Introduction
Breast cancer is one of the most prevalent
malignancies worldwide, accounting for 11.7% of all new cancer
cases in 2020 (1). Breast
malignancies constitute a group of diagnoses that each presents
with different characteristics. Current practice subdivides tumors
according to expression of certain molecular markers, and this
subdivision is used to guide the therapeutic approach (2). Of these general subgroups,
estrogen-receptor-positive (ER+) breast cancers
represent almost 2/3 of all cases (3). ER+ patients are eligible
for endocrine therapy, including some agents such as tamoxifen,
which aims to inhibit tumor growth by interfering with
proliferative estrogen signaling (4). In ER+ patients, disease
progression despite anti-estrogenic treatment may be suggestive of
either intrinsic or acquired endocrine resistance. The underlying
mechanisms of such resistance are not yet fully understood,
although alterations in various signaling pathways, including
estrogen-signaling, seem to be of importance (5). As not all patients benefit from
current treatment regimens, it is crucial to develop novel risk
assessment tools capable of early identification of tumor
characteristics associated with aggressiveness, including endocrine
resistance in hormone-sensitive cases.
One novel protein of interest is named solute
carrier family 7 member 5 (SLC7A5), which is a transmembrane amino
acid transporter also known as large neutral amino acid-transporter
1 (LAT1). SLC7A5 has been described in several types of cancer,
including breast cancer, and functions by facilitating the exchange
of essential amino acids over the cell membrane (6,7). The
connection between overexpression of SLC7A5 and cancer has been
associated with alterations in cell signaling and metabolic
homeostasis, which are both considered as important hallmarks of
cancer (8). SLC7A5 has also been
reported to be induced by hypoxia in several tissues, including
renal cell carcinoma, suggesting an association between SLC7A5
expression and changes in tumor environment and cell demands
(9).
Although largely independent of physiological
restrictions, readily proliferating tumors are still to some degree
constrained by nutrient and oxygen deprivation as they ultimately
outgrow their supporting blood vessels (10). As local tissue hypoxia arises,
malignant cells are forced to rearrange their genetic programming,
and since exogenous amino acids are essential for the synthesis of
various proteins and nucleic acids, tumors might start to
overexpress amino acid exchangers in order to gain access to a
ready supply of building blocks to sustain their proliferation and
existence (11,12). In the case of the amino acid
exchanger SLC7A5, the protein also appears to be directly involved
in proliferative signaling by facilitating the uptake of leucine,
which is an essential activator of the mTORC1 signaling complex
(13). SLC7A5 is therefore likely
to influence tumor cell behavior and may ultimately contribute to
disease progression. Thus, SLC7A5 may be considered as a promising
biomarker candidate and a novel future therapeutic target.
Based upon the nature of SLC7A5, we hypothesized
that upregulation of SLC7A5 may function as a way for breast cancer
cells to prolong their lifespan under conditions that are not
suitable for proliferation, such as during hypoxia or in
conjunction with impaired cell signaling during anti-estrogenic
treatment. To confirm this hypothesis, we aimed to study the gene
and protein expression of SLC7A5 in ER+ breast cancer
using immunohistochemistry and microarray analysis. The present
study discussed the prognostic and biological role of SLC7A5 in
ER+ breast cancer by determining how SLC7A5 protein
expression may be associated with various clinicopathological
characteristics linked to patient outcome. The present study also
explored the correlation between SLC7A5 mRNA expression and the
mRNA levels of certain genes linked to breast cancer biology such
as estrogen receptor 1 (ESR1), Erb-B2 receptor tyrosine kinase 2
(ERBB2), MYC proto-oncogene, BHLH transcription factor (MYC) and
mechanistic target of rapamycin (MTOR), to cell proliferation such
as marker of proliferation Ki-67 (MKI67) and cyclin D1 (CCND1), to
hypoxia such as hypoxia inducible factor 1 subunit alpha (HIF1A),
endothelial PAS domain-containing protein 1 (EPAS1 or HIF2A) and
vascular endothelial growth factor A (VEGFA), and to metabolism
such as lactate dehydrogenase A (LDHA), solute carrier family 2,
facilitated glucose transporter member 1 (SLC2A1 or GLUT1),
carbonic anhydrase 9 (CA9) and NDUFA4 mitochondrial complex
associated like 2 (NDUFA4L2).
Materials and methods
Patients and breast cancer
specimens
The present study included female patients diagnosed
with breast cancer at the Department of Oncology, Örebro University
Hospital, Sweden, between January 2000 and December 2010. Patients
were identified from the registry of the Regional Cancer Centre
Uppsala Örebro, Sweden. Information regarding the patients' primary
tumor characteristics was retrieved from the registry, along with
clinical data. This study was approved by the regional ethics
committee in Uppsala (2011/070) and written consent was obtained
from participants who were still alive. All procedures were
performed in accordance with the ethical standards of the regional
ethics committee and with the 1964 Declaration of Helsinki and its
later amendments.
