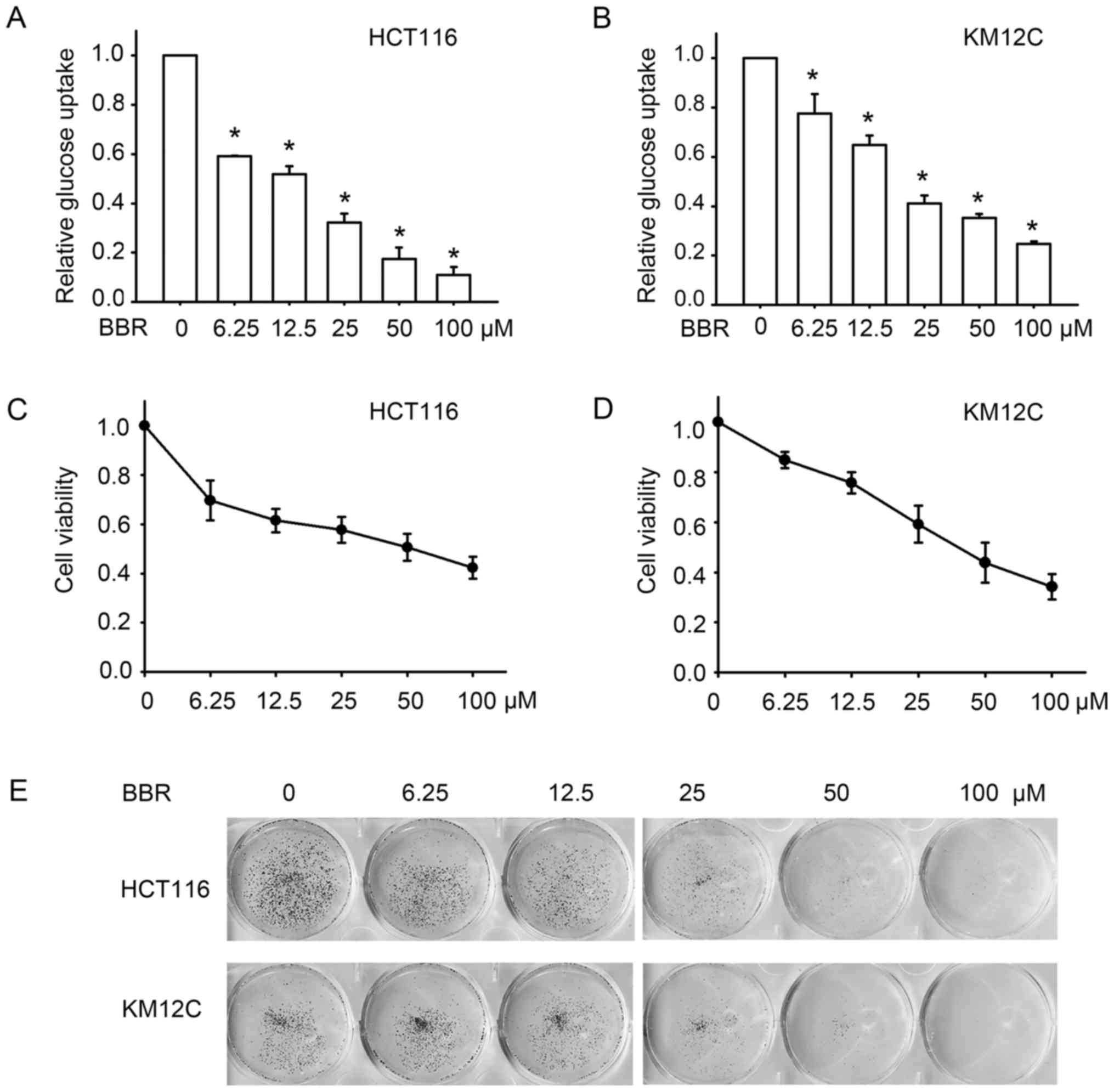
Berberine inhibits glucose uptake and
cell growth in colon cancer cells
Berberine has been shown to enhance glucose uptake
in cell lines including 3T3 adipose cells (5,13) and
L6 myotubes (7). To test its effect
on colon cancer cells, two colon cancer cell lines, HCT116 and
KM12C, were treated with 0, 6.25, 12.5, 25, 50 and 100 µM of
berberine for 24 or 15 h respectively, and the glucose uptake of
these cell lines was assessed. In contrast to berberine's reported
effect of enhancing glucose update, we unexpectedly revealed that
berberine inhibited glucose uptake in the colon cancer cell lines.
