Introduction
Lung cancer is one of the most malignant tumors and
the leading cause of cancer-related deaths worldwide (1). Despite the development of anticancer
therapies, such as chemotherapy, targeted therapies, and
radiotherapy, the 5-year survival rates are still not optimistic.
Primary and acquired resistance has been considered as a key factor
in reducing the efficacy of current cytotoxic therapies in the
treatment of non-small cell lung cancer (NSCLC). Studies have
demonstrated the presence of a subset of cells called cancer stem
cells (CSCs), which have stem-like properties, as observed in lung
cancer (2).
Although epidermal growth factor receptor tyrosine
kinase inhibitors (EGFR-TKIs) are effective for lung adenocarcinoma
patients with EGFR mutations, the patients invariably develop
resistance to these drugs after a period of treatment (3). Recently, various mechanisms of
resistance to EGFR-TKIs have been revealed, such as secondary
mutations (T790M) (4), aberrated
activation of the bypass pathways (c-Met, HGF, AXL) (5,6), and
abnormal downstream pathways (K-RAS mutations, loss of PTEN,
mutations of BRAF and PIK3CA mutation) (3,7).
However, another possible mechanism is associated with targeting
drug resistance, which involves CSCs. Several in vitro and
in vivo experiments have revealed that CSCs are the source
of drug resistance and therapeutic failure for lung cancer
(2,8). The correlation between CSCs and the
efficacy of targeted drugs is still not clear. Stem-related
transcription factors, such as OCT4, SOX2 and NANOG,
are considered as the critical regulators of self-renewal and
pluripotency of stem cells, playing an important role in tumor
proliferation and differentiation (9–11).
Thus, the expression of CSC-related markers can reflect the number
of CSCs in cancer tissues (12,13).
Hence, in the present study the analysis of the
expression of cancer stem cell-related markers were investigated
and two groups of patients with significant differences in clinical
outcomes after treatment with first-generation EGFR-TKIs were
retrospectively analyzed, aiming to explore the relationship
between the efficacy of targeted drug therapy and CSCs and provide
potential targets for the development of new therapeutic
strategies.
Materials and methods
Patients and tissue samples
The pathological tissue samples of patients with
lung cancer were collected from the tissue bank of the Shanghai
Chest Hospital. The pathological tissues of the patients analyzed
in this study were obtained before treatment with EGFR-TKI drugs.
The present study was approved by the Ethics Committee of Shanghai
Chest Hospital. All patients provided their informed consent.
In this retrospective study, patients with
significantly different therapeutic effects after the treatment
with first-generation EGFR-TKIs from January 2014 to December 2016
at Shanghai Chest Hospital were identified. The patients were
divided into a sensitive group [progression-free survival (PFS)
>26 months] and a resistant group (PFS <5 months) according
to the efficacy of first-generation EGFR-TKI treatment. At present,
the average PFS of first-generation targeted drugs is 10–13 months
(14–16). Therefore, the cutoff value of
sensitivity and resistance for targeted drugs should be set as two
times the average upper line and one half the lower limit of the
average. Other eligibility criteria included the following:
EGFR-sensitive mutation (either exon 19 deletion, L858R in exon 21,
or other sensitive mutations) and patients receiving
first-generation EGFR-TKIs (including gefitinib, icotinib and
erlotinib) as the first-line therapy. PFS was calculated from the
date of initiation of EGFR-TKI therapy until the date of
progression or last follow-up. The exclusion criteria were as
follows: Non-first-line EGFR-TKI therapy, incomplete medical
history information, loss to follow-up, history of other
malignancies, and EGFR exon 20 insertion. In the present study,
lung cancer staging was conducted on the baseline status of all
patients enrolled according to the AJCC 8th edition TNM staging
system. Tumor EGFR mutation status was determined by the
amplification refractory mutation system (ARMS). The clinical
characteristics, including age, sex, smoking status [a ‘smoker’ is
defined as a patient who has smoked >100 cigarettes during his
lifetime (17,18)], EGFR mutation status, TNM stage,
tumor differentiation status and type of EGFR-TKIs, are summarized
in Table I.
 | Table I.Epidemiological, clinicopathological
and mutation status data in sensitive and resistant patients
treated with first-generation EGFR-TKIs. |
Table I.
