Introduction
Bladder cancer (BCa) is the most common urogenital
malignant tumor and its incidence is increasing worldwide (1). Despite the advances in diagnostic
approach and treatment strategies, the prognosis of patients with
advanced BCa remains poor (2).
Therefore, further investigations of the molecular mechanisms
involved in the pathogenesis of BCa and identification of potential
therapeutic targets is imperative.
MicroRNAs (miRNAs) are noncoding small RNA molecules
that post-transcriptionally negatively modulate protein expression
via binding to the 3′-UTR of the target genes (3). miRNAs play a critical role in
biological processes, including cell growth, apoptosis,
differentiation, motility and malignant transformation (4,5). In
addition, miRNAs can inhibit or enhance the expression of oncogenes
or tumor suppressor genes, and thereby markedly affect the biology
of cancer (6,7). For example, miR-145 (8), miR-122 (9) and miR-31 (10) have been revealed to influence
tumorigenesis, tumor proliferation, invasion and metastasis.
Several studies have also revealed that miR-154 acts as a tumor
suppressor in a wide variety of human cancers, including prostate
(11), colorectal (12), breast (13), liver (14) and BCa (15). However, the role of miR-154 in BCa
progression has not yet been fully elucidated.
ATG7 (autophagy-related gene 7), an E1-like
activating enzyme, is essential for the autophagy conjugation
system and autophagosome formation (16,17). A
previous study revealed that ATG7 was critical for sustained tumor
cell proliferation and progression of lung tumors to adenomas and
carcinomas (18). Consistently with
this finding, ATG7 overexpression was shown to promote growth of
human BCa both in vitro and in vivo through the
FOXO1/p27 pathway. These findings indicated that ATG7 is important
for BCa development (19). Till
date, several miRNAs, including miR-520b (20), miR-7 (21), miR-375 (22) and miR-217 (23), have been confirmed to suppress cell
growth and survival by targeting ATG7 in tumor cells. Thus,
identification of miRNAs that target ATG7 in BCa will facilitate
the development of ATG7-based therapies for BCa.
In the present study, we aimed to elucidate the role
and the underlying mechanism of miR-154 in BCa. We found
significant downregulation of miR-154 in BCa tissues and cell
lines. We assessed the effect of miR-154 on cell proliferation,
migration and invasion. Furthermore, we examined its effect on
tumor growth in vivo. Our results revealed that miR-154
exerts a critical role in BCa progression and represents a
potential target for BCa treatment.
Materials and methods
Cell lines and human tissues
Human BCa cell lines (J82, T24 and UM-UC-3) and
human bladder urothelial cell line (SV-HUC-1) were purchased from
the Shanghai Institute of Cell Biology at the Chinese Academy of
Sciences (Shanghai, China). All cell lines were authenticated via
STR profiling before running the experiments. The cells were
maintained in RPMI-1640 medium supplemented with 10% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) in a humidified atmosphere with 5% CO2 at 37°C. The
BCa specimens and paired adjacent non-tumor bladder urothelial
tissues were obtained between January 2010 and December 2016 from
patients undergoing a surgical procedure at the Shanghai Tenth
People's Hospital of Tongji University, School of Medicine
(Shanghai, China) and immediately frozen in liquid nitrogen. All
patients provided written consent. The study was approved by the
Ethics Committee of Tongji University and the BCa diagnosis was
based on hematoxylin and eosin and immunohistochemical staining of
tumor tissue sections. None of these patients had received any
preoperative chemotherapy or radiotherapy.
Overexpression or knockdown of
miR-154
The miR-154 mimics (named miR-154) and the negative
control (named NC) were used for transient gain-of-function study.
The miR-154 inhibitor oligo (named miR-154 inhibitor) and inhibitor
negative control oligo (named inhibitor NC) were used for transient
loss-of-function study. All the aforementioned products were
purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China). T24
and UM-UC-3 cells were seeded into 6-well plates in RPMI-1640 media
supplemented with 10% FBS. At 70% confluence, the cells were
transfected with Lipofectamine 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. The
cells were harvested 48 h after transfection and subjected to
analysis by qRT-PCR or western blotting.
Plasmid construction and siRNA
interference assay
The ATG7-coding sequence without the 3′-UTR was
cloned and inserted into the pcDNA3.1 vector by Sangon Biotech Co.,
Ltd. (Shanghai, China). An empty pcDNA3.1(+) served as the negative
control. Three siRNA sequences that targeted different sites of the
human ATG7 cDNA (siATG7) were designed and synthesized by Sangon
Biotech Co., Ltd. A scrambled siRNA that did not target the human
ATG7 cDNA was synthesized and used as a negative control. The siRNA
sequences were as follows: siATG7 #1, 5′-GCCGUGGAAUUGAUGGUAU-3′
(sense); siATG7 #2, 5′-GGAUCCUGGACUCUCUAAA-3′ (sense); and siATG7
#3, 5′-GAAGCUCCCAAGGACAUUA-3′ (sense). Either the ATG7
overexpression plasmid or the ATG7 siRNAs were transfected into the
T24 and UM-UC-3 cells using Lipofectamine 3000 reagent (Invitrogen;
Thermo Fisher Scientific, Inc.). Total RNA and protein were
isolated 24 h post-transfection. The ATG7 protein expression levels
were assessed by western blotting. The siRNA sequence with the
maximal interfering effect (siATG7 #1) was selected and used for
all the subsequent experiments.
Total RNA extraction and quantitative
real-time PCR
Total RNA was extracted from frozen tissues and
cultured cells using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The
concentration and purity of RNA were determined by ND-2000
Spectrophotometer (Thermo Fisher Scientific, Inc.). For miR-154
detection, cDNA was synthesized using 1 µg of total RNA by One Step
PrimeScript miRNA cDNA Synthesis kit (Qiagen, Inc., Valencia, CA,
USA). Quantitative real-time PCR (qRT-PCR) assay was performed
using KAPA SYBR FAST qPCR Kit (Kapa Biosystems, Inc., Wilmington,
MA, USA). The amplification procedure was as follows: 5 min at
95°C, followed by 40 cycles at 95°C for 30 sec and 65°C for 45 sec.