As shown in Fig. 1A and B,
treatment with the different concentrations of berberine
significantly reduced glucose uptake in these two cell lines. At
concentrations between 6.25 and 100 µM, berberine decreased glucose
uptake of HCT116 by 40–90%, and by 10–75% in KM12C cells, which was
slightly less sensitive. We then validated the anticancer effect of
berberine by detecting cell proliferation after berberine
treatment. MTT assay indicated that berberine decreased the
viability of the HCT116 and KM12C cells in a
concentration-dependent manner (Fig. 1C
and D). To further investigate the long-term effect of
berberine on cell growth, colony forming assay was performed.
Similarly, berberine inhibited the growth of colon cancer cells,
manifested as the reduction in the number of colonies of the HCT116
and KM12C cells in a concentration-dependent manner (Fig. 1E). These results indicated that
berberine inhibits glucose uptake in colon cancer cells, and these
effects may contribute to its antitumor effect.
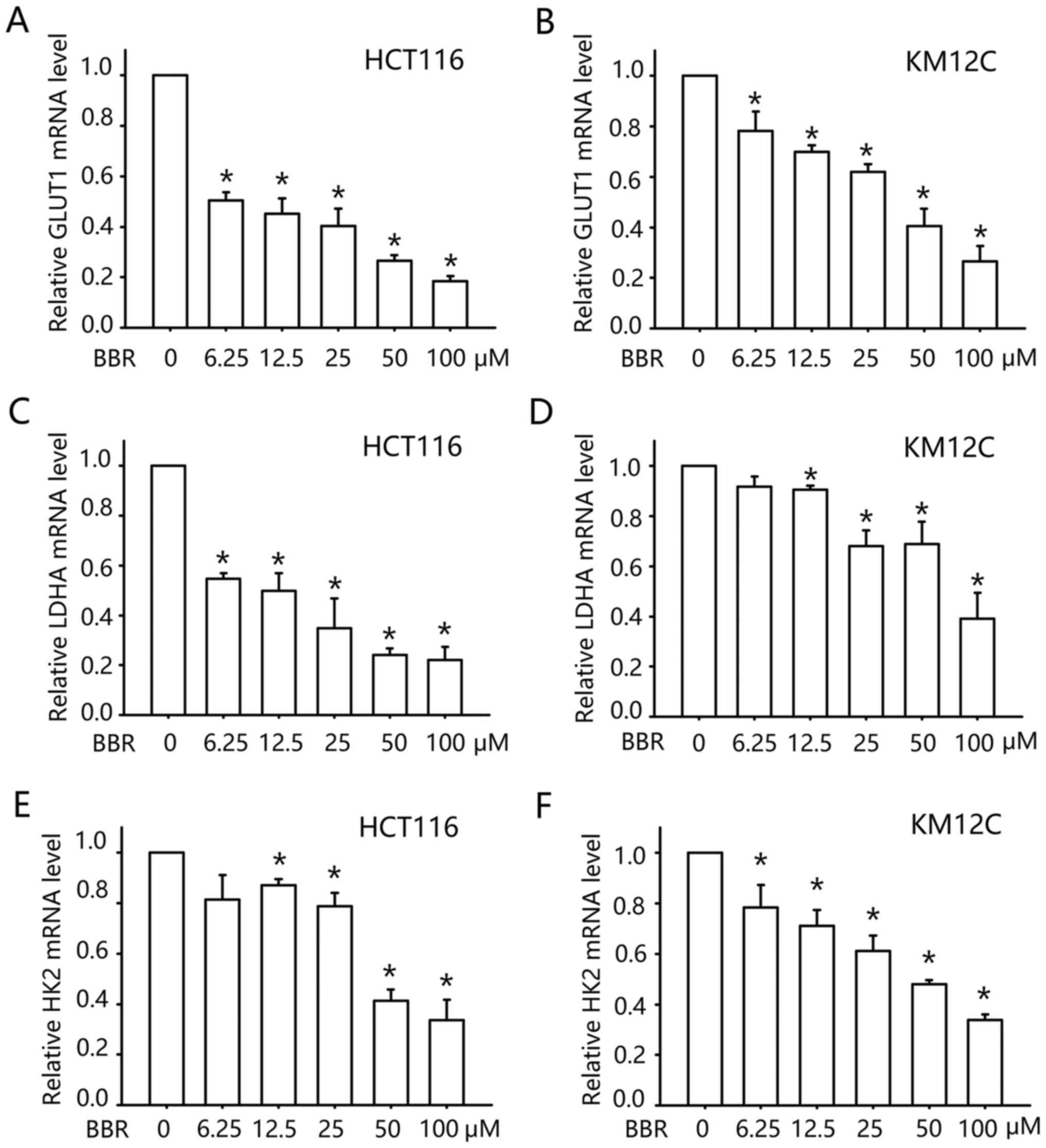
Berberine inhibits the transcription
of glucose metabolism-related gene in colon cancer cells
We then further investigated the mechanism by which
berberine inhibits glucose uptake. Glucose transporter 1 (GLUT1) is
the dominate glucose transport gene in colon cancer cells (14,15),
which facilitates the transport of glucose across the plasma
membranes. We examined the transcription of GLUT1 after berberine
treatment. As shown in Fig. 2A and