Epidemiological, clinicopathological
and mutation status data in sensitive and resistant patients
treated with first-generation EGFR-TKIs.
|
Characteristics | No. of cases
(%) | Sensitive group, n
(%) | Resistant group, n
(%) | P-value |
|---|
| Age (years) |
|
|
|
|
|
≤60 | 42 | 15 (48.4) | 27 (65.9) | 0.137 |
|
>60 | 30 | 16 (51.6) | 14 (34.1) |
|
| Sex |
|
|
|
|
|
Male | 28 | 8 (25.8) | 20 (48.8) | 0.048 |
|
Female | 44 | 23 (74.2) | 21 (51.2) |
|
| Smoking status |
|
|
|
|
|
Smokera | 23 | 4 (12.9) | 19 (46.3) | 0.003 |
|
Never-smoker | 49 | 27 (87.1) | 22 (53.7) |
|
| EGFR mutation
status |
|
|
|
|
|
19del | 35 | 15 (48.4) | 20 (48.8) | 0.288 |
|
21L858R | 34 | 16 (51.6) | 18 (43.9) |
|
|
Others | 3 | 0 (0.0) | 3 (7.3) |
|
| TNM stage |
|
|
|
|
|
IIB | 1 | 1 (3.2) | 0 (0.0) | 0.003 |
|
IIIA/IIIB | 25 | 17 (54.8) | 8 (19.5) |
|
| IV | 46 | 13 (41.9) | 33 (80.5) |
|
|
Differentiation |
|
|
|
|
|
Well | 14 | 10 (32.3) | 4 (9.8) | 0.009 |
|
Moderate | 26 | 13 (41.9) | 13 (31.7) |
|
|
Poor | 32 | 8 (25.8) | 24 (58.5) |
|
| Type of
EGFR-TKIs |
|
|
|
|
|
Gefitinib | 31 | 17 (54.8) | 15 (36.6) | 0.679 |
|
Icotinib | 22 | 8 (25.8) | 15 (36.6) |
|
|
Erlotinib | 17 | 6 (19.4) | 11 (26.8) |
|
Immunohistochemical staining
The formalin-fixed, paraffin-embedded tissues were
sectioned at a thickness of 4 µm using a slicer and baked in an
oven at 65°C for >2 h. The paraffin sections were deparaffinized
using xylene and washed with gradient alcohol, and then distilled
water. The sections were placed in the antigen repair solution,
heated in a microwave oven, and cooled to room temperature for
antigen repair. The endogenous peroxidase was removed with 3%
hydrogen peroxide. Then, the goat serum was used for blocking to
eliminate unspecific staining. The slides were incubated overnight
at 4°C with various primary antibodies. To visualize antigens,
peroxidase-labeled antibodies (code K5007; REAL EnVision
Rabbit/Mouse; Dako) were applied at room temperature for 60 min.
The slices were rinsed in phosphate-buffered saline (PBS), stained
with DAB (liquid DAB + substrate, DAKO), and re-stained with
hematoxylin. Two independent observers examined the stained slides
in a blind fashion.
Immunohistochemical (IHC) staining was evaluated
using the histological score (H score) (19). The intensity of IHC staining was
classified as strongly positive, moderately positive and weakly
positive according to the color depth of the positive cells, and
the corresponding values were 1, 2, and 3. The H score was
calculated as follows: H Score = Staining intensities (SI) ×
percentage of positive cells= [1 × (% cells 1+) + 2 × (% cells 2+)
+ 3 × (% cells 3+)]. This score ranged from 0 to 300 (300 indicated
strong staining in 100% of the tumor cells).
The antibodies used were as follows: Human
anti-SOX2 and anti-OCT4 were purchased from Abclonal
(cat. nos. A0561 and A7920, respectively). Human anti-NANOG
was obtained from Abcam (product code ab109250). Human
anti-SOX2, anti-OCT4, and anti-NANOG were used
at a 1:100 dilution in PBS for IHC staining.
Immunofluorescence staining
The paraffin sections were stained with indirect
immunofluorescence staining after dewaxing and antigen repair. They
were dyed with indirect immunofluorescence dyeing of indirect
method after deparaffinization and antigen repair. The histological
sections were washed with PBS at pH 7.3. Then, polyclonal rabbit
antibodies against SOX2 (dilution of 1:100), OCT4
(dilution of 1:100), and NANOG (dilution of 1:100) were
added to each section. The sections were maintained overnight at
4°C overnight. After washing three times with PBS, the sections
were stained with donkey antibodies against rabbit immunoglobulin G
(Jackson/Alexa Fluor® 488; cat. no 711-545-152; dilution
of 1:100). The nuclei were counterstained with
4′,6-diamidino-2-phenylindole (DAPI; blue). As a control, the
primary antibodies were replaced with PBS.