The expression of miR-154 was normalized to that of U6. To
determine the mRNA level of ATG7, cDNA was synthesized using
PrimeScript RT Reagent kit (Takara Bio, Inc., Otsu, Japan)
according to the manufacturer's instructions. The mRNA expression
level of ATG7 was normalized to that of β-actin (Sangon Biotech
Co., Ltd). qRT-PCR was performed using KAPA SYBR FAST qPCR kit
(Kapa Biosystems, Inc.). The primers for qRT-PCR analysis were as
follows: miR-154 forward, 5′-TAGGTTATCCGTGTTG-3′ and reverse,
5′-ATCCAGTGCAGGGTCCGAGG-3′; U6 forward, 5′-TGCGGGTGCTCGCTTCGCAGC-3′
and reverse, 5′-CCAGTGCAGGGTCCGAGGT-3′; ATG7 forward,
5′-GCTTCCGTGACCGTACCATG-3′ and reverse,
5′-TCCATACATTCACTGAGGTTCACCATC-3′; β-actin forward,
5′-CCTGGCACCCAGCACAAT-3′ and reverse, 5′-GGGCCGGACTCGTCATAC-3′. The
PCR parameters for relative quantification were as follows: 2 min
at 95°C, followed by 40 cycles of 45 sec at 57°C and 45 sec at
72°C. The relative expression of miR-154 and ATG7 were calculated
using the 2−ΔΔCq method (24).
Western blotting
The total protein of cells and tissues was extracted
in RIPA buffer (Beyotime Institute of Biotechnology, Shanghai,
China) supplemented with 1% protease inhibitor cocktail (Thermo
Fisher Scientific, Inc.). The reaction mixture was incubated on ice
for 30 min and centrifuged for 10 min at 12,000 × g at 4°C. The
supernatant was collected, and the protein concentration was
estimated using a Pierce BCA protein assay kit (Thermo Fisher
Scientific, Inc.). Then 20 µg of protein was loaded into 12.5%
SDS-PAGE gel and transferred to nitrocellulose membranes. The
membranes were blocked in 5% non-fat milk for 1 h and then
incubated with primary antibodies: Anti-ATG7 (dilution 1:50,000;
cat. no. ab52472; Abcam, Cambridge, MA, USA) and anti-β-actin
(dilution 1:2,000; Santa Cruz Biotechnology, Inc., Santa Cruz, CA,
USA) overnight at 4°C. After washing with PBST three times, the
membranes were incubated with the corresponding secondary
antibodies at room temperature for 1 h. The protein band was
visualized using the Odyssey scanner (LI-COR Biosciences, Lincoln,
NE, USA).
Cell proliferation assay
Cell proliferation was assessed using Cell Counting
Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto,
Japan). The transfected cells were seeded into 96-well plates at a
density of 1,000 cells/well. Then, 10 µl CCK-8 reagent was added to
each plate at selected time-points and incubated for 2 h at 37°C.
The absorbance was measured at 450 nm with a microplate
spectrophotometer (BioTek, Instruments, Inc., Winooski, VT,
USA).
Wound healing and Transwell
assays
Wound healing assays were performed to detect the
migration capacity of the cells. The transfected cells were seeded
into 6-well plates and cultured until 80% confluence was achieved.
A sterile 200-µl pipette tip was used to scratch the monolayer in
each well. The wound closure was observed and images were captured
at 24 h after making the wound and compared with the 0-h images.
Transwell assays were performed using Transwell chambers (Corning
Inc., Corning, NY, USA) coated with Matrigel (BD Biosciences, San
Jose, CA, USA) on the upper surface. Briefly, 3×104
cells were seeded in the upper chambers in serum-free medium. The
lower chambers were filled with RPMI-1640 medium supplemented with
10% FBS. After incubation at 37°C for 24 h, the cells on the lower
membrane were fixed with 75% ethanol for 20 min and stained with
0.1% crystal violet. Five visual fields (×100 magnification) were
selected randomly, and invaded cells were counted and imaged under
a light microscope (Olympus Corp., Tokyo, Japan).
Tumor formation in nude mice
A lentiviral vector that overexpressed miR-154 was
purchased. (Invitrogen; Thermo Fisher Scientific, Inc.). T24 cells
were transduced with the lentiviruses and stable clones were
selected with puromycin for 2 weeks according to the manufacturer's
instructions. Cells were then harvested for animal experiments. Ten
4-week-old male BALB/c nude mice were purchased from Shanghai
Experimental Animal Center, Chinese Academy of Sciences (Shanghai,
China). The mice were maintained under a 12-h dark/light cycle with
ad libitum access to food in specific pathogen-free
conditions (55% humidity and 22°C). The mice were randomly divided
into two groups (5 per group) and subcutaneously injected with T24
cells (2×106 cells/mouse) that were infected with either
control lentivirus or a lentivirus that overexpressed miR-154. The
tumor volume was calculated by the following formula: Tumor volume
(mm3) = [length (mm)] × [width (mm)]2 × 0.5.
To determine the proliferation of the cells, Ki-67 staining of
tumor tissues obtained from xenograft mice was performed as
previously described (25). The
mice were sacrificed by cervical dislocation after 28 days. All
animal studies were approved by the Institutional Animal Care and
Use Committee of the Shanghai Tenth People's Hospital (Shanghai,
China).
Dual-Luciferase reporter assay
The target genes were predicted using bioinformatics
analysis tools, including TargetScan (http://www.targetscan.org/vert_72/), ComiR (http://www.benoslab.pitt.edu/comir/) and miRANDA
(http://www.microrna.org/). To confirm the
presence of miR-154 binding sites in the ATG7 3′-UTR,
ATG7-wild-type 3′-UTR (ATG7-wt) and ATG7-mutant 3′-UTR (ATG7-mut)
luciferase psiCHECK-2 reporter vectors were constructed. For the
luciferase assay, T24 and UM-UC-3 cells were plated into 24-well
plates and co-transfected with 100 ng luciferase psiCHECK-2
reporter vectors and miR-154/miR-154 inhibitor or negative control.