B, treatment with berberine inhibited the mRNA level of GLUT1
in a concentration-dependent manner. At concentrations between 6.25
and 25 µM, berberine decreased the mRNA level of GLUT1 in the
HCT116 cells by 50–70%, and similar to the data of glucose uptake,
KM12C cells were less sensitive. Berberine (6.25–25 µM) reduced the
mRNA level of GLUT1 by 10–35% in the KM12C cells. These data
indicated that the inhibitory effect of berberine on glucose uptake
may be mediated by the transcription inhibition of GLUT1.
The altered energy metabolism of cancer cells is not
only attained by enhancing glucose uptake, but also a higher rate
of glycolysis. Thus, we examined the transcription of two
glycolytic enzymes following berberine treatment: Lactate
dehydrogenase A (LDHA) and hexokinases 2 (HK2). LDHA catalyzes the
inter-conversion of pyruvate and L-lactate with concomitant
inter-conversion of NADH and NAD+. HK2 phosphorylates
glucose to produce glucose-6-phosphate (G6P), the first step in
most glucose metabolism pathways. As shown in Fig. 2C-F, treatment with 0–100 µM
berberine inhibited the mRNA levels of LDHA and HK2 in a
concentration-dependent manner as assessed by qPCR. Thus, our data
indicated that berberine may downregulate glucose metabolism in
colon cancer cells by inhibition of the transcription of glucose
metabolism-related genes.
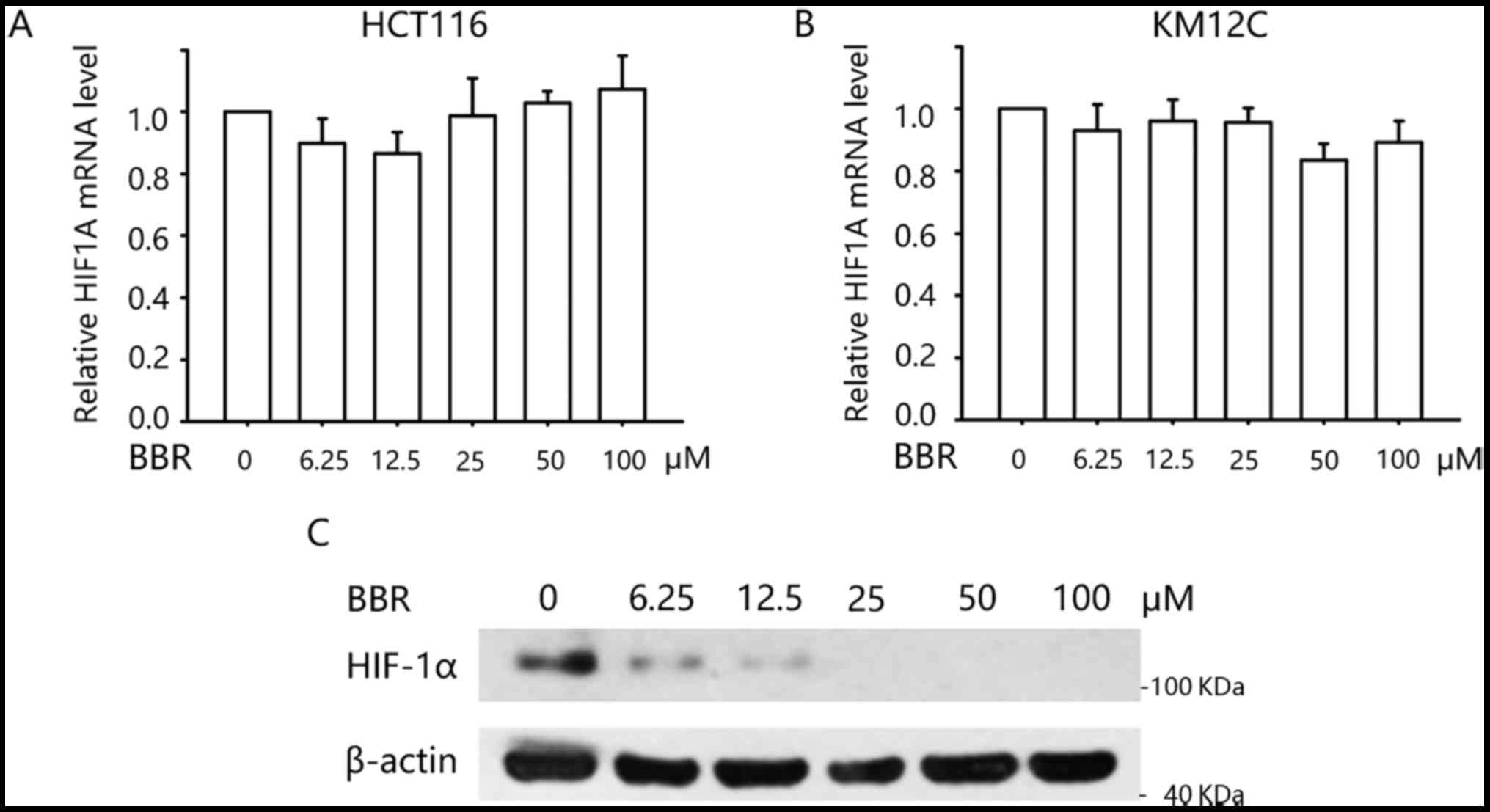
Berberine inhibits HIF-1α expression
at the post-transcriptional level
We next explored how berberine regulates the