Reverse transcriptase-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the paraffin-embedded
tumor tissues using an FFPE RNA Kit (product code R6954-01; Omega
Bio-tek, Inc.). RNA was reverse-transcribed using Hifair II 1st
Strand cDNA Synthesis SuperMix for qPCR (product no. 11123ES60;
Yeasen) according to the manufacturer's protocols. Approximately 1
µg of cDNA sample was used for each PCR analysis. RT-qPCR was
performed with UNICON® qPCR SYBR-Green Master Mix
(product no. 11199ES08; Yeasen) on an AB (Applied Biosystems;
Thermo Fisher Scientific, Inc.) ViiA 7 instrument. The GAPDH
housekeeping gene was used as a control. The thermocycling
conditions of the RT-qPCR consisted of an initial denaturation step
at 95°C for 30 sec, followed by 40 cycles at 95°C for 10 sec and
60°C for 34 sec. The dissolution curve stage is set by default by
the instrument (1 cycle). The expression of tested genes relative
to GAPDH was determined using the method of ΔΔCq (20). The expression levels of CSC-related
markers in each sample were calculated. The primer sequences of
each gene were as follows: PrimerBank ID: 325651854c1, SOX2
forward, 5′-GCCGAGTGGAAACTTTTGTCG-3′ and reverse,
5′-GGCAGCGTGTACTTATCCTTCT-3′; PrimerBank ID: 4505967a1, OCT4
forward, 5′-CTTGAATCCCGAATGGAAAGGG-3′ and reverse,
5′-GTGTATATCCCAGGGTGATCCTC-3′; PrimerBank ID: 153945815c1,
NANOG forward, 5′-TTTGTGGGCCTGAAGAAAACT-3′ and reverse,
5′-AGGGCTGTCCTGAATAAGCAG-3′; PrimerBank ID: 378404907c1,
GAPDH forward, 5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse:
5′-GGCTGTTGTCATACTTCTCATGG-3′.
Statistical analysis
The unpaired nonparametric t-test was used to
detect the differences in the results of IHC and RT-qPCR analyses
between the two groups. The results of t-tests were reported
as the mean ± standard deviation. The chi-square test was used to
detect the differences in the distribution of clinicopathological
factors between the two groups. The correlation of the expression
of OCT4, SOX2, and NANOG with the clinicopathological
features was determined using Pearson's correlation coefficient
test. P<0.05 was considered to indicate a statistically
significant difference. The statistical data were obtained using an
SPSS software package, version 24.0 (IBM Corp.).
Results
Patients and clinical
characteristics
A total of 72 patients who met the inclusion
criteria were included in the analyses. Of the 72 lung
adenocarcinoma patients with EGFR mutation who received
first-generation EGFR-TKI treatment, 31 were sensitive (sensitive
group) and 41 were resistant to first-generation EGFR-TKIs
(resistant group). The median age of the entire cohort was 58
(31–76) years. The demographics of the whole cohort are presented
in Table I. The expression of
SOX2, OCT4 and NANOG in 72 lung adenocarcinoma
samples was analyzed by IHC staining, immunofluorescence staining
and RT-qPCR.
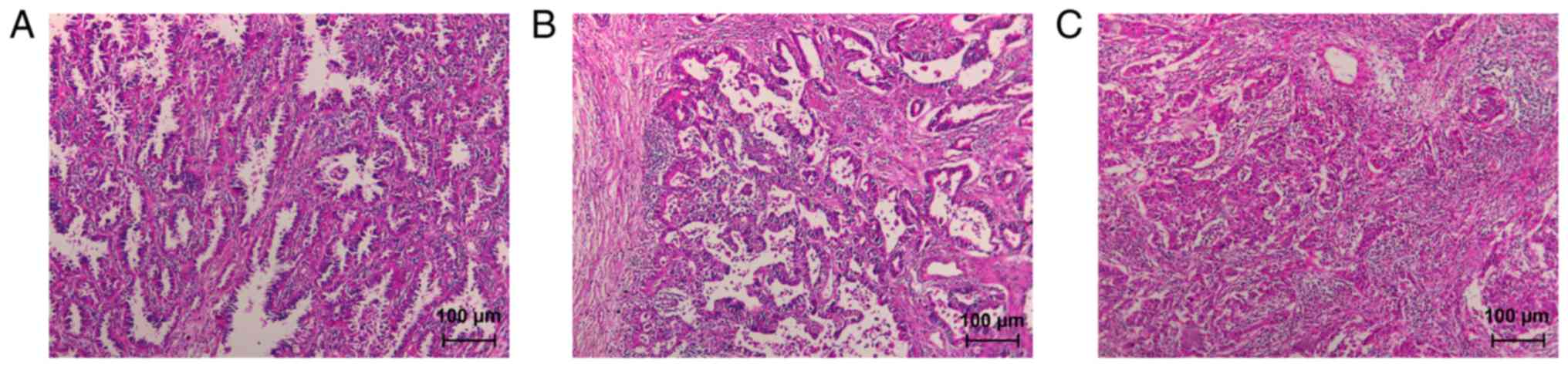
According to the integrity of glandular structures,
cellular heterogeneity, and mucus secretion status, lung
adenocarcinomas were classified as well, moderately, and poorly
differentiated. In well-differentiated tumors, the malignant glands
were composed of tall columnar or mucous epithelium, with large,
round tumor nuclei and prominent nucleoli; the glandular structures
were easily demonstrared under routine microscopy (Fig. 1A). In moderately differentiated
tumors, glandular development was not as good as that in
well-differentiated tumors (Fig.