All plasmid vectors were purchased from Promega Corp. (Madison, WI,
USA). After 48 h of incubation the luciferase activity was assessed
using a Luciferase Reporter Assay System (Promega Corp.), according
to the manufacturer's instructions.
Statistical analysis
Data were analyzed using SPSS 15.0 software (SPSS,
Inc., Chicago, IL, USA). Results are presented as the mean ±
standard deviation (SD) from at least three independent
experiments. The Student's t-test was used to assess between-group
differences, and one-way analysis of variance (ANOVA) plus post hoc
Bonferroni test was used when comparing more than two groups. The
association between the characteristics of patients and miR-154
expression was evaluated by Chi-square test or Fisher's exact test.
The relationship between ATG7 and miR-154 expression was quantified
using Spearman's correlation. Survival analysis was performed by
Kaplan-Meier method and log-rank t-test. P-values <0.05
were considered to indicate a statistically significant
difference.
Results
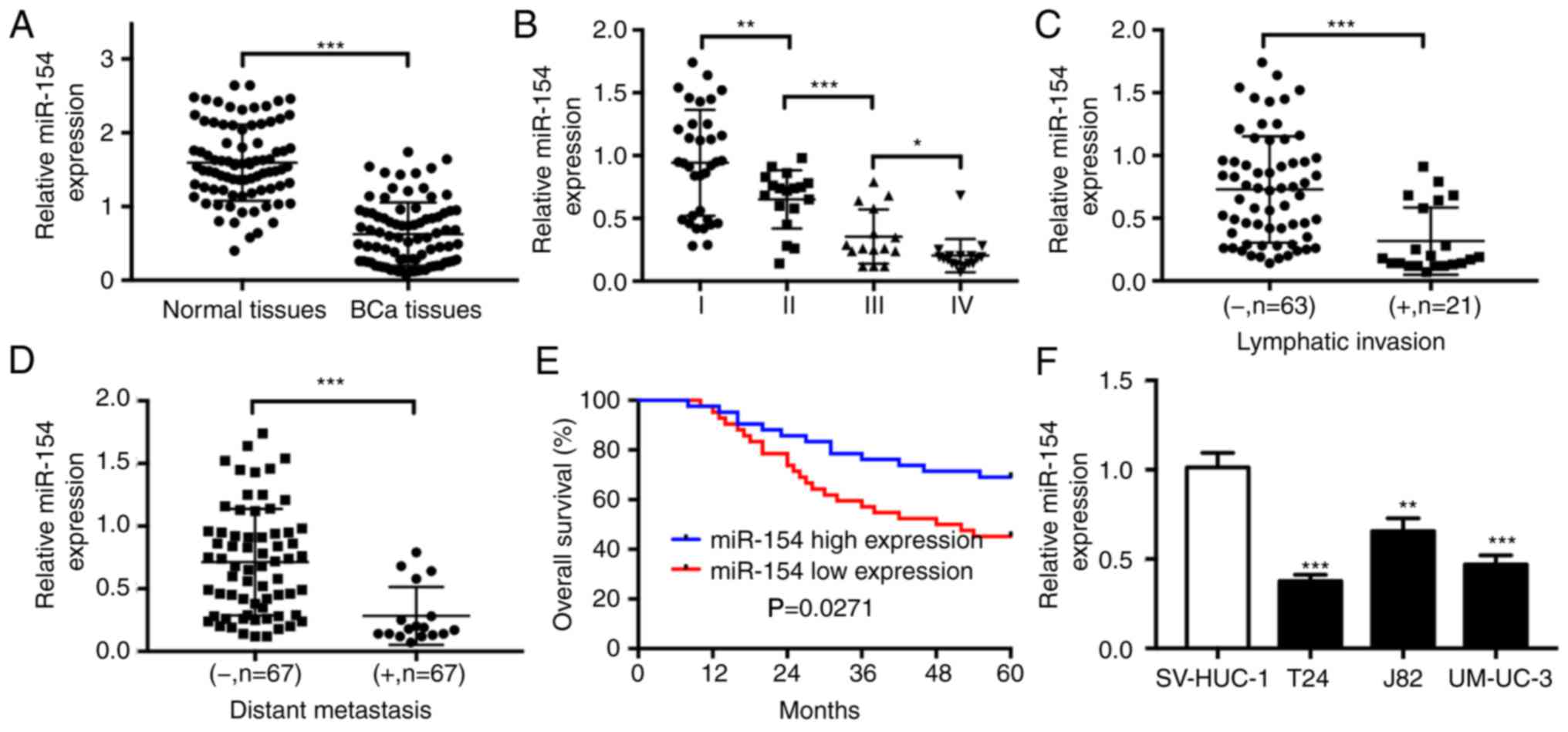
miR-154 is downregulated in BCa
By utilizing real-time PCR, the expression of
miR-154 in 84 BCa tissue samples was found to be significantly
decreased in comparison with the corresponding adjacent normal
bladder urothelial tissues (Fig.
1A). Next, the 84 clinical samples were divided into two groups
according to the median relative expression levels of miR-154. The
correlation between miR-154 levels and clinicopathological features
of BCa is summarized in Table I.
miR-154 expression was significantly related with T stage
(P=0.008), lymphatic invasion (P=0.023) and distant metastasis
(P=0.015). The results revealed a gradual decrease in miR-154
levels with progression of T stage (Fig. 1B). In addition, miR-154 expression
in BCa tissues with lymphatic invasion was significantly
downregulated when compared to that in tissues without lymphatic
invasion (Fig. 1C). Further
analysis revealed that miR-154 expression was also significantly
decreased in metastatic BCa tissues (Fig. 1D). We further evaluated the impact
of miR-154 on the survival outcomes of BCa patients. The overall
survival (OS) of patients with low miR-154 expression was poorer
than that of patients with high miR-154 expression (Fig. 1E). Next, we also detected
significant downregulation of miR-154 in BCa cell lines compared
with the human bladder urothelial cell line SV-HUC-1 (Fig. 1F). Collectively, these data
confirmed the low expression level of miR-154 in BCa tissues and
cell lines.
 | Table I.Correlation of miR-154 expression
with clinicopathological factors in 84 BCa patients. |
Table I.