transcription of the above glucose metabolism-related genes. It is
well known that the enhanced glucose uptake and glycolysis of
cancer cells is partly due to highly expressed HIF-1α. HIF-1α is
upregulated in low O2 concentrations but also by
oncogene activation or loss of tumor suppressors (16). Upregulated HIF-1α would further
facilitate the transcription of genes involved in glucose uptake
and glycolysis-related genes, including GLUT1, LDHA and HK2
(17,18). Therefore, we next examined the
effect of berberine on the mRNA and protein level of HIF-1α. As
determined by qPCR, we found that treatment with berberine did not
alter the mRNA level of HIF1A in the HCT116 and KM12C cells
(Fig. 3A and B). Western blotting
demonstrated that KM12C cells expressed HIF-1α protein even in
normoxic condition. Treatment with 0–100 µM berberine decreased the
protein level in a concentration-dependent manner (Fig. 3C). These data indicated that
berberine negatively modulates glucose metabolism through
regulation of HIF-1α at the post-transcriptional level.
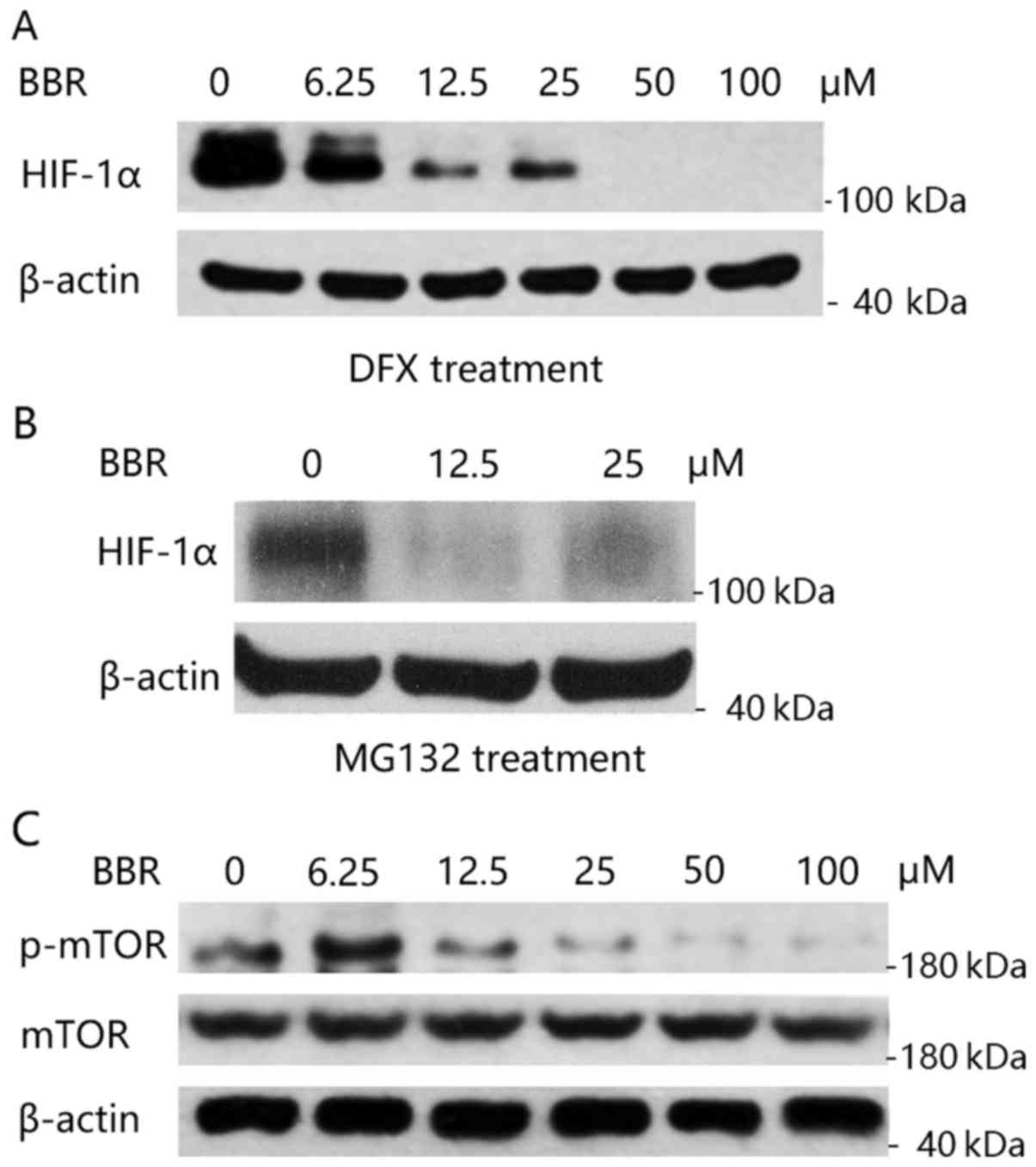
Berberine suppresses HIF-1α protein
synthesis by inhibition of the mTOR pathway
We then aimed to resolve whether berberine affects
HIF-1α degradation. It is known that HIF-1α protein degradation is
regulated by prolyl hydroxylation and then the ubiquitin-dependent
proteasome pathway (19). Briefly,
HIF-1α is hydroxylated by prolyl hydroxylases (PHD1-3) at proline
(Pro)-402 and −564. Hydroxylated HIF-1α then binds to the von