1B). In poorly differentiated tumors, the adenocarcinoma was
composed of a neoplastic cell population with very poor glandular
development. The gland structure was unclear under routine
microscopy, and the tumor cells had obvious abnormalities (Fig. 1C).
Correlations between clinical features
and efficacy of EGFR-TKIs and the expression of SOX2, OCT4 and
NANOG
The association between clinicopathological features
of the 72 patients with lung adenocarcinoma and the sensitivity of
first-generation EGFR-TKIs are presented in Table I. The chi-square test revealed that
men (P=0.048), smokers (P=0.003), patients with stage IV (P=0.003),
and patients with poor differentiation (P=0.009) were more likely
to be in the resistant group, with statistically significant
differences. No obvious correlation was revealed between the
sensitivity of first-generation EGFR-TKIs and age, EGFR mutation
status, and type of EGFR-TKIs of patients (Table I).
The correlation of the expression of SOX2,
OCT4 and NANOG with clinicopathological features of
patients with lung adenocarcinoma treated with first-generation
EGFR-TKIs was evaluated. As revealed in Table II, the expression of SOX2
was significantly correlated with PFS (R=−0.256, P=0.030), TNM
stage (R=0.257, P=0.029), and differentiation (R=0.347, P=0.003).
The expression of OCT4 was positively associated with PFS
(R=−0.242, P=0.041), TNM stage (R=−0.359, P=0.002), and
differentiation (R=0.269, P=0.022). The expression of NANOG
was significantly asscoiated to PFS (R=−0.289, P=0.014) and
differentiation (R=0.258, P=0.029).
 | Table II.Correlation of OCT4, SOX2 and
NANOG expression with clinicopathological features
determined by Pearson's correlation test. |
Table II.
Correlation of OCT4, SOX2 and
NANOG expression with clinicopathological features
determined by Pearson's correlation test.
| Parameters | PFS | Age | Sex | Smoking status | EGFR mutation
status | TNM stage |
Differentiation | Type of
EGFR-TKIs |
|---|
| SOX2
expression |
|
|
|
|
|
|
|
|
| R | −0.256a | 0.064 | −0.104 | 0.036 | 0.047 | 0.257a | 0.347a | 0.004 |
|
P-value | 0.030 | 0.595 | 0.385 | 0.767 | 0.692 | 0.029 | 0.003 | 0.976 |
| OCT4
expression |
|
|
|
|
|
|
|
|
| R | −0.242a | −0.077 | −0.175 | 0.170 | 0.139 | 0.359a | 0.269a | 0.183 |
|
P-value | 0.041 | 0.520 | 0.141 | 0.152 | 0.244 | 0.002 | 0.022 | 0.125 |
| NANOG
expression |
|
|
|
|
|
|
|
|
| R | −0.289a | 0.057 | 0.027 | −0.128 | 0.012 | −0.074 | 0.258a | 0.054 |
|
P-value | 0.014 | 0.634 | 0.819 | 0.286 | 0.923 | 0.539 | 0.029 | 0.655 |
The association between clinicopathological features
of the 72 patients with lung adenocarcinoma and the expression of
SOX2, OCT4 and NANOG analyzed using the chi-square
test is presented in Table III.
An association was observed between the efficacy of EGFR-TKI
therapy and the expression of OCT4 and NANOG (P=0.039
and P=0.004, respectively), TNM stage and the expression of
SOX2 and OCT4 (P=0.034 and P=0.007, respectively),
differentiation and the expression of SOX2 and NANOG
(P<0.001 and P=0.008, respectively).
 | Table III.Association between SOX2, OCT4 and
NANOG expressiona and
clinicopathological factors in 72 patients with lung adenocarcinoma
analyzed by chi-square test. |
Table III.
Association between SOX2, OCT4 and
NANOG expressiona and
clinicopathological factors in 72 patients with lung adenocarcinoma
analyzed by chi-square test.