Correlation of miR-154 expression
with clinicopathological factors in 84 BCa patients.
| Parameters | No. of
patients | High
expression | Low expression | P-value |
|---|
| Sex |
|
|
| 0.637 |
|
Male | 58 | 28 | 30 |
|
|
Female | 26 | 14 | 12 |
|
| Age (years) |
|
|
| 0.498 |
|
≥60 | 53 | 25 | 28 |
|
|
<60 | 31 | 17 | 14 |
|
| Tumor size |
|
|
| 0.126 |
| ≤3 | 39 | 23 | 16 |
|
|
>3 | 45 | 19 | 26 |
|
| Histological
grade |
|
|
| 0.434 |
|
High | 65 | 31 | 34 |
|
|
Low | 19 | 11 | 8 |
|
| Tumor stage |
|
|
| 0.008 |
| I | 34 | 23 | 11 |
|
|
≥II | 50 | 19 | 31 |
|
| Lymphatic
invasion |
|
|
| 0.023 |
|
Positive | 21 | 6 | 15 |
|
|
Negative | 63 | 36 | 27 |
|
| Distant
metastasis |
|
|
| 0.015 |
|
Positive | 17 | 4 | 13 |
|
|
Negative | 67 | 38 | 29 |
|
miR-154 suppresses BCa cell
proliferation, migration, and invasion in vitro
Owing to the marked downregulation of miR-154 in
human BCa tissues and cells, miR-154 may function as a putative
tumor suppressor in BCa. To study the tumor suppressive role of
miR-154 in BCa, we examined the proliferation, migration and
invasion of transfected BCa cells. First, the mRNA expression level
of miR-154 was assessed by qRT-PCR (Fig. 2A). When compared to the group
transfected with the negative control, T24 and UM-UC-3 cells
transfected with miR-154 mimics exhibited a significantly reduced
proliferation ability (Fig. 2B).
The opposite effect was observed in BCa cells transfected with
miR-154 inhibitor (Fig. 2B). The
effect of miR-154 on the migration and invasion of BCa cells was
next assessed. Wound healing assays revealed that T24 and UM-UC-3
cells transfected with miR-154 mimics exhibited a slower recovery
capacity when compared to the control cells; however, decreased
miR-154 levels in BCa cells transfected with the miR-154 inhibitor
led to faster wound closure when compared to the control cells
(Fig. 2C and D). Transwell assays
revealed a similar result in that miR-154 overexpression inhibited
invasion, and decreased miR-154 levels accelerated the invasion of
T24 and UM-UC-3 cells (Fig. 2E and
F). Collectively, these results indicated that miR-154
suppressed the biological behavior of BCa in vitro.
 | Figure 2.miR-154 inhibits the proliferation,
migration and invasion of BCa cells. (A) The mRNA expression level
of miR-154 was assessed by qRT-PCR in T24 and UM-UC-3 cells
transfected with miR-154, miR-154 inhibitor, or negative control,
respectively. (B) T24 and UM-UC-3 cell lines were transfected with
miR-154, miR-154 inhibitor, or negative control, and cell
proliferation was assessed by the CCK-8 assay. (C and D) Wound
healing assays assessed the effect of miR-154 on BCa cell motility.
(E and F) Transwell assays assessed the effect of miR-154 on BCa
cell invasion. *P<0.05, **P<0.01, ***P<0.001. BCa, bladder
cancer. |
miR-154 directly targets ATG7
To explore the potential mechanisms that underlie
the miR-154-mediated BCa cell biological behaviors, three common
bioinformatic databases (TargetScan, ComiR and miRANDA) were used
to predict the mRNA targets of miR-154. ATG7 gene was selected as a
potential target of miR-154 (Fig.
3A). We conducted a luciferase reporter assay to determine
whether
ATG7 was regulated by miR-154 in BCa
cells
The results revealed that miR-154 overexpression by
mimics significantly inhibited and miR-154 inhibitor significantly
increased the reporter activity of the wild-type ATG7 3′-UTR but
not that of the mutant ATG7 3′-UTR (Fig. 3B). Western blot analysis
demonstrated that miR-154 mimics significantly inhibited, while the
miR-154 inhibitor increased the endogenous ATG7 protein expression
in both T24 and UM-UC-3 cells (Fig.
3C).
To assess the potential association between the
expressions of ATG7 and miR-154 in BCa, the expression of ATG7 was
investigated in the same pairs of BCa tissues. The results revealed
that ATG7 expression was generally higher in BCa tissues (Fig. 3D). Further analysis revealed a
strong inverse correlation between miR-154 and ATG7 in BCa tissues
(Fig. 3E; P<0.0001, r=−0.5516).
Additionally, the results of qRT-PCR assays and western blotting
revealed a higher expression of ATG7 in BCa cell lines when
compared to that in SV-HUC-1 cells (Fig. 3F).