Hippel-Lindau tumor-suppressor protein (VHL). VHL is the
recognition component of an E3 ubiquitin-protein and ubiquitylated
HIF-1α is rapidly degraded by the proteasome (20). We first treated KM12C cells with
desferrioxamine (DFX), an iron chelator, which is known to inhibit
hydroxylation of HIF-1α by chelating the iron required for activity
of the HIF-1α-specific proline hydroxylases, and thus blocks HIF-1α
degradation (21). As shown in
Fig. 4A, when HIF-1α degradation
was blocked by DFX, berberine was still able to decrease the
expression of HIF-1α. Similarly, treatment with proteasome
inhibitor, MG132, was unable to inhibit berberine's effect on the
reduction of HIF-1α expression (Fig.
4B). These results demonstrated that the negative
post-transcriptional regulation of berberine on HIF-1α expression
was not by proteasomal degradation.
We then aimed to ascertain whether berberine
interrupts HIF-1α protein translation. Mammalian target of
rapamycin (mTOR) activity is a major determinant of the rate of
HIF-1α protein synthesis (22,23).
We then detected the activity of the mTOR pathway in KM12C cells
after the treatment of berberine. As shown in Fig. 4C, berberine significantly inhibited
the phosphorylation of mTOR (p-mTOR) while not altering the total
expression level of mTOR protein. Taken together, our data
indicated that berberine inhibits HIF-1α protein expression by
inhibition of the mTOR pathway, and thereby interruption of HIF-1α
protein synthesis.
