Introduction
Osteosarcoma (OS) is a primary malignant bone tumor
that typically affects the long tubular bones of children and
adults worldwide (1). OS is
characterized by the direct formation of immature bone or osteoid
tissue by tumor cells (2), as well
as poor prognosis (3,4). Despite medical advances and
development of early diagnosis, the prognosis of patients with
metastatic osteosarcoma remains poor, with a cure rate of ~30%
(5–7). OS remains a leading cause of
cancer-associated mortality among children and teenagers (8–10).
Therefore, it is urgently necessary to develop novel targets and
research the underlying mechanisms of bone carcinogenesis, which is
critical for the treatment and prognosis of OS in the future.
Long non-coding RNAs (lncRNAs) are sequences >200
nt in length and without protein-coding capacity (11). Previous studies have provided
evidence that lncRNAs serve significant roles in the biological
behavior of tumors, including proliferation, invasion, metastasis,
differentiation and apoptosis (12–15).
Notably, a previous study has confirmed the dysregulation of
lncRNAs in human OS (16). lncRNA
FGFR3-AS1 promotes OS growth by regulating fibroblast growth factor
receptor 3 (17). In addition,
lncRNA small nucleolar RNA host gene 12 promotes OS cell
proliferation and migration in vitro (18). Therefore, further investigations are
required to evaluate the expression pattern, biological role and
functional mechanisms of lncRNAs in OS. Long non-coding RNA gastric
carcinoma proliferation enhancing transcript 1 (lncGHET1; AK123072)
has been demonstrated to be abnormally expressed in gastric,
bladder and colorectal cancer (19–22).
However, the role and underlying mechanisms of lncGHET1 in OS
remain unclear.
The present study analyzed the lncGHET1 expression
pattern in OS cell lines using reverse transcription-quantitative
PCR (RT-qPCR). In addition, loss-of-function experiments were
performed to investigate the biological roles of lncGHET1 in OS.
The results revealed that lncGHET1 expression was upregulated in OS
cell lines. Additionally, the results of in vitro and in
vivo assays revealed that silenced lncGHET1 inhibited cell
proliferation, tumor growth, migration, invasion and epithelial-
to-mesenchymal transition (EMT), and promoted apoptosis, partly via
the Wnt/β-catenin signaling pathway. The findings suggested that
lncGHET1 functions as an oncogene, which may contribute to the
development for diagnosis and treatment of OS.
Materials and methods
Cell culture and transfection
Human OS U2OS, MG-63 and SaOs-2 cell lines, and the
epithelial hFOB cell line were purchased from American Type Culture
Collection. All cells were cultured in RPMI-1640 medium
(Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10%
FBS (Gibco; Thermo Fisher Scientific, Inc.) and 100 U/ml penicillin
and streptomycin (Thermo Fisher Scientific, Inc.) and maintained at
37°C with 5% CO2. Small interfering RNA
(si/siRNA)-lncGHET1 (5′-CGGCAGGCATTAGAGATGAACAGCA-3′) and negative
control (si-NC) (5′-CGGCAGGCAUUAGAGAUGAACAGCA-3′) were designed and
synthesized by Shanghai GenePharma Co., Ltd. For cell transfection,
50 nM siRNA or si-NC was transfected into U2OS and MG-63 cells
(1×106) using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. After 48 h, the cells were collected for
further experimentation. The transfection efficiency was evaluated
by GFP and RT-qPCR analyses.
Cell Counting Kit-8 (CCK-8) assay
A CCK-8 (Sigma-Aldrich; Merck KGaA) assay was
performed to examine cell viability according to the manufacturer's
protocol. Briefly, MG-63 and U2OS cells were seeded at a density of
1×104 per well and incubated in a humidified incubator
at 37°C for 24, 48 and 72 h. Subsequently, the cells were incubated
with 10 µl CCK-8 solution for another 2 h at 37°C. Optical density
was determined at a wavelength of 450 nm.
Colony formation and
5-ethynyl-20-deoxyuridine (EdU) assay
For the colony formation assay, transfected cells
(1,000 cells/well) were seeded and cultured for 14 days.
Subsequently, the cells were fixed with 4% polyoxymethylene for 20
min at room temperature and stained with 10% crystal violet for 30
min at room temperature. The colonies were counted and analyzed
under a light microscope. For the EdU assay, 1×106
transfected cells were seeded and cultured for 24 h, and then fixed
with 4% paraformaldehyde at room temperature for 15 min, followed
by 100 µl 1X Hoechst 33342 (5 µg/ml) staining for 30 min at room
temperature. Cells were visualized under a fluorescence microscope
(Leica Microsystems GmbH).
RT-qPCR analysis
Total RNAs from U2OS and MG-63 cells with different
treatments were extracted using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Reverse transcription was performed with
SuperScriptTM II reverse transcriptase (Invitrogen; Thermo Fisher
Scientific, Inc.). The protocol was as follows: 42°C for 60 min,
70°C for 15 min and chilling at 4°C. The relative levels of genes
were detected by RT-qPCR using SYBR Premix Ex Taq™ (Takara Bio,
Inc.). PCR cycling conditions were as follows: 95°C for 5 min,
followed by denaturation for 10 sec at 95°C and extension for 30
sec at 60°C for 40 cycles, and 60°C for 5 min. GAPDH was used as an
internal loading control. All reactions were performed in
triplicate. Fold changes were calculated using the
2−ΔΔCq method (23). The
primers were as follows: GHET1 forward, 5′-CCCCACAAATGAAGACACT-3′
and reverse, 5′-TTCCCAACACCCTATAAGAT-3′; snail forward,
5′-TGTTGCAGTGAGGGCAAGAA-3′ and reverse, 5′-GACCCTGGTTGCTTCAAGGA-3′;
N-cadherin forward, 5′-CGAGCCGCCTGCGCTGCCAC-3′ and reverse,
5′-CGCTGCTCTCCGCTCCCCGC-3′; E-cadherin forward,
5′-TACGCCTGGGACTCCACCTA-3′ and reverse, 5′-CCAGAAACGGAGGCCTGAT-3′;
vimentin forward, 5′-TACAGGAAGCTGCTGGAAGG-3′ and reverse,
5′-ACCAGAGGGAGTGAATCCAG-3′; and GAPDH forward,
5′-CGGAGTCAACGGATTTGGTCGTAT-3′ and reverse,
5′-AGCCTTCTCCATGGTGGTGAAGAC-3′.
Western blot analysis
Total proteins were extracted using RIPA buffer
(Cell Signaling Technology, Inc.) and the protein concentrations
were determined using the BCA Protein Assay kit (Beyotime Institute
of Biotechnology). Proteins (30 µg/lane) were separated by 10%
SDS-PAGE and transferred to PVDF membranes (EMD Millipore).