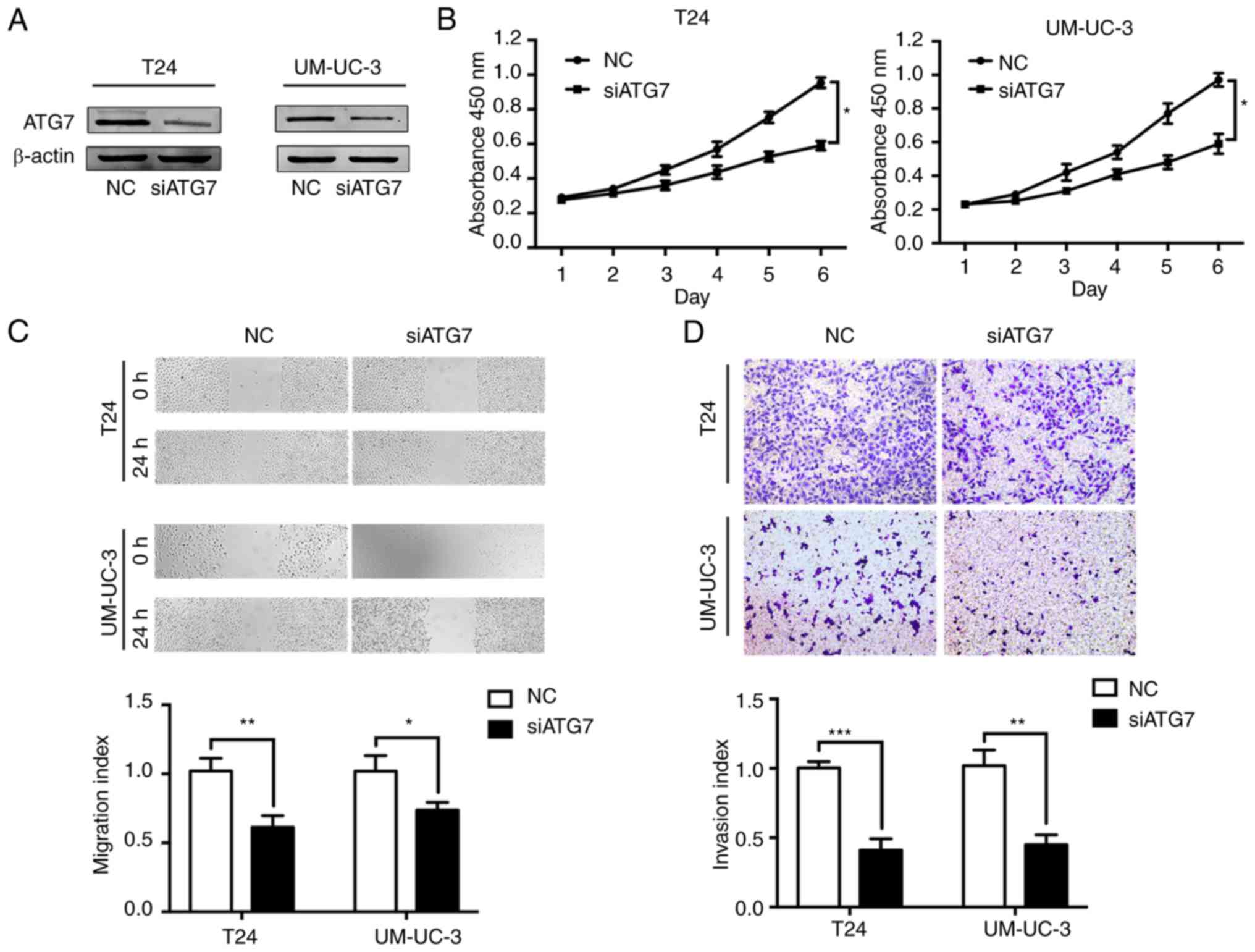
ATG7 functions to suppress BCa
It was hypothesized that ATG7 may act as the vital
effector of miR-154 in the progression of BCa. To determine whether
ATG7 was involved in miR-154-suppressed BCa progression, ATG7 was
knocked down by siRNA (siATG7) in T24 and UM-UC-3 cell lines
(Fig. 4A) and BCa cellular
functions were monitored. The results of CCK-8 assays revealed that
the proliferation capacity of BCa cells was significantly inhibited
after treatment with siATG7 (Fig.
4B). Similarly, the BCa cells with lower ATG7 expression
exhibited suppressed cell migration and invasion capacities
(Fig. 4C and D). Collectively,
consistent with miR-154, siATG7 suppressed BCa progression by
inhibiting tumor cell proliferation, migration, and invasion.
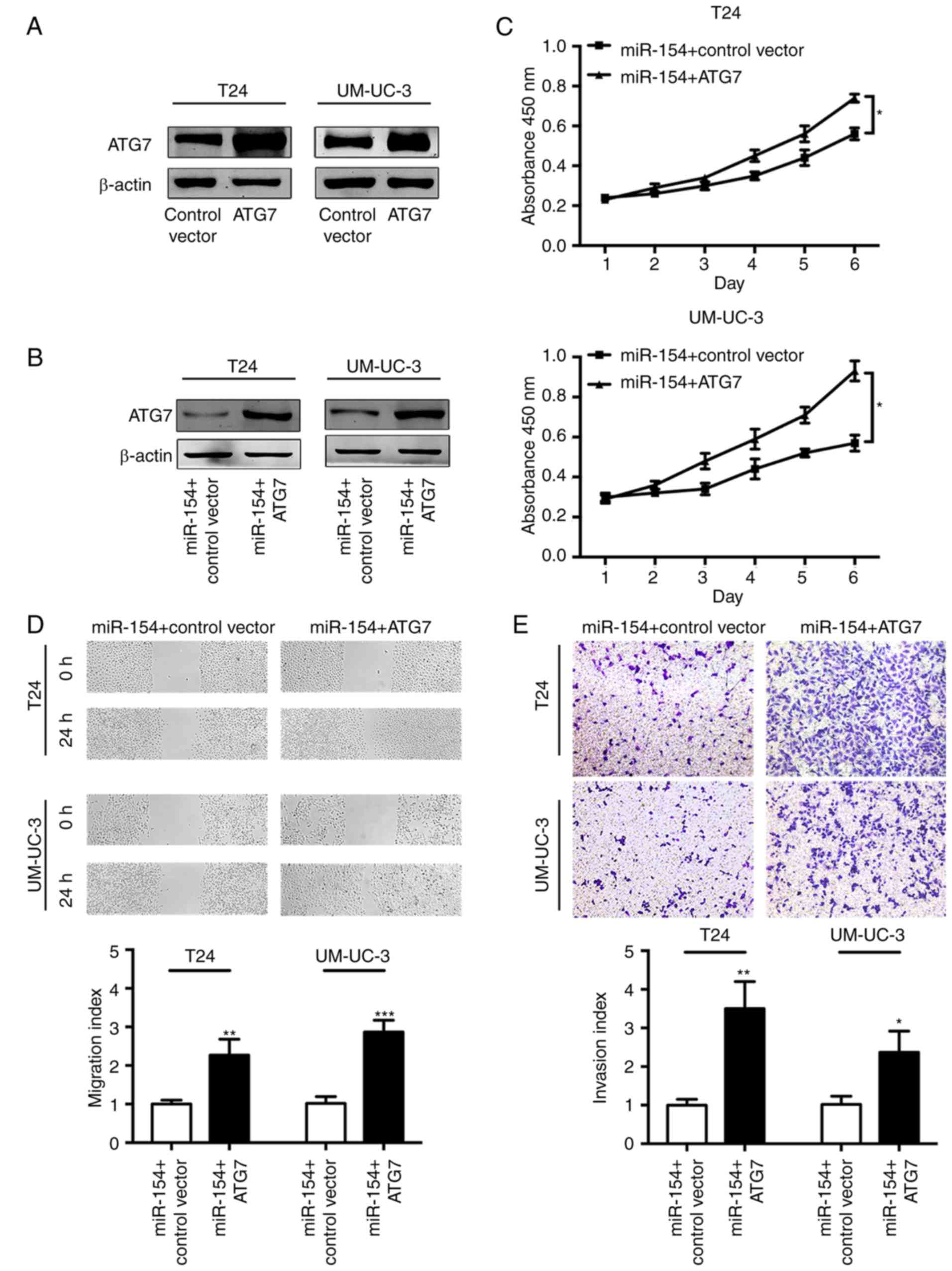
To better understand the reliance of miR-154 on ATG7
for modulation of the biological behavior of BCa cells, we
overexpressed ATG7 in BCa cells (Fig.
5A). The transfection of the ATG7 overexpression vector
restored ATG7 protein levels when compared to the control group of
BCa cells pretreated with the miR-154 mimics (Fig. 5B). Notably, subsequent CCK-8 assays
revealed that reintroducing ATG7 restored the miR-154-mediated
growth suppression of BCa cells (Fig.
5C). Likewise, the suppressive effect of miR-154 on BCa cell
migration and invasion could be reversed by reconstitution of ATG7
(Fig. 5D and E). Collectively,
these findings implied that miR-154 partially suppressed the
progression of BCa cells by inhibiting ATG7.
miR-154 inhibits BCa proliferation and
tumorigenesis in vivo
BCa xenograft mouse models were then used to
determine the function of miR-154 in vivo. The miR-154
levels in the stably transfected cell line was 200-fold higher than
that in the control cell line (Fig.
6A). Consistent with the in vitro findings, the
overexpression of miR-154 caused significant inhibition of tumor
growth in vivo (Fig. 6B).
The miR-154 level in the xenograft tumor tissues was 100-fold
higher than that in the control group (Fig. 6C). Furthermore, the
miR-154-overexpressed group revealed low ATG7 levels when compared
to that in the control group (Fig.
6D). In addition, Ki-67 staining revealed less proliferation in