Discussion
In the present study, we demonstrated that berberine
inhibits glucose uptake and cell growth in colon cancer cells. This
effect is mediated by the inhibition of the mTOR pathway, which
leads to the suppression of HIF-1α protein synthesis. The reduction
in the HIF-1α expression level may attenuate the transcription
activity of HIF-1α and decrease the transcription of glucose
metabolism-related genes, GLUT1, LDHA and HK2.
The main mechanism by which berberine exerts
antidiabetic effects relates to its glucose-lowering activity. It
has been reported that berberine improves glucose uptake in
glucose-consuming tissues, such as adipose, liver or muscle cells
(5,6,13,24,25).
In the present study, we revealed that berberine inhibited colon
cancer cell glucose uptake and metabolism, which is in contrast
from what has been reported in normal cells. Berberine has been
considered as an anticancer drug with low toxicity to normal cells
(26–28). For example, Inoue et al
reported that berberine showed higher cytotoxicity against five
human oral squamous cell carcinomas (HSC-2, HSC-3, HSC-4, NA and
CA9-22) and one human promyelocytic leukemia (HL-60) cell line,
when compared with normal human oral tissue-derived cells (gingival
fibroblasts, pulp cells and periodontal ligament fibroblasts)
(26). Chidambara et al
reported that berberine inhibited proliferation of colon cancer
cells (SW480) but showed low toxicity to normal colon cells
(CCD-CoN112) (29). The mechanisms
for this tumor-specific selective toxicity of berberine remains
elusive. Since tumor cells exhibit higher levels of glucose uptake
and glycolytic metabolism as compared to normal cells owing to
upregulated HIF-1α expression, the differential effect of berberine
on glucose uptake between normal and cancer cells may attribute to
the selectivity of berberine for cancer-targeted treatment.
HIF-1α was initially identified due to its response
to low O2 concentrations, but it can also be regulated
by oncogene activation or loss of tumor suppressors. Therefore,
upregulation of HIF-1α is quite common in many human cancers.
HIF-1α overexpression is associated with increased patient
mortality in many different types of cancer (16). Inhibition of HIF-1α activity has
marked effects on tumor growth and is considered as an anticancer
therapeutic strategy (30,31). In the present study, we identified
berberine as a novel HIF-1α inhibitor that could be used for colon
cancer treatment. Berberine reduced HIF-1α expression in normoxia
and also hypoxia conditions (DFX treatment is considered as a
hypoxia mimic). Given the different expression of HIF-1α in tumor
and normal cells, the effect of berberine on HIF-1α may account for
its selective toxicity in cancer. Consistent with our research,
there are several studies that have reported the inhibitory effect
of berberine on HIF-1α expression in other types of cancers, such
as esophageal squamous cancer (32), gastric adenocarcinoma cell line
SC-M1 (33), nasopharyngeal
carcinoma (34) and prostate cancer
(35).
The mTOR kinase integrates and transmits signals
from a diverse array of signaling pathways to regulate cell
survival and growth. Activated mTOR stimulates protein synthesis
and cell growth through phosphorylation of ribosomal protein S6
kinase (p70S6K), eukaryotic initiation factor 4E binding protein 1
(4E-BP1) and eukaryotic elongation factor 2 kinase (EEF2K)
(36). The mTOR pathway consists of
the downstream effectors of PI3K-AKT signaling (37). It is reported that berberine
inhibits PI3K-AKT signaling in colorectal cancer cells and melanoma
cells (38,39). Our finding that berberine suppresses
mTOR activity may be due to its effects on the PI3K/AKT pathway.
Between the two colon cancer cell lines that we tested, HCT116
cells were much more sensitive to berberine than KM12C cells. One
of the differences between HCT116 and KM12C cells is that KM12C
cells are PTEN-loss, while HCT116 cells are PTEN-competent. PTEN is
an upstream inhibitory mediator to mTOR. The loss of PTEN in KM12C
cells may lead to increased mTOR expression, which may affect the
sensitivity to berberine (40).
In conclusion, the present study reveals a new role
of berberine in the inhibition of tumor glucose uptake. The results
not only suggest a possible mechanism involved in berberine's tumor
selectivity but also disclose a promising therapeutic effect of
berberine in colon cancer treatment.
Acknowledgements
Not applicable.