|
| SOX2 | OCT4 | NANOG |
|---|
|
|
|
|
|
|---|
|
Characteristics | Positive | Negative | P-value | Positive | Negative | P-value | Positive | Negative | P-value |
|---|
| Efficacy |
|
|
|
|
|
|
|
|
|
|
Resistant (PFS <5
months) | 36 | 5 | 0.074 | 35 | 6 | 0.039c | 34 | 7 | 0.004d |
|
Sensitive (PFS >26
months) | 22 | 9 |
| 20 | 11 |
| 16 | 15 |
|
| Age (years) |
|
|
|
|
|
|
|
|
|
|
≤60 | 34 | 8 | 0.920 | 33 | 9 | 0.606 | 26 | 16 | 0.100 |
|
>60 | 24 | 6 |
| 22 | 8 |
| 24 | 6 |
|
| Sex |
|
|
|
|
|
|
|
|
|
|
Male | 24 | 4 | 0.378 | 24 | 4 | 0.137 | 19 | 9 | 0.816 |
|
Female | 34 | 10 |
| 31 | 13 |
| 31 | 13 |
|
| Smoking status |
|
|
|
|
|
|
|
|
|
|
Smokerb | 19 | 4 | 0.763 | 20 | 3 | 0.234 | 14 | 9 | 0.289 |
|
Never-smoker | 39 | 10 |
| 35 | 14 |
| 36 | 13 |
|
| EGFR mutation
status |
|
|
|
|
|
|
|
|
|
|
19del | 27 | 8 | 0.572 | 25 | 10 | 0.455 | 24 | 11 | 0.978 |
|
21L858R | 29 | 5 |
| 27 | 7 |
| 24 | 10 |
|
|
Others | 2 | 1 |
| 3 | 0 |
| 2 | 1 |
|
| TNM stage |
|
|
|
|
|
|
|
|
|
|
IIB | 1 | 0 | 0.034c | 0 | 1 | 0.007d | 0 | 1 | 0.137 |
|
IIIA/IIIB | 16 | 9 |
| 15 | 10 |
| 20 | 5 |
|
| IV | 41 | 5 |
| 40 | 6 |
| 30 | 16 |
|
|
Differentiation |
|
|
|
|
|
|
|
|
|
|
Well | 6 | 8 |
P<0.001d | 8 | 7 | 0.059 | 5 | 9 | 0.008c |
|
Moderate | 24 | 2 |
| 21 | 5 |
| 21 | 5 |
|
|
Poor | 28 | 4 |
| 26 | 5 |
| 24 | 8 |
|
| Type of
EGFR-TKIs |
|
|
|
|
|
|
|
|
|
|
Gefitinib | 25 | 7 | 0.637 | 21 | 10 | 0.236 | 22 | 10 | 0.742 |
|
Icotinib | 20 | 3 |
| 18 | 5 |
| 15 | 8 |
|
|
Erlotinib | 13 | 4 |
| 16 | 2 |
| 13 | 4 |
|
IHC and immunofluorescence
staining
The expression levels of SOX2, OCT4 and
NANOG were examined in the tissue samples from the two
groups of patients with lung adenocarcinoma having significantly
different prognosis to elucidate whether the expression of lung
CSC-related markers was associated with the efficacy of EGFR-TKIs
in lung adenocarcinomas. The representative confocal images of
SOX2, OCT4 and NANOG with immunofluorescence staining
in lung adenocarcinoma specimens are presented in Fig. 2. IHC staining revealed the
expression of SOX2 in 58 lung adenocarcinoma cases; 55 cases
exhibited cells positive for OCT4, and 50 cases exhibited
the expression of NANOG. The staining intensities of
SOX2, OCT4 and NANOG are presented in Fig. 3. The color reactions were observed
in the nucleus and cytoplasm of the tumor cells. The staining
intensity was divided into four grades based on the staining depth
of positive cells: Unstained (negative), weakly positive (+),
moderately positive (++), and strongly positive (+++). The tissue
sections of each sample were stained by IHC staining, and the
H-score was calculated. The results of the t-test revealed
that the expression levels of SOX2 (Fig. 4A, P=0.003), OCT4 (Fig. 4B, P=0.036), and NANOG
(Fig. 4C, P=0.032) were
significantly higher in the resistant group than in the sensitive
group.
PCR detection for SOX2, OCT4 and
NANOG
The relative expression levels of SOX2, OCT4
and NANOG were detected using RT-qPCR. RT-qPCR could not be
performed in five patients due to the small tissue sample. The
GAPDH housekeeping gene was used as a control. The
expression level of each tested gene relative to the expression of
GAPDH was calculated using the ΔΔCq method. Significant
differences in the expression level of each tested gene in the two
groups were identified using the unpaired nonparametric
t-test. The results revealed that the relative levels of
SOX2 (Fig. 5A, P=0.018),
OCT4 (Fig. 5B, P=0.035) and
NANOG (Fig. 5C, P=0.044)
were significantly higher in the resistant group than in the
sensitive group. The RT-qPCR results were consistent with the IHC
analysis results.
Discussion
Lung cancer is one of the most common malignant
tumors, accounting for nearly 30% of all cancer-related deaths
(21). The clinical use of
first-generation EGFR-TKIs has revealed significant efficacy for
advanced lung adenocarcinoma patients with EGFR-sensitive mutation
and an increased survival rate. However, two groups of patients had
significantly different therapeutic effects after using the
targeted drugs. These included patients with rapid progression or a
long period of disease stabilization without progression. At
present, the resistance to EGFR-TKI therapy, either intrinsic or
acquired, is a major barrier to a complete cure. The median PFS for
patients treated with EGFR-TKIs is only ~10-13 months (14–16).