the miR-154 group (Fig. 6E).
Collectively, the results indicated a critical role of miR-154 in
suppressing BCa growth in vivo.
Discussion
Emerging evidence suggests that miRNAs play a
critical role in carcinogenesis and cancer progression (26). Therefore, a better understanding of
the biological functions of miRNAs may help identify novel
molecular markers for BCa and facilitate the development of novel
therapeutic strategies. Aberrantly expressed miRNAs have been
reported in several human cancers, including BCa (27–29).
Previous studies have revealed that aberrant expression of miR-154
is involved in the initiation and tumor progression of various
human cancers. Xu et al found that miR-154 was frequently
downregulated in breast cancer tissues and that it functioned as a
tumor suppressor in breast cancer by targeting E2F5 (13). Chen and Gao demonstrated that
miR-154 inhibited the growth of skin squamous cell carcinoma cells
by targeting the p53 signaling pathway (30). Additionally, miR-154 was revealed to
be downregulated in human hepatocellular carcinoma tissues and to
inhibit cell proliferation, migration and invasion via suppression
of ZEB2 (14). However, the role of
miR-154 in BCa remains unclear.
In the present study, we found marked downregulation
of miR-154 in BCa tissues and cell lines, which was consistent with
the results of a previous study (15). Moreover, a decreased miR-154 level
was correlated with aggressive clinicopathological features,
including advanced T stage, lymphatic invasion, and distant
metastasis. Decreased miR-154 expression predicted unfavorable OS
of BCa patients. To better characterize the role of miR-154 in BCa,
functional studies were conducted. miR-154 overexpression inhibited
the proliferation, migration, and invasion of BCa cells, while
knockdown of miR-154 yielded the opposite effects in vitro.
These findings indicated that miR-154 functioned as a tumor
suppressor in BCa.
Zhao et al revealed miR-154 inhibited BC cell
proliferation, migration, and invasion by regulating RSF1 and RUNX2
expression (15). Actually, it has
been well established that one specific microRNA is able to target
multiple downstream genes and thus determine its biological role.