References
|
1
|
Hsu PP and Sabatini DM: Cancer cell
metabolism: Warburg and beyond. Cell. 134:703–707. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Denko NC: Hypoxia, HIF1 and glucose
metabolism in the solid tumour. Nat Rev Cancer. 8:705–713. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cicero AF and Tartagni E: Antidiabetic
properties of berberine: From cellular pharmacology to clinical
effects. Hosp Pract. 40:56–63. 2012. View Article : Google Scholar
|
|
4
|
Pang B, Zhao LH, Zhou Q, Zhao TY, Wang H,
Gu CJ and Tong XL: Application of berberine on treating type 2
diabetes mellitus. Int J Endocrinol. 15:9057492015.
|
|
5
|
Cok A, Plaisier C, Salie MJ, Oram DS,
Chenge J and Louters LL: Berberine acutely activates the glucose
transport activity of GLUT1. Biochimie. 93:1187–1192. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhou L, Yang Y, Wang X, Liu S, Shang W,
Yuan G, Li F, Tang J, Chen M and Chen J: Berberine stimulates
glucose transport through a mechanism distinct from insulin.
Metabolism. 56:405–412. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cheng Z, Pang T, Gu M, Gao AH, Xie CM, Li
JY, Nan FJ and Li J: Berberine-stimulated glucose uptake in L6
myotubes involves both AMPK and p38 MAPK. Biochim Biophys Acta.
1760:1682–1689. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ortiz LM, Lombardi P, Tillhon M and
Scovassi AI: Berberine, an epiphany against cancer. Molecules.
19:12349–12367. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang N, Tan HY, Li L, Yuen MF and Feng Y:
Berberine and Coptidis Rhizoma as potential anticancer agents:
Recent updates and future perspectives. J Ethnopharmacol.
176:35–48. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tan W, Li Y, Chen M and Wang Y: Berberine
hydrochloride: Anticancer activity and nanoparticulate delivery
system. Int J Nanomedicine. 6:1773–1777. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zou C, Wang Y and Shen Z: 2-NBDG as a
fluorescent indicator for direct glucose uptake measurement. J
Biochem Biophys Methods. 64:207–215. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim SH, Shin EJ, Kim ED, Bayaraa T, Frost
SC and Hyun CK: Berberine activates GLUT1-mediated glucose uptake
in 3T3-L1 adipocytes. Biol Pharm Bull. 30:2120–2125. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Haber RS, Rathan A, Weiser KR, Pritsker A,
Itzkowitz SH, Bodian C, Slater G, Weiss A and Burstein DE: GLUT1
glucose transporter expression in colorectal carcinoma: A marker
for poor prognosis. Cancer. 83:34–40. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Saigusa S, Toiyama Y, Tanaka K, Okugawa Y,
Fujikawa H, Matsushita K, Uchida K, Inoue Y and Kusunoki M:
Prognostic significance of glucose transporter-1 (GLUT1) gene
expression in rectal cancer after preoperative chemoradiotherapy.
Surg Today. 42:460–469. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Semenza GL: Targeting HIF-1 for cancer
therapy. Nat Rev Cancer. 3:721–732. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Koppenol WH, Bounds PL and Dang CV: Otto
Warburg's contributions to current concepts of cancer metabolism.
Nat Rev Cancer. 11:325–337. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Semenza GL: Regulation of cancer cell
metabolism by hypoxia-inducible factor 1. Semin Cancer Biol.
19:12–16. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kallio PJ, Wilson WJ, O'Brien S, Makino Y
and Poellinger L: Regulation of the hypoxia-inducible transcription
factor 1alpha by the ubiquitin-proteasome pathway. J Biol Chem.
274:6519–6525. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee JW, Bae SH, Jeong JW, Kim SH and Kim
KW: Hypoxia-inducible factor (HIF-1)alpha: Its protein stability
and biological functions. Exp Mol Med. 36:1–12. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Demidenko ZN, Rapisarda A, Garayoa M,
Giannakakou P, Melillo G and Blagosklonny MV: Accumulation of
hypoxia-inducible factor-1alpha is limited by
transcription-dependent depletion. Oncogene. 24:4829–4838. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Abraham RT: mTOR as a positive regulator
of tumor cell responses to hypoxia. Curr Top Microbiol Immunol.
279:299–319. 2004.PubMed/NCBI
|
|
23
|
Masoud GN and Li W: HIF-1α pathway: Role,
regulation and intervention for cancer therapy. Acta Pharm Sin B.