The patients included in the present study (n=154)
were all diagnosed with an ER+ tumor and treated with
endocrine therapy, and had no distant metastasis at the time of
diagnosis. Records from registry were first retrieved in 2013, when
a total of 116 eligible patients were included (patients with
recurrence, n=27; recurrence-free patients, n=89). The selection
procedure for these patients has been previously described in
detail (14). To minimize the risk
of including patients with undetected metastasis at the time of
primary diagnosis, a cut-off of 24 months was used to distinguish
recurrence. Updated records were retrieved from the registry in
2017, and additional patients (patients with recurrence, n=12;
recurrence-free patients, n=26) were selected and included in the
study based on the same inclusion criteria. The 154 patients were
subsequently referred to as the Örebro cohort.
Immunohistochemistry (IHC)
Formalin-fixed paraffin-embedded primary breast
tumor tissues were available from 152 out of the 154 patients. The
protein expression of SLC7A5 was evaluated using IHC on 4-µm
sections mounted on glass slides. These tissues were stained as
previously described (15).
Briefly, tissues were deparaffinized and rehydrated using
Tissue-Clear® (Sakura Finetek Europe) followed by a
series of ethanol (100, 95 and 70%) and deionized water.
Heat-induced epitope retrieval was performed in DIVA Decloaker
buffer (Biocare Medical) in a decloaking chamber (Biocare Medical)
for 10 min at 110°C. The staining procedure was then performed
using the automated slide stainer IntelliPATH FLX™ (Biocare
Medical). For protein detection, a rabbit monoclonal anti-SLC7A5
antibody (cat. no. ab208776; 1:500; 1 h incubation at room
temperature; Abcam) was used together with the MACH 1 Universal
HRP-Polymer Detection system (Biocare Medical) and Betazoid DAB
chromogen according to the manufacturer's instructions. The slides
were washed with TBS Automation Wash Buffer (Biocare Medical) in
between each reaction. Following subsequent counterstaining with
Mayer's haematoxylin, the tissues were dehydrated in ethanol (70,
95 and 100%) and xylene and mounted with coverslips using
Pertex® mounting medium. For each run of the staining
process, a tissue sample (testis) stained for SLC7A5 protein
expression was included as a positive control.
Evaluation of IHC staining
The slides were digitally scanned using the
Pannoramic 250 Flash II scanner (3DHistech Ltd.) and the resulting
images were analyzed using the CaseViewer software package version
2.4 (3DHistech Ltd.). SLC7A5 protein staining was graded according
to HercepTest™ (Dako) scoring guidelines, originally intended for
assessment of human epidermal growth factor receptor 2
(HER2)-positivity (Table I).
Staining was graded as 0, 1+, 2+ or 3+ according to membrane
staining intensity, whether the staining was complete or
incomplete, and the amount of positively stained tumor cells in the
tissue specimen. Scores of 0 or 1+ were considered as negative
while scores of 2+ and 3+ were considered as positive. Grading was
performed by two separate observers and final grades were based on
consensus scores.
 | Table I.Immunohistochemistry SLC7A5 protein
expression scoring guidelines adapted from the HercepTest™ (Dako)
guidelines for human epidermal growth factor receptor 2 protein
expression assessment. |
Table I.
Immunohistochemistry SLC7A5 protein
expression scoring guidelines adapted from the HercepTest™ (Dako)
guidelines for human epidermal growth factor receptor 2 protein
expression assessment.
| Score to
report | Protein
overexpression assessment | Staining
Pattern |
|---|
| 0 | Negative | No staining is
observed, or membrane staining is observed in < 10% of the tumor
cells. |
| 1+ | Negative | A faint/barely
perceptible incomplete membrane staining is detected in > 10% of
tumor cells. |
| 2+ | Weakly
positive | A weak to moderate
complete membrane staining is observed in > 10% tumor cells |
| 3+ | Strongly
positive | A strong complete
membrane staining is observed in > 10% of the tumor cells |
Microarray
For 80 patients, mRNA gene expression data were
obtained through microarray analysis performed on a previous
occasion (data not shown). Of these, 78 specimens were also
available for IHC staining of SLC7A5. Briefly, freshly frozen
primary tumor tissues (stored at −80°C) with confirmed tumor cell
content were homogenized using a TissueLyser II (Qiagen AB) with 5
mm steel beads (Qiagen AB) for 2×2 min at 30 Hz. Lysing and RNA
extraction was performed using the Allprep DNA/RNA/Protein mini kit
(Qiagen AB). RNA concentrations were evaluated and purity
determined according to the absorbance ratio at A260, A280 and A230
nm using a NanoDrop Spectrophotometer ND-1000 (NanoDrop
Technologies; Thermo Fisher Scientific, Inc.). Values of A260/A280
between 1.8 and 2.1 and A260/A230>2.0 were considered as
acceptable purity for microarray. RNA integrity number (RIN) values
were determined using the Agilent RNA 6000 Nano Kit for the 2100
Bioanalyzer Instrument (Agilent Technologies, Inc.), with RIN>8
considered as an acceptable quality for subsequent microarray
analysis. The RNA was stored at −80°C.