To explore the potential molecular mechanisms by which miR-154
functioned as a tumor suppressor in BCa, we performed
bioinformatics analysis using TargetScan, ComiR, and miRANDA and
predicted ATG7 as a candidate target of miR-154. Subsequently, dual
luciferase reporter assays identified ATG7 as a direct target of
miR-154 in BCa. ATG7, an E1-like activating enzyme, is essential
for the biogenesis of autophagosomes (17). Recent studies have demonstrated that
ATG7 is upregulated in some human malignancies, such as lung cancer
(31), neuroblastoma (32) and BCa (19). ATG7 was reported to be upregulated
in human colorectal cancer and was shown to promote the oncogenic
transformation of cells with deregulated WNT/β-catenin signaling
(33). In addition, Piya et
al revealed that ATG7 knockdown in acute myeloid leukemia cells
resulted in a proapoptotic phenotype that showed increased
chemosensitivity (34). In a recent
study, deletion of ATG7 was revealed to suppress a
carcinogen-induced pro-tumorigenic inflammatory microenvironment
and tumorigenesis in epithelial cells (35). All these findings indicated that
ATG7 is an important oncogene. In the present study, western blot
analysis demonstrated that the expression level of ATG7 in T24 and
UM-UC-3 cells transfected with miR-154 mimics was significantly
lower than that in the negative control. Additionally, ATG7 was
significantly increased in BCa tissues and cell lines. Further
analysis revealed a strong inverse correlation between miR-154 and
ATG7 in BCa tissues. Based on these results, we hypothesized that
miR-154-mediated ATG7 inhibition is a promising potential
therapeutic option for BCa.
Furthermore, ATG7-siRNA knockdown inhibited the
proliferation, migration, and invasion ability of BCa cells, which
was similar to the effects of miR-154 overexpression. Notably, we
further observed that the restoration of ATG7 expression
successfully attenuated the inhibitory effects of miR-154 on BCa
cell proliferation, migration, and invasion. Collectively, these
findings indicated that the tumor-suppressive effects of miR-154
may be mediated via downregulation of ATG7 in BCa cells. Future
research is required to understand the precise mechanism of action
of the miR-154/ATG7 pathway and its role in regulating other
signaling pathways in BCa.
To summarize, our data supports the assumption that
miR-154 may function as a tumor suppressor in BCa by attenuating
the expression of ATG7. These findings improved our understanding
of the molecular mechanisms that underlie BCa and provide novel
therapeutic targets.
Acknowledgements
The authors would like to thank the personnel of the
Central Laboratory of the Shanghai Tenth People's Hospital for
their assistance and support.
Funding
The present study was supported by the National
Natural Science Foundation of China (nos. 81472389 and
81272836).
Availability of data and materials
The analyzed datasets generated during the study are
available from the corresponding author on reasonable request.
Authors' contributions
JZ, FY and XY made substantial contributions to the
conception and design of the study; JZ and FY drafted the
manuscript; JZ, FY and XY made substantial contributions to the
acquisition, analysis and interpretation of the data for the study;
SM, LW, WZ, ZZ, YW and YG contributed to the acquisition and
analysis of the data for the study; JZ, FY and XY revised the
manuscript critically for important intellectual content. All
authors gave the final approval of the manuscript to be published
and agree to be accountable for all aspects of the study in
ensuring that questions related to the accuracy or integrity of any
part of the study are appropriately investigated and resolved.
Ethics approval and consent to
participate
The patient study was approved by the Ethics
Committee of Shanghai Tenth People's Hospital of Tongji University
(Inner Mongolia, China). Informed consent was obtained from all
patients or their relatives. The animal experiments were approved
by the Animal Care and Use Committee of Tongji University (Inner
Mongolia, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kamat AM, Hahn NM, Efstathiou JA, Lerner
SP, Malmstrom PU, Choi W, Guo CC, Lotan Y and Kassouf W: Bladder
cancer. Lancet. 388:2796–2810. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Antoni S, Ferlay J, Soerjomataram I, Znaor
A, Jemal A and Bray F: Bladder cancer incidence and mortality: A
global overview and recent trends. Eur Urol. 71:96–108. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Babashah S and Soleimani M: The oncogenic
and tumour suppressive roles of microRNAs in cancer and apoptosis.
Eur J Cancer. 47:1127–1137. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Robb T, Reid G and Blenkiron C: Exploiting
microRNAs As cancer therapeutics. Target Oncol. 12:163–178. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gandellini P, Doldi V and Zaffaroni N:
microRNAs as players and signals in the metastatic cascade:
Implications for the development of novel anti-metastatic
therapies. Semin Cancer Biol. 44:132–140. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wong CM, Tsang FH and Ng IO: Non-coding
RNAs in hepatocellular carcinoma: Molecular functions and
pathological implications. Nat Rev Gastroenterol Hepatol.
15:137–151. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sathyanarayanan A, Chandrasekaran KS and
Karunagaran D: microRNA-145 downregulates
SIP1-expression but differentially regulates proliferation,
migration, invasion and Wnt signaling in SW480 and SW620 cells. J
Cell Biochem. 119:2022–2035. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fan Y, Ma X, Li H, Gao Y, Huang Q, Zhang
Y, Bao X, Du Q, Luo G, Liu K, et al: miR-122 promotes metastasis of
clear-cell renal cell carcinoma by downregulating Dicer. Int J
Cancer. 142:547–560. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lv C, Li F, Li X, Tian Y, Zhang Y, Sheng
X, Song Y, Meng Q, Yuan S, Luan L, et al: MiR-31 promotes
mammary stem cell expansion and breast tumorigenesis by suppressing
Wnt signaling antagonists. Nat Commun. 8:10362017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Formosa A, Markert EK, Lena AM, Italiano
D, Finazzi-Agro E, Levine AJ, Bernardini S, Garabadgiu AV, Melino G
and Candi E: MicroRNAs, miR-154, miR-299-5p, miR-376a, miR-376c,
miR-377, miR-381, miR-487b, miR-485-3p, miR-495 and miR-654-3p,
mapped to the 14q32.31 locus, regulate proliferation, apoptosis,
migration and invasion in metastatic prostate cancer cells.