Membranes were blocked with 5% non-fat milk for 2 h at room
temperature. Subsequently, the membranes were incubated with
primary antibody at 4°C overnight. After washing with TBS with 1%
Tween-20, the membranes were probed with HRP-conjugated rabbit
anti-mouse IgG secondary antibodies (cat. nos. ab6721 and ab6728;
dilution, 1:5,000; Abcam) for 1 h at room temperature. GAPDH was
used as a loading control. The protein bands were visualized using
an Enhanced Chemiluminescence Detection system (Thermo Fisher
Scientific, Inc.). The data were analyzed using Image-Pro plus
software v6.0 (National Institutes of Health). Antibodies for p21
(cat. no. 64016; dilution, 1:1,000), p27 (cat. no. 3688; dilution,
1:2,000), cyclin D1 (cat. no. 55506; dilution, 1:2,000), GAPDH
(cat. no. 5174; dilution, 1:5,000), Bax (cat. no. 2772; dilution,
1:2,000), Bcl-2 (cat. no. 3498; dilution, 1:2,000), caspase-3 (cat.
no. 9664; dilution, 1:1,000), caspase-9 (cat. no. 9509; dilution,
1:2,000) and MMP-2 (cat. no. 87809; dilution, 1:1,000) were
purchased from Cell Signaling Technology, Inc. Antibodies for MMP-9
(cat. no. ab38898; dilution, 1:2,000), Snail (cat. no. ab53519;
dilution, 1:2,000), N-cadherin (cat. no. ab18203; dilution,
1:2,000), E-cadherin (cat. no. ab231303; dilution, 1:2,000),
vimentin (cat. no. ab92547; dilution, 1:1,000), Wnt (cat. no.
ab219412; dilution, 1:1,000) and β-catenin (cat. no. ab16051;
dilution, 1:1,000). All of these antibodies were purchased from
Abcam.
Wound healing assay
In order to evaluate the role of lncGHET1 in
migration of OS cells, 1×106 MG-63 and U2OS cells were
seeded in a 6-well plate, and 2 ml culture medium supplemented with
10% FBS were added. When cells were grown to 90% confluence, cells
were synchronized in serum-free medium for 24 h, followed by a
standard wound (<3 mm) created on the cell monolayer (time set
as 0 h). A PBS solution was used to remove floating cells.
Subsequently, cells were incubated in fresh complete medium (1%
FBS) for 0, 24 and 48 h and the number of migrated cells was
observed and counted under a light microscope. The results were
quantified by Image-Pro plus software 6.0 (National Institutes of
Health).
Transwell assay
The migration and invasion abilities of MG-63 and
U2OS cells were measured using a transwell assay (cat. no. 3422;
6.5-mm insert; 8-µm polycarbonate membrane; Costar; Corning, Inc.).
Briefly, 1×105 transfected cells were suspended in
serum-free medium and seeded into the upper chamber without (for
the migration assay) Matrigel (BD Biosciences) or with a porous
membrane coated with Matrigel (BD Biosciences) (for the transwell
invasion assay). The Matrigel was incubated at 37°C for 4 h before
testing. Subsequently, serum-free medium was added into the upper
chambers. Medium containing 10% FBS was added into the lower
chambers. Following incubation for 48 h, the cells from the upper
compartments were scraped off with cotton swabs, whereas the cells
that migrated or invaded to the lower surface of the membrane were
fixed with 4% paraformaldehyde at room temperature for 10 min and
stained with 10% crystal violet at room temperature. The numbers of
migrated or invasive cells were counted in five random fields under
a light microscope at ×200 magnification. Experiments were
independently repeated three times.
Cell cycle and apoptosis analyses
The effect of lncGHET1 on cell cycle and apoptosis
was evaluated by flow cytometry using the Annexin V Apoptosis
Detection kit (BD Biosciences) and the Cell Cycle kit (BD
Biosciences). Briefly, for cell cycle analysis, MG-63 and U2OS
cells were harvested and washed with cold PBS twice. Following
fixation with 70% ethanol at 4°C overnight and labelling with
propidium iodide (PI) solution (0.1 µg/µl) in the presence of
Ribonuclease A (Takara Bio, Inc.) at 37°C for 30 min in the dark,
cells were assessed by flow cytometry (FACScan; BD Biosciences).
For cell apoptosis analysis, cells were washed with cold PBS twice.
Subsequently, cells were incubated with Annexin V-FITC and PI (BD
Biosciences) for 15 min in the dark at room temperature. Finally,
cell apoptosis was assessed using flow cytometry (FACScan; BD
Biosciences) using FlowJo 10.06 software (FlowJo LLC). The data of
these experiments were analyzed by FlowJo v10 software (Tree Star,
Inc.).
Xenograft tumors in nude mice
Female BALB/c nude mice (SPF, n=6; age, 6 weeks;
weight, 18–22 g) were provided by Nanjing Medical University
(Nanjing, China). The mice were housed in 12-h light/dark cycle in
a temperature-controlled (22-26°C) and humidity-controlled (40-70%)
room with freely accessible chow and tap water. To detect the
effect of tumor growth in vivo, 5×105 transfected
MG-63 and U2OS cells were suspended in PBS and subcutaneously
injected into the right flank of nude mice, and the mice of the
control group were treated with PBS. Following a 36-day period,
nude mice were euthanized by a high dose of anesthesia (150 mg/kg
pentobarbital; intraperitoneal injection) according to protocols
set by the Ethical Committee of the Ethical Committee of Nanjing
Medical University (approval no. SYXK(SU)2018-0114). After nude
mice were sacrificed, neoplasms were isolated for further analyses.
The tumor volumes were recorded by slide calipers every week and
calculated using the following formula: Volume=0.5 × length × width
× width.
Hematoxylin and eosin (H&E)
staining
The tumor tissues were fixed in 4% paraformaldehyde
at room temperature for 10 min and then embedded in paraffin. The
sections were immersed in 0.5% hematoxylin for 5 min, followed by
staining with 0.5% eosin solution for 1 min at room temperature. At
least three different sections of tumor tissues were examined for
each group using a light microscope to assess the histopathological
alterations.
Tunel assay
The in situ Cell Death Detection kit was
purchased from Roche Diagnostics. All the steps were preformed
according to the manufacturer's protocol. The sections were dewaxed
in xylene and hydrated by graded ethanol solution. Samples were
fixed in 10% formalin for 24 h at 4°C, embedded in paraffin and
assessed using TUNEL staining (Roche Diagnostics GmbH) for 1 h at
room temperature according to the manufacturer's protocol. After
washing with PBS for several times, sections (5-µm thick) were
incubated with 20 µg/ml proteinase K for 30 min at 37°C.
Subsequently, the sections were incubated with a terminal
deoxynucleotidyl transferase, followed by treatment with 3%
hydrogen peroxide for 5 min. After that, the sections were
incubated with peroxidase-conjugated antibody (cat. no. ab197034;
dilution, 1:1,000; Abcam) for 10 min at room temperature. After
washing with PBS for several times, the DAB solution with 3%
hydrogen peroxide was added, and the methyl green was added.
Finally, the sections were treated with Mayer's hematoxylin. The
mounting medium was neutral resin. The images were observed in five
fields under a fluorescence microscope.
Statistical analysis
All experiments were performed at least three times.
The results are presented as the mean ± SD. Statistical analyses
were performed using SPSS v19.0 software (IBM Corp.). Student's
t-test was performed to evaluate significant differences between
two independent groups of samples. One-way ANOVA followed by
Tukey's post hoc test was applied to compare differences among
multiple groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
lncGHET1 is upregulated in OS cell
lines
The present study first examined the expression
levels of lncGHET1 in the U2OS, MG63, SaOs-2 and hFOB cell lines.