In the present study, IHC and RT-qPCR analyses were
used to detect and analyze the expression of CSC-related markers in
patients with lung adenocarcinoma having significantly different
effects after treatment with EGFR-TKIs. The results revealed that
the level of CSC markers was higher in the EGFR-TKI resistant group
than in the EGFR-TKI sensitive group, and the difference was
statistically significant. In addition, the expression of
SOX2 and OCT4 was significantly correlated with PFS,
TNM stage, and differentiation, whereas the expression of
NANOG was significantly associated to PFS and
differentiation.
CSCs play an important role in tumor development,
infiltration, metastasis and recurrence (2,22).
SOX2, OCT4 and NANOG are the core transcriptional
regulators of CSCs and constitute the core transcriptional network
to maintain pluripotency and self-renewal ability of CSCs (9,23,24).
These transcription factors can reprogram somatic cells into
pluripotent stem cells and contribute to drug resistance and poor
prognosis (25,26). Several previous studies indicated
that OCT4, SOX2 and NANOG were involved in the
occurrence, drug resistance, and development of tumors (11,23,24,27).
Shien et al investigated the molecular and cellular profiles
of cells with acquired resistant to EGFR-TKI in EGFR-mutant lung
cancers. In their study, gefitinib-resistant sublines were
established by exposing EGFR mutant cell lines to gefitinib using
stepwise escalation and high-concentration exposure methods. They
found that the sublines established by the high-concentration
exposure methods had CSC-like properties (28). The targeted drugs could kill some of
the rapidly dividing cancer cells with sensitive EGFR mutations.
However, CSCs not sensitive to various treatments could be further
replicated, self-renewed, and evolved into more drug-resistant lung
cancer cells after a period of EGFR-TKI therapy.
In the present study, it was revealed that the
number of male patients, patients who smoked, patients with stage
IV, and patients with poor differentiation was significantly higher
in the resistant group than in the sensitive group. These results
were consistent with the findings of other previous studies
(29–34). Since a majority of the smokers were
men in the present study, it is possible that smoking may make the
interaction between sex and the efficacy of EGFR-TKIs confusing. In
addition, some studies have demonstrated that the expression of CSC
markers was related to clinical and pathological parameters in
patients with lung cancer disease (35,36).
For instance, Li et al revealed that the high expression
level of CSC markers was significantly associated with poorer
differentiation, higher TNM stage and worse prognosis of lung
cancer (35).
The present study also had some limitations and
biases that should be acknowledged. First, the sample size of this
study was relatively small. The follow-up studies should have a
larger sample size for further verification. Second, this study
suffered from selection bias due to its retrospective nature. In
addition, the mechanisms discussed in this study required further
experiments.
Therefore, the present study concluded that the
therapeutic effect of first-generation EGFR-TKIs was highly
correlated with the levels of CSC-related markers. The markers of
CSCs were highly expressed in EGFR-TKI resistant patients.
Acknowledgements
The authors are grateful to all patients enrolled in
this study.
Funding
The present study was supported by the Science and
Technology Commission of Shanghai Municipality, China (no.
18441904700) and the Nurture projects for Basic Research of
Shanghai Chest Hospital (no. 2018YNJCM05).
Availability of data and materials
The data used and/or analyzed in this study can be
obtained from the corresponding author on reasonable request.
Authors' contributions
XZha and FH designed and supervised the study, and
edited the manuscript. BH and CL designed the experiments and
analyzed the data. FH, LZ and XY performed all experimental work.
HZ, XZhe and YS supported administration and drafted the work or
revised it critically for important intellectual content. All
authors read, approved the final manuscript and agree to be
accountable for all aspects of the work in ensuring that questions
related to the accuracy or integrity of any part of the work are
appropriately investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Shanghai Chest Hospital. All patients provided their
informed consent. Investigations involving human patients in this
study were performed according to the principles of the Declaration
of Helsinki and the ethical standards of the national research
committee.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
EGFR
|
epidermal growth factor receptor
|
|
TKIs
|
tyrosine kinase inhibitors
|
|
IHC
|
immunohistochemistry
|
|
PFS
|
progression-free survival
|
|
RT-qPCR
|
reverse transcriptase-quantitative
polymerase chain reaction
|
|
NSCLC
|
non-small cell lung cancer
|
|
CSCs
|
cancer stem cells
|
|
TNM
|
tumor size, lymph node and distant
metastasis
|
|
SOX2
|
sex-determining region Y-box 2
|
|
OCT4
|
octamer-binding transcription factor
4
|
|
NANOG
|
NANOG homeobox
|
|
ARMS
|
amplification refractory mutation
system
|
References
|
1
|
Spira A and Ettinger DS: Multidisciplinary
management of lung cancer. N Engl J Med. 350:379–392. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Leon G, MacDonagh L, Finn SP, Cuffe S and
Barr MP: Cancer stem cells in drug resistant lung cancer: Targeting
cell surface markers and signaling pathways. Pharmacol Ther.