Oncogene. 33:5173–5182. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xin C, Zhang H and Liu Z: miR-154
suppresses colorectal cancer cell growth and motility by targeting
TLR2. Mol Cell Biochem. 387:271–277. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu H, Fei D, Zong S and Fan Z:
MicroRNA-154 inhibits growth and invasion of breast cancer cells
through targeting E2F5. Am J Transl Res. 8:2620–2630.
2016.PubMed/NCBI
|
|
14
|
Pang X, Huang K, Zhang Q, Zhang Y and Niu
J: miR-154 targeting ZEB2 in hepatocellular carcinoma functions as
a potential tumor suppressor. Oncol Rep. 34:3272–3279. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao X, Ji Z, Xie Y, Liu G and Li H:
MicroRNA-154 as a prognostic factor in bladder cancer inhibits
cellular malignancy by targeting RSF1 and RUNX2. Oncol Rep.
38:2727–2734. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Karvela M, Baquero P, Kuntz EM,
Mukhopadhyay A, Mitchell R, Allan EK, Chan E, Kranc KR, Calabretta
B, Salomoni P, et al: ATG7 regulates energy metabolism,
differentiation and survival of Philadelphia-chromosome-positive
cells. Autophagy. 12:936–948. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kimmelman AC and White E: Autophagy and
tumor metabolism. Cell Metab. 25:1037–1043. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guo JY, Karsli-Uzunbas G, Mathew R, Aisner
SC, Kamphorst JJ, Strohecker AM, Chen G, Price S, Lu W, Teng X, et
al: Autophagy suppresses progression of K-ras-induced lung tumors
to oncocytomas and maintains lipid homeostasis. Genes Dev.
27:1447–1461. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhu J, Li Y, Tian Z, Hua X, Gu J, Li J,
Liu C, Jin H, Wang Y, Jiang G, et al: ATG7 overexpression is
crucial for tumorigenic growth of bladder cancer in vitro and in
vivo by targeting the ETS2/miRNA196b/FOXO1/p27 Axis. Mol Ther
Nucleic Acids. 7:299–313. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gao AM, Zhang XY, Hu JN and Ke ZP:
Apigenin sensitizes hepatocellular carcinoma cells to doxorubic
through regulating miR-520b/ATG7 axis. Chem Biol Interact.
280:45–50. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gu DN, Jiang MJ, Mei Z, Dai JJ, Dai CY,
Fang C, Huang Q and Tian L: microRNA-7 impairs autophagy-derived
pools of glucose to suppress pancreatic cancer progression. Cancer
Lett. 400:69–78. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chang Y, Yan W, He X, Zhang L, Li C, Huang
H, Nace G, Geller DA, Lin J and Tsung A: miR-375 inhibits autophagy
and reduces viability of hepatocellular carcinoma cells under
hypoxic conditions. Gastroenterology. 143:177–187.e8. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zeng Y, Huo G, Mo Y, Wang W and Chen H:
MIR137 regulates starvation-induced autophagy by targeting ATG7. J
Mol Neurosci. 56:815–821. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2ΔΔCT method. Methods.
25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Margulis V, Shariat SF, Ashfaq R,
Sagalowsky AI and Lotan Y: Ki-67 is an independent predictor of
bladder cancer outcome in patients treated with radical cystectomy
for organ-confined disease. Clin Cancer Res. 12:7369–7373. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang W, Zhao E, Yu Y, Geng B, Zhang W and
Li X: MiR-216a exerts tumor-suppressing functions in renal cell
carcinoma by targeting TLR4. Am J Cancer Res. 8:476–488.
2018.PubMed/NCBI
|
|
28
|
Xu M, Li J, Wang X, Meng S, Shen J, Wang
S, Xu X, Xie B, Liu B and Xie L: MiR-22 suppresses
epithelial-mesenchymal transition in bladder cancer by inhibiting
Snail and MAPK1/Slug/vimentin feedback loop. Cell Death Dis.
9:2092018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Juracek J, Peltanova B, Dolezel J, Fedorko
M, Pacik D, Radova L, Vesela P, Svoboda M, Slaby O and Stanik M:
Genome-wide identification of urinary cell-free microRNAs for
non-invasive detection of bladder cancer. J Cell Mol Med.
22:2033–2038. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen HQ and Gao D: Inhibitory effect of
microRNA-154 targeting WHSC1 on cell proliferation of human skin
squamous cell carcinoma through mediating the P53 signaling
pathway. Int J Biochem Cell Biol. 100:22–29. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sun S, Wang Z, Tang F, Hu P, Yang Z, Xue
C, Gong J, Shi L and Xie C: ATG7 promotes the tumorigenesis of lung
cancer but might be dispensable for prognosis predication: A
clinicopathologic study. Onco Targets Ther. 9:4975–4981. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yu Y, Zhang J, Jin Y, Yang Y, Shi J, Chen
F, Han S, Chu P, Lu J, Wang H, et al: MiR-20a-5p suppresses tumor
proliferation by targeting autophagy-related gene 7 in
neuroblastoma. Cancer Cell Int. 18:52018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Levy J, Cacheux W, Bara MA, L'Hermitte A,
Lepage P, Fraudeau M, Trentesaux C, Lemarchand J, Durand A, Crain
AM, et al: Intestinal inhibition of Atg7 prevents tumour initiation
through a microbiome-influenced immune response and suppresses
tumour growth. Nat Cell Biol. 17:1062–1073. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Piya S, Kornblau SM, Ruvolo VR, Mu H,
Ruvolo PP, McQueen T, Davis RE, Hail N Jr, Kantarjian H, Andreeff
M, et al: Atg7 suppression enhances chemotherapeutic agent
sensitivity and overcomes stroma-mediated chemoresistance in acute
myeloid leukemia. Blood. 128:1260–1269. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Qiang L, Sample A, Shea CR, Soltani K,
Macleod KF and He YY: Autophagy gene ATG7 regulates
ultraviolet radiation-induced inflammation and skin tumorigenesis.
Autophagy. 13:2086–2103. 2017. View Article : Google Scholar : PubMed/NCBI
|