Microarray data acquisition
High-quality RNA (100 ng) was used to prepare
Cy3-labeled and amplified cRNA with the One Color Low Input Quick
Amp Labeling Kit (Agilent Technologies, Inc.) according to the
manufacturer's instructions. The cRNA concentration and specific
activity (pmol Cy3/µg cRNA) was determined using NanoDrop.
Following the Gene Expression Hybridization Kit (Agilent
Technologies, Inc.) protocol, the samples were hybridized to
SurePrint G3 Human Gene Expression 8×60k v2 Microarrays (Agilent
Technologies, Inc.) for 17 h at 65°C in a Hybridization Oven
(Agilent Technologies, Inc.). The microarrays were scanned with a
G2565CA Microarray Scanner (Agilent Technologies, Inc.) and image
analysis and data extraction were assessed with version 10.7.3.1 of
Agilent's Feature Extraction Software package (Agilent
Technologies, Inc.). All microarrays were verified and passed
quality check parameters included in the software package prior to
data analysis. Before extracting gene expression data for SLC7A5,
ESR1, ERBB2, CCND1, MYC, HIF1A, MKI67, VEGFA, MTOR, EPAS1, LDHA,
SLC2A1, CA9 and NDUFA4L2, the arrays underwent quantile
normalization and ComBat correction to account for batch variations
dependent of sample distributions on different slides (SVA R
package) prior to log2-transformation. The normalized fluorescence
intensity values for each gene were further transformed into
z-values prior to statistical analysis.
METABRIC data
The METABRIC dataset, with both clinical and gene
expression data, was downloaded from cBioPortal (16,17).
Collection of the METABRIC study specimens was ethically approved
by the institutional review board and the METABRIC study protocol
was approved by ethics committees in Vancouver and Cambridge. The
study was previously described in detail (18). Briefly, tumor RNA from >2,000
breast cancer patients was analyzed using the Illumina HT-12 v3
platform (Illumina_Human_WG-v3). Gene expression data (mRNA
expression z-values relative to diploid samples) were available
from 1,904 breast cancer patients in total, although one patient
with breast sarcoma was excluded, leaving a remaining pool of 1,903
patients.
Statistical analysis
For examining the associations between SLC7A5
protein expression and the clinicopathological characteristics in
the Örebro patient cohort, the χ2 or Fisher's exact
tests were used. The Mann-Whitney U test was used for continuous
variables. Survival curves were generated by Kaplan-Meier
estimation using the log-rank test with distant recurrence set as
primary patient outcome. The hazard ratio with 95% confidence
interval (CI) was calculated using Cox regression.
Prior to gene correlation analysis, the mRNA
microarray data (fluorescence intensity values) were z-transformed,
with a z-value of 0 representing the mean and scores of ±1 equaling
1 standard deviation from the mean. Normal distribution of SLC7A5
expression was evaluated using Shapiro-Wilk test, arguing for the
use of non-parametric tests for analysis of SLC7A5 correlation to
other assessed genes as well as clinical data from both the Örebro
cohort and METABRIC data. The correlation between z-values (gene
expressions) was estimated using Spearman's rank correlation
coefficient (ρ). The Mann Whitney U-test was used to compare SLC7A5
gene expression with the corresponding protein expression and
various clinical variables. One-way ANOVA and Kruskal-Wallis test
were used to compare multiple categorical clinical variables. Data
analysis was performed using SPSS software version 7.03 (SPSS,
Inc.) and GraphPad Prism version 7.03 (GraphPad Software, Inc.).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Characterization of SLC7A5 expression
in ER+ breast cancer tissues
Immunohistochemical staining of SLC7A5 was evaluated
in all available cases (n=152). The overall protein expression was
found to be heterogeneous, and staining varied in intensity within
and between tumors. Representative images of protein staining are
provided in Fig. 1, where ~15% of
the cases were found to be positive (2+ and 3+) for SLC7A5 protein
expression (Table SI).