158:71–90. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Huang L and Fu L: Mechanisms of resistance
to EGFR tyrosine kinase inhibitors. Acta Pharm Sin B. 5:390–401.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kobayashi S, Boggon TJ, Dayaram T, Jänne
PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG and Halmos
B: EGFR mutation and resistance of non-small-cell lung cancer to
gefitinib. N Engl J Med. 352:786–792. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Engelman JA, Zejnullahu K, Mitsudomi T,
Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen
J, et al: MET amplification leads to gefitinib resistance in lung
cancer by activating ERBB3 signaling. Science. 316:1039–1043. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rho JK, Choi YJ, Kim SY, Kim TW, Choi EK,
Yoon SJ, Park BM, Park E, Bae JH, Choi CM and Lee JC: MET and AXL
inhibitor NPS-1034 exerts efficacy against lung cancer cells
resistant to EGFR kinase inhibitors because of MET or AXL
activation. Cancer Res. 74:253–262. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sequist LV, Waltman BA, Dias-Santagata D,
Digumarthy S, Turke AB, Fidias P, Bergethon K, Shaw AT, Gettinger
S, Cosper AK, et al: Genotypic and histological evolution of lung
cancers acquiring resistance to EGFR inhibitors. Sci Transl Med.
3:75ra262011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
MacDonagh L, Gray SG, Breen E, Cuffe S,
Finn SP, O'Byrne KJ and Barr MP: Lung cancer stem cells: The root
of resistance. Cancer Lett. 372:147–156. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Loh YH, Wu Q, Chew JL, Vega VB, Zhang W,
Chen X, Bourque G, George J, Leong B, Liu J, et al: The Oct4 and
Nanog transcription network regulates pluripotency in mouse
embryonic stem cells. Nat Genet. 38:431–440. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang Z, Oron E, Nelson B, Razis S and
Ivanova N: Distinct lineage specification roles for NANOG, OCT4,
and SOX2 in human embryonic stem cells. Cell Stem Cell. 10:440–454.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cai L, Ye Z, Zhou BY, Mali P, Zhou C and
Cheng L: Promoting human embryonic stem cell renewal or
differentiation by modulating Wnt signal and culture conditions.
Cell Res. 17:62–72. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lazarevic M, Milosevic M, Trisic D, Toljic
B, Simonovic J, Nikolic N, Mikovic N, Jelovac D, Petrovic M,
Vukadinovic M and Milasin J: Putative cancer stem cells are present
in surgical margins of oral squamous cell carcinoma. J BUON.
23:1686–1692. 2018.PubMed/NCBI
|
|
13
|
Milosevic M, Lazarevic M, Toljic B,
Simonovic J, Trisic D, Nikolic N, Petrovic M and Milasin J:
Characterization of stem-like cancer cells in basal cell carcinoma
and its surgical margins. Exp Dermatol. 27:1160–1165. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tan CS, Gilligan D and Pacey S: Treatment
approaches for EGFR-inhibitor-resistant patients with
non-small-cell lung cancer. Lancet Oncol. 16:e447–e459. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cho BC, Chewaskulyong B, Lee KH,
Dechaphunkul A, Sriuranpong V, Imamura F, Nogami N, Kurata T,
Okamoto I, Zhou C, et al: Osimertinib versus standard of care EGFR
TKI as first-line treatment in patients with EGFRm advanced NSCLC:
FLAURA asian subset. J Thorac Oncol. 14:99–106. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kelly RJ, Shepherd FA, Krivoshik A, Jie F
and Horn L: A phase 3, randomized, open-label study of asp8273
versus erlotinib or gefitinib in patients with advanced stage
iiib/iv non-small cell lung cancer. Ann Oncol. 30:1127–1133. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cha YK, Lee HY, Ahn MJ, Choi YL, Lee JH,
Park K and Lee KS: Survival outcome assessed according to tumor
burden and progression patterns in patients with epidermal growth
factor receptor mutant lung adenocarcinoma undergoing epidermal
growth factor receptor tyrosine kinase inhibitor therapy. Clin Lung
Cancer. 16:228–236. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gu D, Su S, Ge D, Chen S, Huang J, Li B,
Chen R and Qiang B: Association study with 33 single-nucleotide
polymorphisms in 11 candidate genes for hypertension in Chinese.