Compared with the hFOB cell line, lncGHET1 expression was markedly
higher in the OS cell lines (Fig.
1A). As shown in Fig. 1B, the
cell proliferation abilities of U2OS, SaOs-2 and MG-63 cells were
significantly higher than those in the hFOB group at 72 h.
Furthermore, colony formation assays showed that there were more
colonies in the former three cell lines compared with in the hFOB
group (Fig. 1C). In addition, an
increased migration and invasion rate was observed in the U2OS,
SaOs-2 and MG-63 groups (Fig. 1D).
These data indicated that lncGHET1 may function as a regulator of
OS progression.
Effects of lncGHET1 on proliferation
of OS cells
To assess the role of lncGHET1 in MG-63 and U2OS
cell proliferation, siRNA was transfected to silence lncGHET1
expression. Following transfection for 48 h, lncGHET1 expression
was detected using fluorescence microscopy and RT-qPCR. As shown in
Fig. 2A and B, the results revealed
that the siRNA transfection decreased lncGHET1 expression, whereas
there was no significant difference observed between the si-NC and
control groups. Subsequently, cell proliferation was assessed using
CCK-8, colony formation and EdU assays. As demonstrated by the
result of the CCK-8 assay, cell growth was suppressed in MG-63 and
U2OS cells which were transfected with si-lncGHET1 compared with
the cells transfected with si-NC and the control group (Fig. 2C). In addition, the colony formation
ability of si-lncGHET1-transfected cells was decreased (Fig. 2D). An EdU assay was conducted to
further confirm the effect of lncGHET1 on OS cells. The results
revealed that downregulation of lncGHET1 markedly decreased the
percentage of EdU-positive cells in MG-63 and U2OS cells (Fig. 3A).
To study the effect of lncGHET1 on the OS cell
cycle, flow cytometry was performed. As shown in Fig. 3B, a significantly greater proportion
of G1 phase cells and a markedly lesser proportion of S
phase were observed in the si-lncGHET1 group compared with in the
si-NC and control groups in both MG-63 and U2OS cells.
Additionally, there was no significant difference among the three
groups in G2 phase. Furthermore, western blot analysis
was performed to determine the protein levels of relative factors,
including cyclin 1, p21 and p27, involved in cell cycle regulation.
As shown in Fig. 3C, compared with
the si-NC and control groups, the expression levels of p21 and p27
protein were identified to be increased, whereas cyclin D1
expression was revealed to be decreased in the si-lncGHET1 group.
Therefore, these data suggested that knockdown of lncGHET1
inhibited cell proliferation in MG-63 and U2OS cells.
Effect of lncGHET1 on development of
xenograft tumors in vivo in OS