5:378–389. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yi P, Lu FE, Xu LJ, Chen G, Dong H and
Wang KF: Berberine reverses free-fatty-acid-induced insulin
resistance in 3T3-L1 adipocytes through targeting IKKbeta. World J
Gastroenterol. 14:876–883. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang CH, Yu RY, Liu YH, Tu XY, Tu J, Wang
YS and Xu GL: Interaction of baicalin with berberine for glucose
uptake in 3T3-L1 adipocytes and HepG2 hepatocytes. J Ethnopharmaco.
151:864–872. 2014. View Article : Google Scholar
|
|
26
|
Inoue K, Kulsum U, Chowdhury SA, Fujisawa
S, Ishihara M, Yokoe I and Sakagami H: Tumor-specific cytotoxicity
and apoptosis-inducing activity of berberines. Anticancer Res.
25:4053–4059. 2005.PubMed/NCBI
|
|
27
|
Liu B, Wang G, Yang J, Pan X, Yang Z and
Zang L: Berberine inhibits human hepatoma cell invasion without
cytotoxicity in healthy hepatocytes. PLoS One. 6:e214162011.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang L, Liu L, Shi Y, Cao H, Chaturvedi R,
Calcutt MW, Hu T, Ren X, Wilson KT, Polk DB and Yan F: Berberine
induces caspase-independent cell death in colon tumor cells through
activation of apoptosis-inducing factor. PLoS One. 7:e364182012.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Murthy Chidambara KN, Jayaprakasha GK and
Patil BS: The natural alkaloid berberine targets multiple pathways
to induce cell death in cultured human colon cancer cells. Eur J
Pharmacol. 688:14–21. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yu T, Tang B and Sun X: Development of
Inhibitors Targeting hypoxia-inducible factor 1 and 2 for cancer
therapy. Yonsei Med J. 58:489–496. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Melillo G: Targeting hypoxia cell
signaling for cancer therapy. Cancer Metastasis Rev. 26:341–352.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yang X, Yang B, Cai J, Zhang C, Zhang Q,
Xu L, Qin Q, Zhu H, Ma J, Tao G, et al: Berberine enhances
radiosensitivity of esophageal squamous cancer by targeting HIF-1α
in vitro and in vivo. Cancer Biol Ther. 14:1068–1073. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lin S, Tsai SC, Lee CC, Wang BW, Liou JY
and Shyu KG: Berberine inhibits HIF-1 alpha expression via enhanced
proteolysis. Mol Pharmacol. 66:612–619. 2004.PubMed/NCBI
|
|
34
|
Zhang C, Yang X, Zhang Q, Yang B, Xu L,
Qin Q, Zhu H, Liu J, Cai J, Tao G, et al: Berberine radiosensitizes
human nasopharyngeal carcinoma by suppressing hypoxia-inducible
factor-1α expression. Acta Otolaryngol. 134:185–192. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang Q, Zhang C, Yang X, Yang B, Wang J,
Kang Y, Wang Z, Li D, Huang G, Ma Z, et al: Berberine inhibits the
expression of hypoxia induction factor-1alpha and increases the
radiosensitivity of prostate cancer. Diagn Pathol. 9:982014.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dazert E and Hall MN: mTOR signaling in
disease. Curr Opin Cell Biol. 23:744–755. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Engelman JA: Targeting PI3K signalling in
cancer: Opportunities, challenges and limitations. Nat Rev Cancer.
9:550–562. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen ZZ: Berberine induced apoptosis of
human osteosarcoma cells by inhibiting phosphoinositide 3
kinase/protein kinase B (PI3K/Akt) signal pathway activation. Iran
J Public Health. 45:578–585. 2016.PubMed/NCBI
|
|
39
|
Kou Y, Li L, Li H, Tan Y, Li B, Wang K and
Du B: Berberine suppressed epithelial mesenchymal transition
through cross-talk regulation of PI3K/AKT and RARα/RARβ in melanoma
cells. Biochem Biophys Res Commun. 479:290–296. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Milella M, Falcone I, Conciatori F,
Matteoni S, Sacconi A, De Luca T, Bazzichetto C, Corbo V, Simbolo
M, Sperduti I, et al: PTEN status is a crucial determinant of the
functional outcome of combined MEK and mTOR inhibition in cancer.
Sci Rep. 7:430132017. View Article : Google Scholar : PubMed/NCBI
|