Hypertension. 47:1147–1154. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
McClelland RA, Finlay P, Walker KJ,
Nicholson D, Robertson JF, Blamey RW and Nicholson I: Automated
quantitation of immunocytochemically localized estrogen receptors
in human breast cancer. Cancer Res. 50:3545–3550. 1990.PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA: Cancer J Clin. 60:277–300. 2010.PubMed/NCBI
|
|
22
|
Cojoc M, Mabert K, Muders MH and Dubrovska
A: A role for cancer stem cells in therapy resistance: Cellular and
molecular mechanisms. Semin Cancer Biol. 31:16–27. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nakatsugawa M, Takahashi A, Hirohashi Y,
Torigoe T, Inoda S, Murase M, Asanuma H, Tamura Y, Morita R,
Michifuri Y, et al: SOX2 is overexpressed in stem-like cells of
human lung adenocarcinoma and augments the tumorigenicity. Lab
Invest. 91:1796–1804. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang X, Hu F, Li C, Zheng X, Zhang B,
Wang H, Tao G, Xu J, Zhang Y and Han B: OCT4&SOX2-specific
cytotoxic T lymphocytes plus programmed cell death protein 1
inhibitor presented with synergistic effect on killing lung cancer
stem-like cells in vitro and treating drug-resistant lung cancer
mice in vivo. J Cell Physiol. 234:6758–6768. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rousseaux S, Debernardi A, Jacquiau B,
Vitte AL, Vesin A, Nagy-Mignotte H, Moro-Sibilot D, Brichon PY,
Lantuejoul S, Hainaut P, et al: Ectopic activation of germline and
placental genes identifies aggressive metastasis-prone lung
cancers. Sci Transl Med. 5:186ra662013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sharma SV, Lee DY, Li B, Quinlan MP,
Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach
MA, et al: A chromatin-mediated reversible drug-tolerant state in
cancer cell subpopulations. Cell. 141:69–80. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chiou SH, Wang ML, Chou YT, Chen CJ, Hong
CF, Hsieh WJ, Chang HT, Chen YS, Lin TW, Hsu HS and Wu CW:
Coexpression of Oct4 and Nanog enhances malignancy in lung
adenocarcinoma by inducing cancer stem cell-like properties and
epithelial-mesenchymal transdifferentiation. Cancer Res.
70:10433–10444. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shien K, Toyooka S, Yamamoto H, Soh J,
Jida M, Thu KL, Hashida S, Maki Y, Ichihara E, Asano H, et al:
Acquired resistance to EGFR inhibitors is associated with a
manifestation of stem cell-like properties in cancer cells. Cancer
Res. 73:3051–3061. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hassan KA, Chen G, Kalemkerian GP, Wicha
MS and Beer DG: An embryonic stem cell-like signature identifies
poorly differentiated lung adenocarcinoma but not squamous cell
carcinoma. Clin Cancer Res. 15:6386–6390. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang Y, Kang S, Fang W, Hong S, Liang W,
Yan Y, Qin T, Tang Y, Sheng J and Zhang L: Impact of smoking status
on EGFR-TKI efficacy for advanced non-small-cell lung cancer in
EGFR mutants: A meta-analysis. Clin Lung Cancer. 16:144–51.e1.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hasegawa Y, Ando M, Maemondo M, Yamamoto
S, Isa S, Saka H, Kubo A, Kawaguchi T, Takada M, Rosell R, et al:
The role of smoking status on the progression-free survival of
non-small cell lung cancer patients harboring activating epidermal
growth factor receptor (EGFR) mutations receiving first-line EGFR
tyrosine kinase inhibitor versus platinum doublet chemotherapy: A
meta-analysis of prospective randomized trials. Oncologist.
20:307–315. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bria E, Milella M, Cuppone F, Novello S,
Ceribelli A, Vaccaro V, Sperduti I, Gelibter A, Scagliotti GV,
Cognetti and Giannarelli D: Outcome of advanced NSCLC patients
harboring sensitizing EGFR mutations randomized to EGFR tyrosine
kinase inhibitors or chemotherapy as first-line treatment: A
meta-analysis. Ann Oncol. 22:2277–2285. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhou J and Ben S: Comparison of
therapeutic effects of EGFR-tyrosine kinase inhibitors on 19Del and
L858R mutations in advanced lung adenocarcinoma and effect on
cellular immune function. Thoracic Cancer. 9:228–233. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lee CK, Wu YL, Ding PN, Lord SJ, Inoue A,
Zhou C, Mitsudomi T, Rosell R, Pavlakis N, Links M, et al: Impact
of specific epidermal growth factor receptor (egfr) mutations and
clinical characteristics on outcomes after treatment with egfr
tyrosine kinase inhibitors versus chemotherapy in egfr-mutant lung
cancer: A meta-analysis. J Clin Oncol. 33:1958–1965. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li X, Wang J, Xu Z, Ahmad A, Li E, Wang Y,
Qin S and Wang Q: Expression of Sox2 and Oct4 and their clinical
significance in human non-small-cell lung cancer. Int J Mol Sci.
13:7663–7675. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wu Y, Du X, Xue C, Li D, Zheng Q, Li X and
Chen H: Quantification of serum SOX2 DNA with FQ-PCR potentially
provides a diagnostic biomarker for lung cancer. Med Oncol.
30:7372013. View Article : Google Scholar : PubMed/NCBI
|