To further confirm the effect of lncGHET1 on the
growth of tumors in vivo, MG-63 and U2OS cells transfected
with si-lncGHET1 or si-NC were injected into nude mice. As shown in
Fig. 4A-D, a significant decrease
in tumor volume and weight was observed in the si-lncGHET1 group
compared with in the control group and no significant change in
body weight was observed. Additionally, H&E staining was
performed to explore the histological alterations of tumors and the
results demonstrated that the tissue structure was clearer and more
complete in the si-lncGHET1 group than in the control group
(Fig. 4E). These results suggested
that silencing lncGHET1 expression inhibited the xenograft tumor
growth of MG-63 and U2OS cells in vivo.
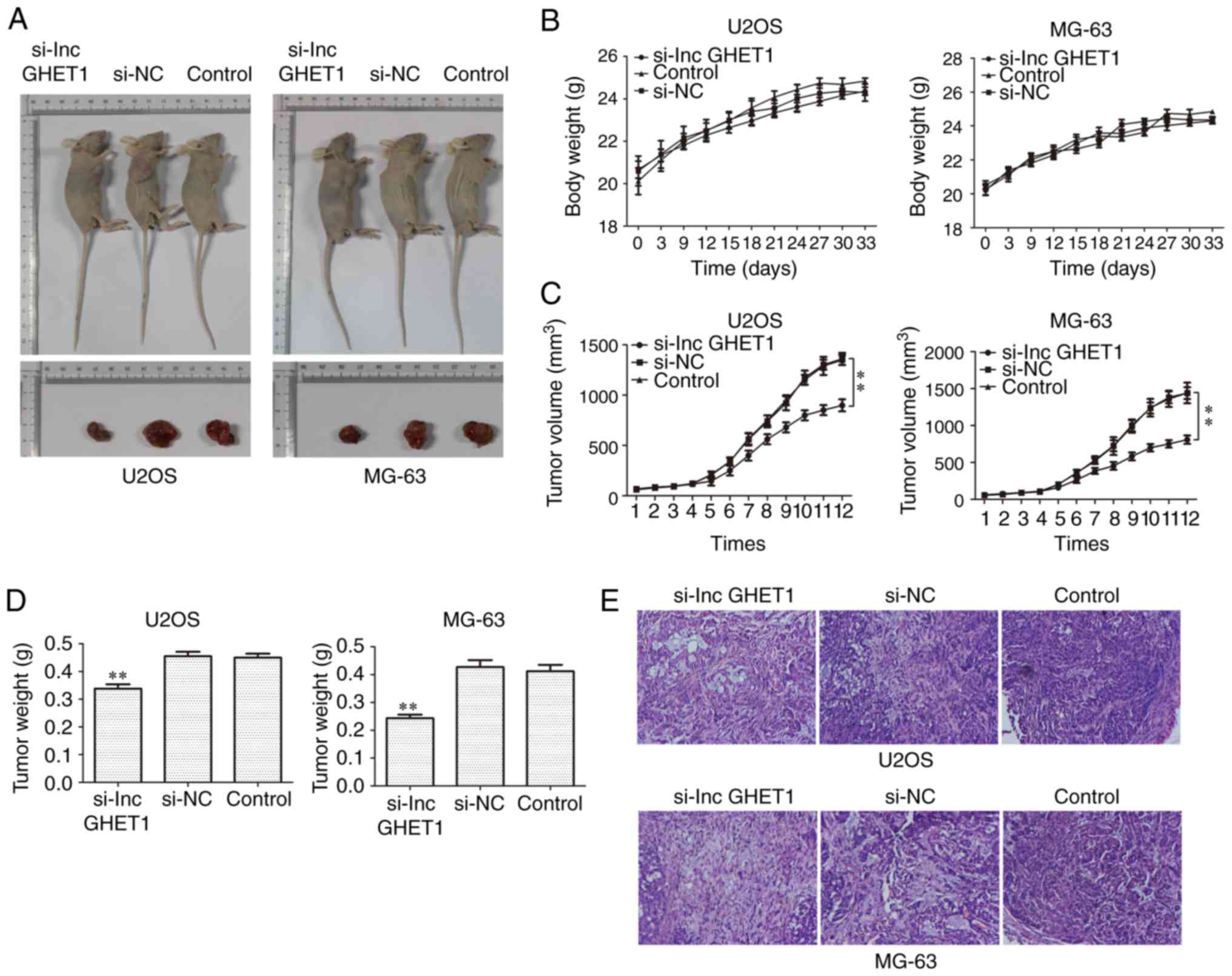 | Figure 4.Effect of lncGHET1 on cell growth
in vivo. (A) Images of nude mice, (B) bodyweight, (C) tumor
volume and (D) tumor weight are shown. The longest and shortest
diameters of the tumor size in U2OS infected nude mice were 13.9,
20.1, 19.0 mm; 7.1, 13.4, 12.5 mm, successively. And these
parameters of tumor size in the MG-63 infected nude mice were 15.1,
17.2, 20.0 mm; 10.9, 11.5, 11.3 mm, successively. (E) Hematoxylin
and eosin staining was performed and the images were captured at a
magnification of ×200. **P<0.01 vs. control group. lncGHET1,
long non-coding RNA gastric carcinoma proliferation enhancing
transcript 1; NC, negative control; si, small interfering RNA. |
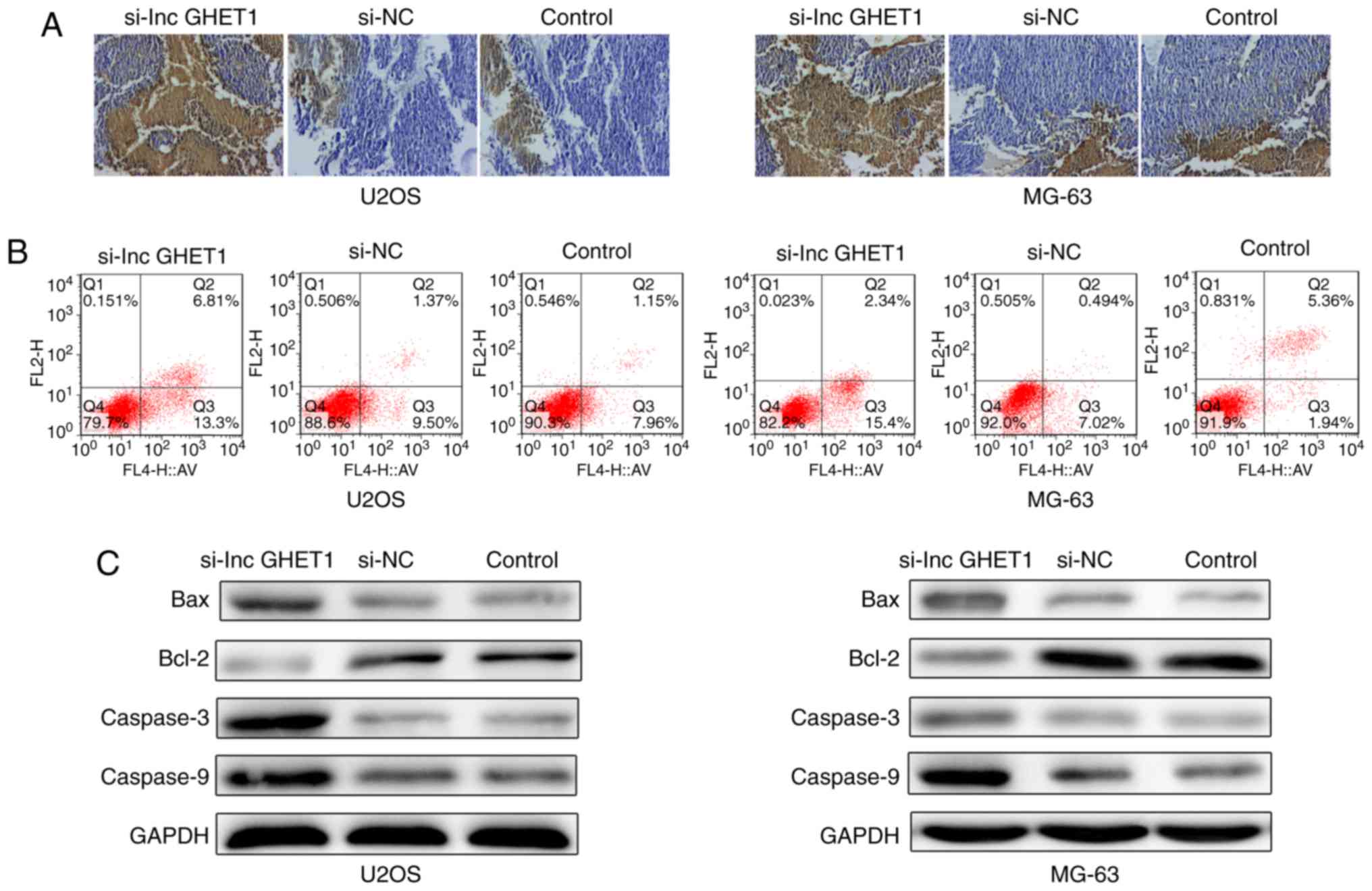
Effect of lncGHET1 on apoptosis of OS
cells in vivo and in vitro
The present study investigated whether lncGHET1
knockdown affected cell apoptosis. The results of a Tunel assay
revealed that the number of apoptotic cells was higher in nude mice
in the si-lncGHET1 group compared with in mice in the control group
(Fig. 5A). Flow cytometric analysis
revealed that the percentage of apoptotic U2OS cells was notably
increased in the si-lncGHET1 group, with 20.11% in the si-lncGHET1
group, 10.87% in the si-NC group and 9.11% in the control group of
U2OS cells. Furthermore, a similar result was observed in MG-63
cells. A notable increase was identified in the si-lncGHET1 group
(17.74%) compared with in the si-NC group (7.51%) and the control
group (7.3%; Fig. 5B).
Additionally, the present study examined several key apoptotic
players in MG-63 and U2OS cells by western blot analysis (Fig. 5C). Compared with the control group,
downregulation of lncGHET1 notably increased protein expression
levels of Bax, caspase-3 and caspase-9, and decreased the
expression levels of Bcl-2. These data indicated that reduced
lncGHET1expression promoted cell apoptosis in MG-63 and U2OS
cells.
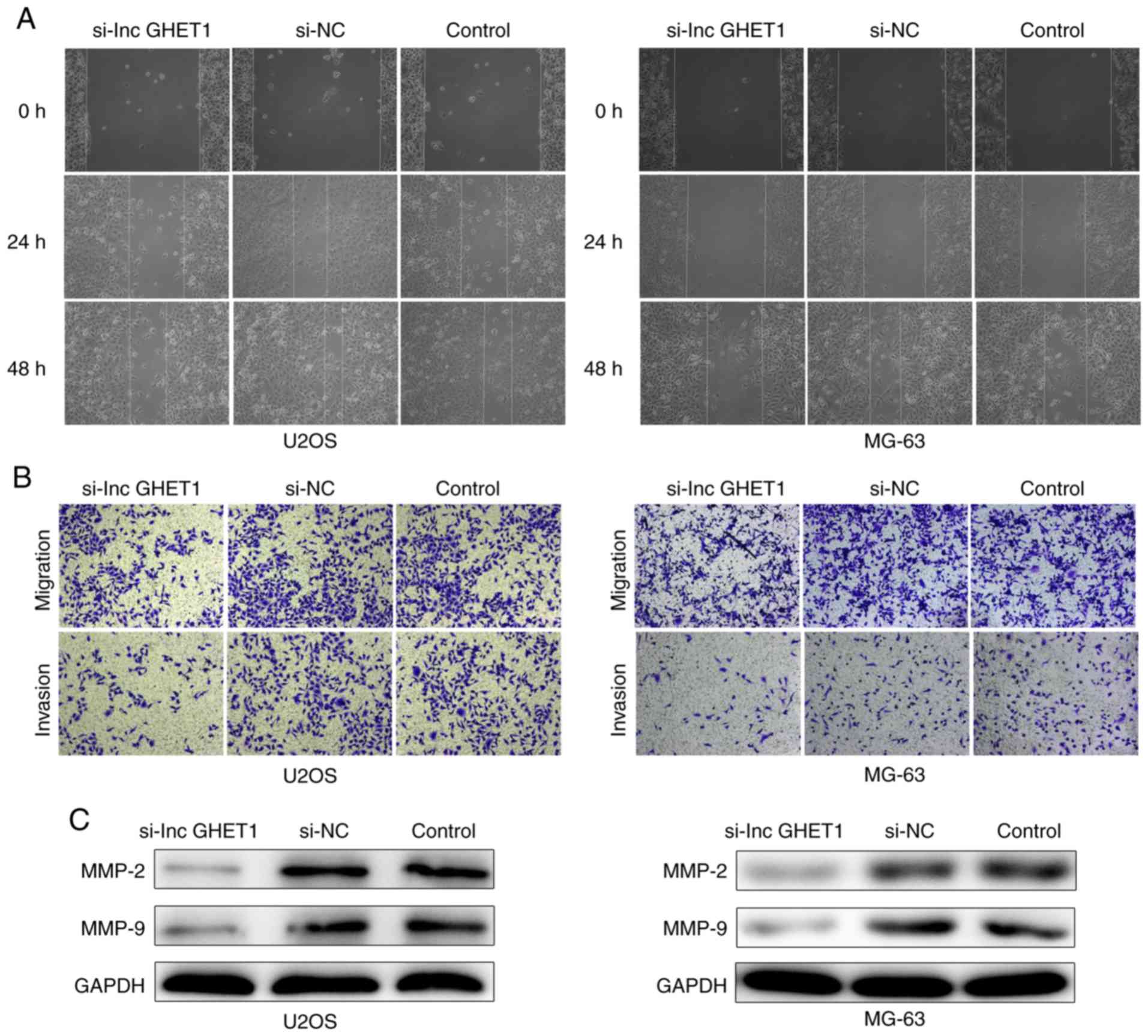
Effect of lncGHET1 on cell migration
and invasion in OS
In order to verify the potential role of lncGHET1 in
OS cell migration and invasion, wound healing, and transwell
migration and invasion assays were conducted. The results of the
wound healing assay revealed that cells migrated towards the wound
at a slower rate in the si-lncGHET1 group compared with in the
control group (Fig. 6A). In
addition, transwell migration assay results indicated that
knockdown of lncGHET1 markedly suppressed the migration ability of
MG-63 and U2OS cells compared with that of cells in the control
group, indicating a decreased migratory ability following lncGHET1
downregulation (Fig. 6B).
Similarly, transwell invasion assays revealed that the number of
invaded cells in the si-lncGHET1 group was lower than that in the
control group (Fig. 6B).
Furthermore, western blotting was applied to
evaluate the matrix metalloproteinase (MMP)-2 and MMP-9 protein
levels. As shown in Fig. 6C,
silencing lncGHET1 decreased the expression levels of MMP-2 and
MMP-9 in MG-63 and U2OS cells compared with the control group.
Therefore, the results suggested that the migration and invasion
abilities of MG-63 and U2OS cells were suppressed following
lncGHET1 knockdown.
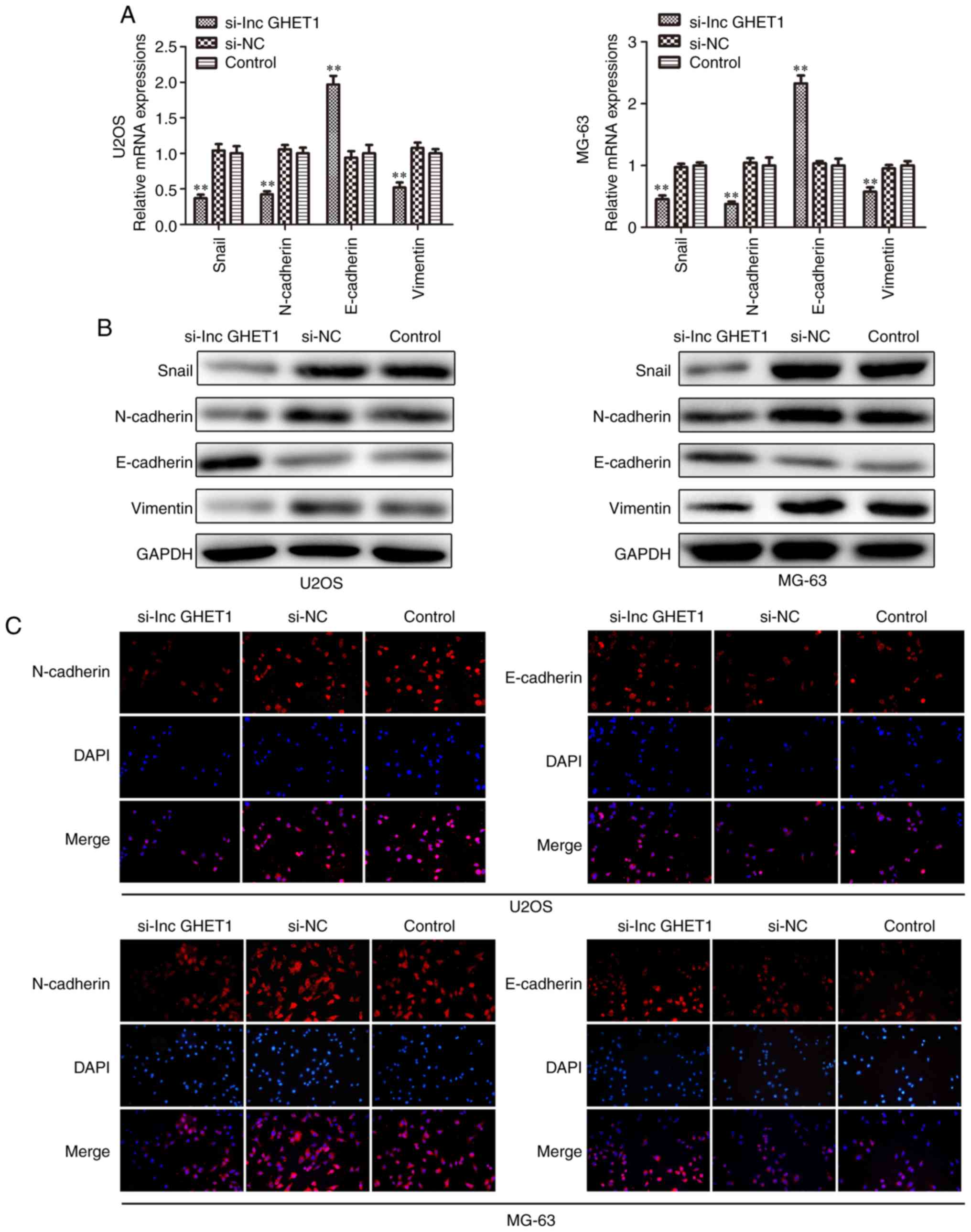
Effect of lncGHET1 on cell EMT of
OS
Increasing studies have demonstrated that EMT acts
as an important step in metastatic dissemination of cancer cells
(24–26) In the present study, the mRNA and
protein expression levels of Snail, N-cadherin, E-cadherin and
Vimentin were examined by RT-qPCR, western blotting and
immunofluorescence analyses. The results revealed that E-cadherin
mRNA expression was significantly upregulated. The levels of
protein expression were also increased, whereas the expression
levels of Snail, N-cadherin and Vimentin were downregulated in
MG-63 and U2OS cells which were transfected with si-lncGHET1
compared with in cells in the control group (Fig. 7A and B). As shown in Fig. 7C, compared with the si-NC and
control group, knockdown of lncGHET1 in the MG-63 and U2OS cells
was associated with lower expression of N-cadherin, while the
expression levels of E-cadherin remained high.
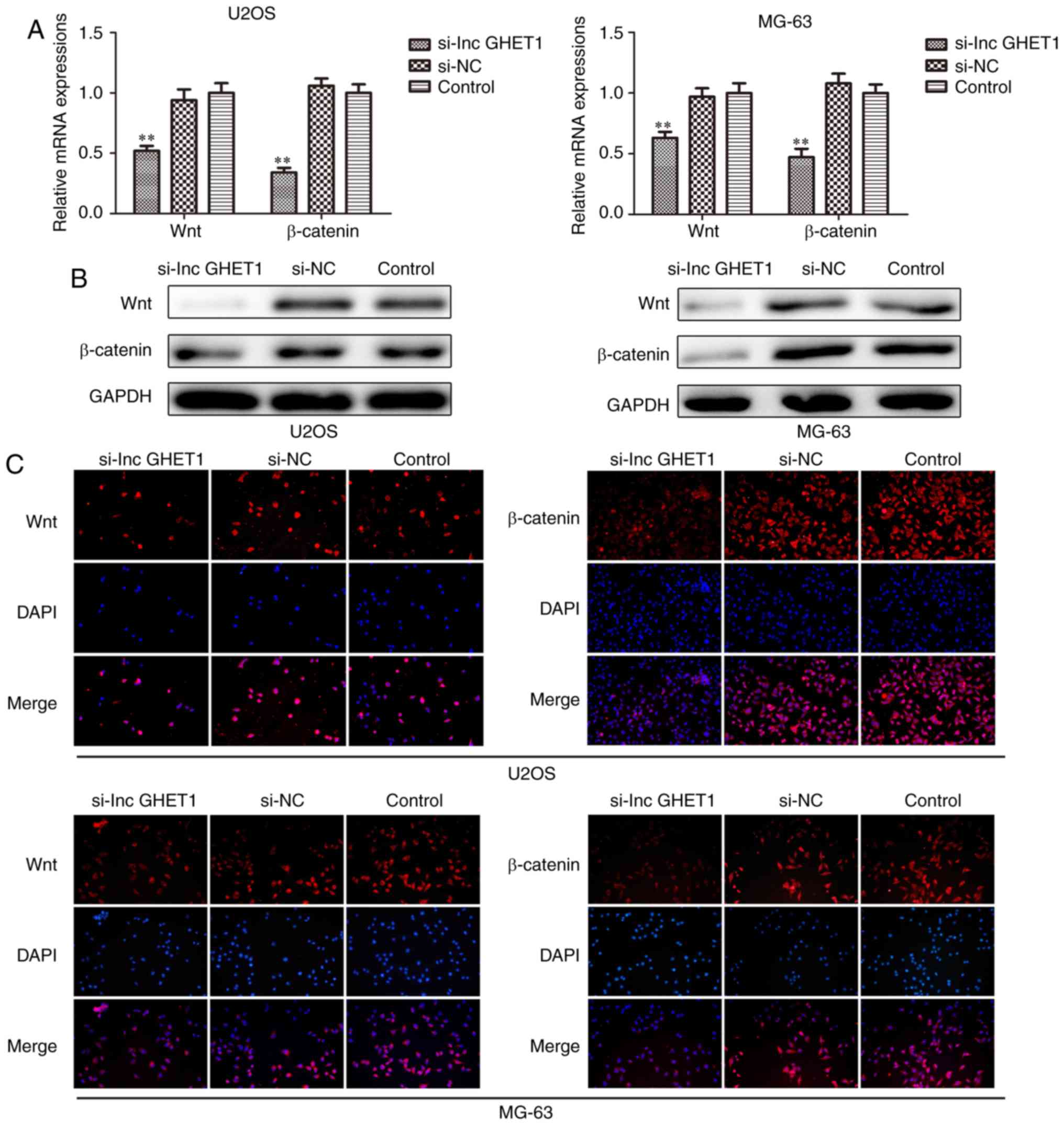
Knockdown of lncGHET1 inhibits cell
proliferation and EMT via the Wnt/β-catenin signaling pathway
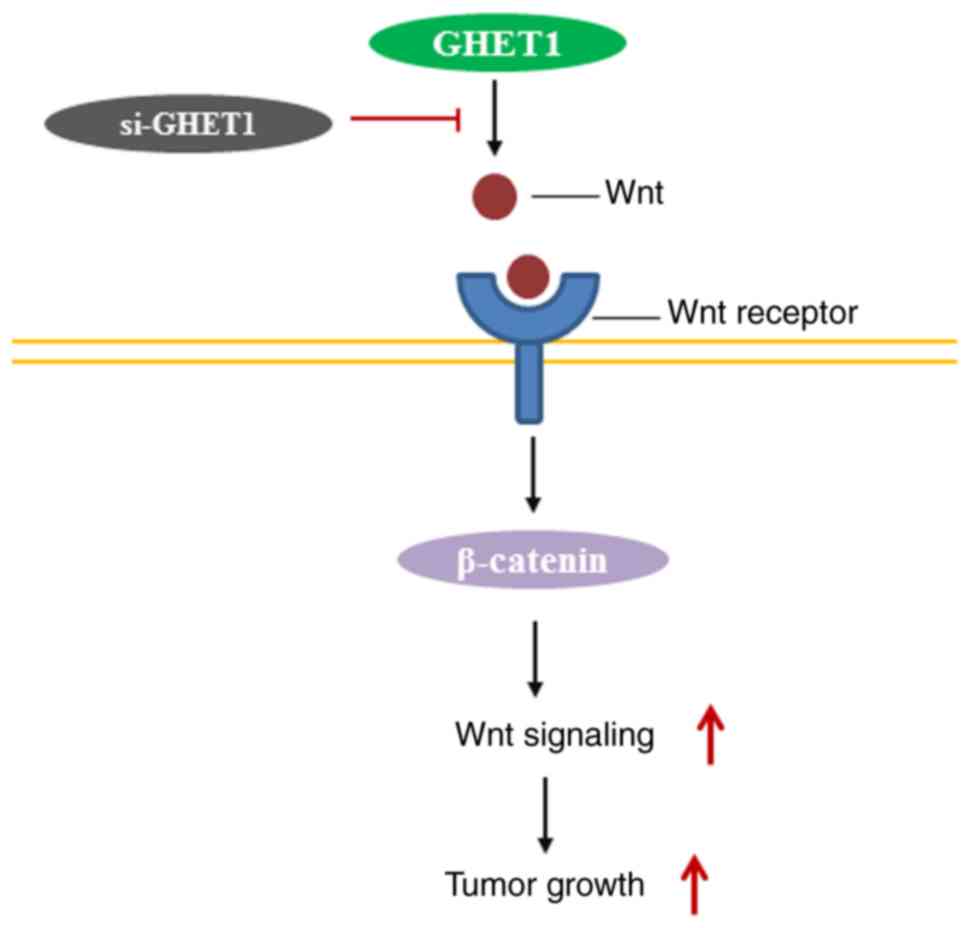
Furthermore, the underlying mechanism of the role
lncGHET1 serves in OS biology was explored. The present study
investigated whether lncGHET1 could regulate the proliferation and
EMT of OS cells via the Wnt/β-catenin signaling pathway. The
present study silenced the expression of lncGHET1 in MG-63 and U2OS
cells. Subsequently, the mRNA and protein expression levels of Wnt
and β-catenin were detected using RT-qPCR, western blotting and
immunofluorescence assays. The results demonstrated that expression
levels of Wnt and β-catenin were decreased following lncGHET1
knockdown when compared with controls (Fig. 8). These data indicated that lncGHET1
exerted its effects in OS partly via the Wnt/β-catenin signaling
pathway.
Discussion
OS is the most prevalent primary pediatric bone
malignancy in the world (3,27–29).
In recent years, advances of modern treatments have been used in
the treatment of OS, such as multiagent chemotherapy, surgery and
Chinese medical treatments (30,31).
However, due to limitations of the effectiveness, the prognosis of
OS is still unsatisfactory. The molecular mechanisms underlying the
pathogenesis of OS have not been fully explored. Therefore, it is
important to elucidate the predictive markers and underlying
regulation process in OS.
In recent years, the roles of lncRNAs have attracted
immense research interests worldwide. Aberrant expression of
lncRNAs has been revealed to be associated with pathogenesis of
cancer and other diseases (32–36).
Previous studies have revealed that they serve important roles in
physiological or pathology processes, particularly in cell growth,
tumorigenesis, differentiation and development (15,37–42).
However, to the best of our knowledge, the effect of lncGHET1 on
the progression of OS remains undetermined.
To investigate the role of lncGHET1 in the
regulation of OS, the present study first examined the expression
levels of lncGHET1 in OS cell lines. The results revealed that all
OS cell lines exhibited relatively high levels of lncGHET1 compared
with hFOB cells. The results indicated that lncGHET1 may serve an
important role in OS. In the present study, the functional role of
lncGHET1 was analyzed in MG-63 and U2OS cells using
loss-of-function approaches in vitro and in vivo. The
results demonstrated that inhibition of lncGHET1 expression
inhibited proliferation, migration and invasion, and suppressed
cell cycle progression, whereas it promoted apoptosis of MG-63 and
U2OS cells in vitro and in vivo, indicating lncGHET1
could be an oncogene in OS.
The EMT process contributes to the tumorigenesis
(39). Increasing studies have
demonstrated the vital role of EMT in cancer invasion and
metastasis (25,43). It should be noted that EMT is
characterized by loss of the epithelial characteristics, including
loss of E-cadherin expression and upregulation of mesenchymal
markers, such as vimentin and N-cadherin (44,45).
Furthermore, a previous study has suggested that the EMT process is
regulated by a set of transcription factors, including Snail, Slug
and Twist (46). The present study
suggested that suppressing lncGHET1 expression increased the
expression levels of E-cadherin. However, Snail, N-cadherin and
Vimentin expression was markedly decreased when lncGHET1 was
silenced. Additionally, the present study explored the molecular
mechanism by which lncGHET1 contributes to tumor progression, and
indicated that lncGHET1 exerted an oncogenic effect, partly through
regulating the Wnt/β-catenin signaling pathway (Fig. 9).
In conclusion, the present study demonstrated that
inhibition of lncGHET1 attenuated cell proliferation, migration,
invasion and EMT, and promoted apoptosis, partly through regulating
the Wnt/β-catenin signaling pathway in OS cells. These findings
indicated that lncGHET1 may serve as an oncogenic lncRNA in OS
progression, and could be a promising molecular marker for
diagnosis and treatment of OS.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WF designed the experiments. XC and WZ performed the
experiments, and were the major contributors in writing the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
All animal experiments were approved by the Ethical
Committee of Nanjing Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cortini M, Avnet S and Baldini N:
Mesenchymal stroma: Role in osteosarcoma progression. Cancer Lett.
405:90–99. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Picci P: Osteosarcoma (Osteogenic
sarcoma). Orphanet J Rare Dis. 2:62007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mckenna WG, Barnes MM, Kinsella TJ,
Rosenberg SA, Lack EE and Glatstein E: Combined modality treatment
of adult soft tissue sarcomas of the head and neck. Int J Radiat
Oncol Biol Phys. 13:1127–1133. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen X, Bahrami A, Pappo A, Easton J,
Dalton J, Hedlund E, Ellison D, Shurtleff S, Wu G, Wei L, et al:
Recurrent somatic structural variations contribute to tumorigenesis
in pediatric osteosarcoma. Cell Rep. 7:104–112. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lian H, Xie P, Yin N, Zhang J, Zhang X, Li
J and Zhang C: Linc00460 promotes osteosarcoma progression via
miR-1224-5p/FADS1 axis. Life Sci. 233:1167572019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim W, Han I, Lee JS, Cho HS, Park JW and
Kim HS: Postmetastasis survival in high-grade extremity
osteosarcoma: A retrospective analysis of prognostic factors in 126
patients. J Surg Oncol. 117:1223–1231. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fellenberg J, Bernd L, Delling G, Witte D
and Zahlten- Hinguranage A: Prognostic significance of
drug-regulated genes in high-grade osteosarcoma. Mod Pathol.
20:1085–1094. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Piperno-Neumann S, Ray-Coquard I, Occean
BV, Laurence V, Cupissol D, Perrin C, Penel N, Bompas E, Rios M, Le
Cesne A, et al: Results of API-AI based regimen in osteosarcoma
adult patients included in the French OS2006/Sarcome-09 study. Int
J Cancer. 146:413–423. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wen J, Qin Y, Li C, Dai X, Wu T and Yin W:
Mangiferin suppresses human metastatic osteosarcoma cell growth by
down-regulating the expression of metalloproteinases-1/2 and
parathyroid hormone receptor 1. AMB Express. 10:132020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang Z, Zhao M and Wang G:
Hsa_circ_0051079 functions as an oncogene by regulating
miR-26a-5p/TGF-β1 in osteosarcoma. Cell Biosci. 9:942019.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang X, Xie X, Xiao YF, Xie R, Hu CJ, Tang
B, Li BS and Yang SM: The emergence of long non-coding RNAs in the
tumorigenesis of hepatocellular carcinoma. Cancer Lett.
360:119–124. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu Y, Zhang R and Ying K: Long non-coding
RNAs: Novel links in respiratory diseases (review). Mol Med Rep.
11:4025–4031. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yi S, Xiao-jiang Y, Zhen-jun L, Gang L,
Wu-jin X and Wei-xia J: UCA1, a long noncoding RNA, promotes the
proliferation of CRC cells via p53/p21 signaling. Open Life Sci.
11:206–210. 2016. View Article : Google Scholar
|
|
15
|
Maruyama R and Suzuki H: Long noncoding
RNA involvement in cancer. BMB Rep. 45:604–611. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li Z, Yu X and Shen J: Long non-coding
RNAs: Emerging players in osteosarcoma. Tumor Biol. 37:2811–2816.
2016. View Article : Google Scholar
|
|
17
|
Sun J, Wang X, Fu C, Wang X, Zou J, Hua H
and Bi Z: Long noncoding RNA FGFR3-AS1 promotes osteosarcoma growth
through regulating its natural antisense transcript FGFR3. Mol Biol
Rep. 43:427–436. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ruan W, Wang P, Feng S, Xue Y and Li Y:
Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12)
promotes cell proliferation and migration by upregulating
angiomotin gene expression in human osteosarcoma cells. Tumor Biol.
37:4065–4073. 2016. View Article : Google Scholar
|
|
19
|
Li T, Mo X, Fu L, Xiao B and Guo J:
Molecular mechanisms of long noncoding RNAs on gastric cancer.
Oncotarget. 7:8601–8612. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li LJ, Zhu JL, Bao WS, Chen DK, Huang WW
and Weng ZL: Long noncoding RNA GHET1 promotes the development of
bladder cancer. Int J Clin Exp Pathol. 7:7196–7205. 2014.PubMed/NCBI
|
|
21
|
Yang F, Xue X, Zheng L, Bi J, Zhou Y, Zhi
K, Gu Y and Fang G: Long non-coding RNA GHET1 promotes gastric
carcinoma cell proliferation by increasing c-Myc mRNA stability.
FEBS J. 281:802–813. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhou J, Li X, Wu M, Lin C, Guo Y and Tian
B: Knockdown of long noncoding RNA GHET1 inhibits cell
proliferation and invasion of colorectal cancer. Oncol Res.
23:303–309. 2016. View Article : Google Scholar
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pastushenko I and Blanpain C: EMT
transition states during tumor progression and metastasis. Trends
Cell Biol. 29:212–226. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang R and Zong X: Aberrant cancer
metabolism in epithelial- mesenchymal transition and cancer
metastasis: Mechanisms in cancer progression. Crit Rev Oncol
Hematol. 115:13–22. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mittal V: Epithelial mesenchymal
transition in tumor metastasis. Annu Rev Pathol. 13:395–412. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wittig JC, Bickels J, Priebat D, Jelinek
J, Kellar-Graney K, Shmookler B and Malawer MM: Osteosarcoma: A
multidisciplinary approach to diagnosis and treatment. Am Fam
Physician. 65:1123–1132. 2002.PubMed/NCBI
|
|
28
|
Isakoff MS, Bielack SS, Meltzer P and
Gorlick R: Osteosarcoma: Current treatment and a collaborative
pathway to success. J Clin Oncol. 33:3029–3035. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang T, Li J, Yin F, Lin B, Wang Z, Xu J,
Wang H, Zuo D, Wang G, Hua Y and Cai Z: Toosendanin demonstrates
promising antitumor efficacy in osteosarcoma by targeting STAT3.
Oncogene. 36:6627–6639. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu X, Xiu LJ, Jiao JP, Zhao J, Zhao Y, Lu
Y, Shi J, Li YJ, Ye M, Gu YF, et al: Traditional Chinese medicine
integrated with chemotherapy for stage IV non-surgical gastric
cancer: A retrospective clinical analysis. J Integr Med.
15:469–475. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zang QQ, Zhang L, Gao N and Huang C:
Ophiopogonin D inhibits cell proliferation, causes cell cycle
arrest at G2/M, and induces apoptosis in human breast carcinoma
MCF-7 cells. J Integr Med. 14:51–59. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen LL and Zhao JC: Functional analysis
of long noncoding RNAs in development and disease. Adv Exp Med
Biol. 825:129–158. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hajjari M, Khoshnevisan A and Shin YK:
Molecular function and regulation of long non-coding RNAs:
Paradigms with potential roles in cancer. Tumour Biol.
35:10645–10663. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gupta RA, Nilay S, Wang KC, Kim J,
Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al:
Long non-coding RNA HOTAIR reprograms chromatin state to promote
cancer metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Takayama K, Horie-Inoue K, Katayama S,
Suzuki T, Tsutsumi S, Ikeda K, Urano T, Fujimura T, Takagi K,
Takahashi S, et al: Androgen-responsive long noncoding RNA CTBP1-AS
promotes prostate cancer. EMBO J. 32:1665–1680. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhao H, Zhang K, Wang T, Cui J, Xi H, Wang
Y, Song Y, Zhao X, Wei B and Chen L: Long non-coding RNA
AFAP1-antisense RNA 1 promotes the proliferation, migration and
invasion of gastric cancer cells and is associated with poor
patient survival. Oncol Lett. 15:8620–8626. 2018.PubMed/NCBI
|
|
37
|
Ernst C and Morton CC: Identification and
function of long non-coding RNA. Front Cell Neurosci. 7:1682013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tam WL and Weinberg RA: The epigenetics of
epithelial-mesenchymal plasticity in cancer. Nat Med. 19:1438–1449.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Van Der Steen N, Honeywell RJ, Dekker H,
Van Meerloo J, Kole J, Musters R, Ruijtenbeek R, Rolfo C, Pauwels
P, Peters GJ and Giovannetti G: Resistance to crizotinib in a cMET
gene amplified tumor cell line is associated with impaired
sequestration of crizotinib in lysosomes. J Mol Clin Med. 1:99–106.
2018.
|
|
41
|
Liu H, Zhao J and Lv J: Inhibitory effects
of miR-101 overexpression on cervical cancer SiHa cells. Eur J
Gynaecol Oncol. 38:236–240. 2017.PubMed/NCBI
|
|
42
|
Hu H, Zhang G, Tian G, Lv G and Jin Y:
miRNA profiling reveals the upregulation of osteogenesis-associated
miRNAs in ovariectomy osteoporosis mice. Clin Exp Obstet Gynecol.
45:817–822. 2018.
|
|
43
|
Pan JJ and Yang MH: The role of
epithelial-mesenchymal transition in pancreatic cancer. J
Gastrointest Oncol. 2:151–156. 2011.PubMed/NCBI
|
|
44
|
Gonzalez DM and Damian M: Signaling
mechanisms of the epithelial-mesenchymal transition. Sci Signal.
7:re82014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ye X and Weinberg RA:
Epithelial-mesenchymal plasticity: A central regulator of cancer
progression. Trends Cell Biol. 25:675–686. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Nieto MA: Epithelial plasticity: A common
theme in embryonic and cancer cells. Science. 342:12348502013.
View Article : Google Scholar : PubMed/NCBI
|