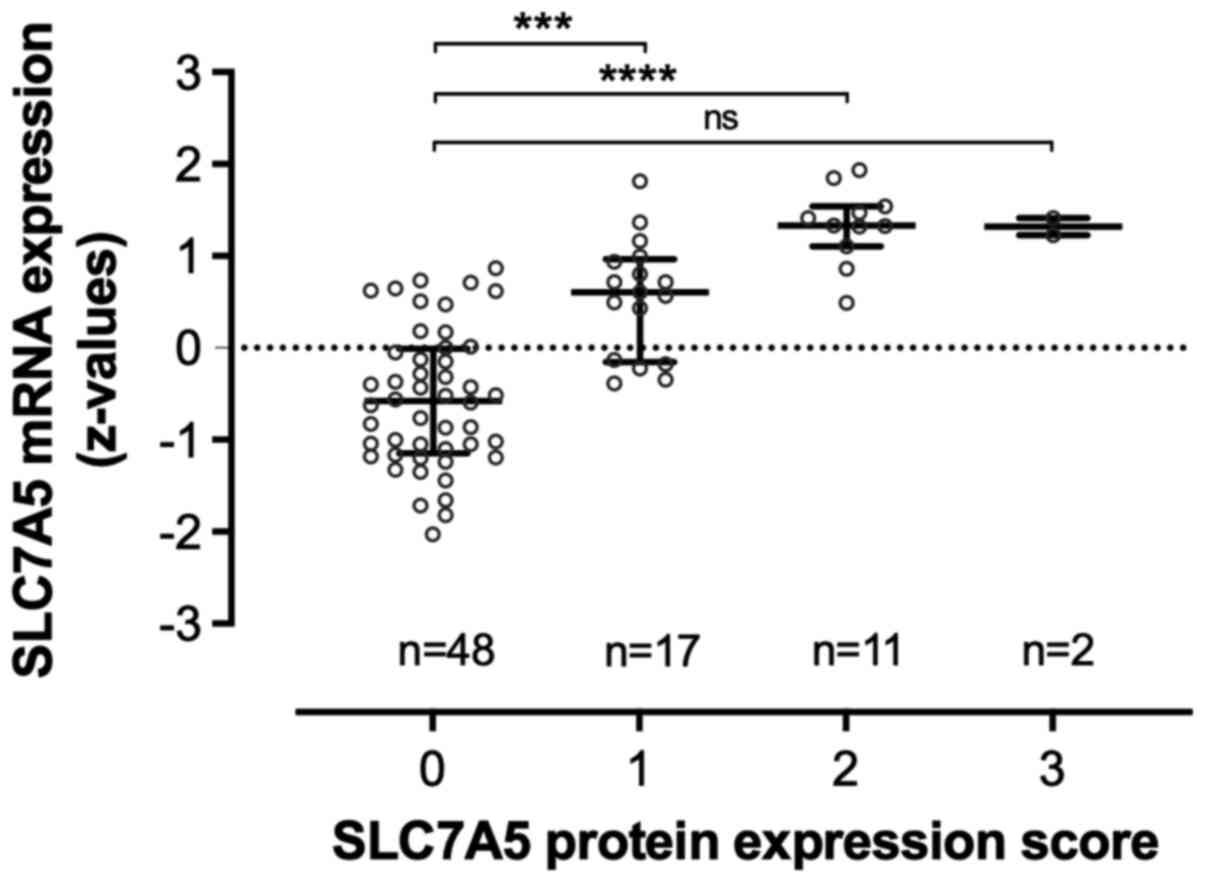
SLC7A5 mRNA z-values from the microarray analysis
(n=80, of which 78 cases were available for IHC staining) were
found to be normally distributed (Table SII). The gene expression of SLC7A5
was significantly higher in tumors with high protein expression
score (Fig. 2).
SLC7A5 protein expression in
association with clinicopathological variables
Associations between SLC7A5 expression and tumor
characteristics in ER+ breast cancer are presented in
Table SI. The results
demonstrated that the positive protein expression of SLC7A5 was
significantly associated with histopathological grade in the form
of Nottingham Histologic Score (P=0.014).
To further evaluate the prognostic value of tumor
SLC7A5 protein expression, survival analysis was performed with
regards to distant recurrence. When comparing tumors that were
positive and negative for SLC7A5, a trend could be observed in
which positive protein expression concurred with lower survival
rate and earlier distant recurrence; however, this association was
not statistically significant (Fig.
3). The estimated hazard ratio for patients with a SLC7A5-
positive tumor was 1.85 (95% CI 0.88-3.90; P=0.11).
SLC7A5 mRNA expression and
clinicopathological tumor characteristics
Microarray data with z-value conversion were
available from 80 patients in the Örebro cohort. In these patients,
increased SLC7A5 mRNA expression was significantly associated with
histopathological grade (Fig. 4A),
thus adhering with the association between increased expression of
SLC7A5 protein and histopathological grade (Table SI). There was no significant
association between SLC7A5 mRNA expression and tumor size, lymph
node infiltration, menopausal status or PR-status or HER2-status in
this cohort (Fig. 4B-F).
For validation of the results obtained from the
Örebro cohort, SLC7A5 mRNA expression was also assessed using data
on breast cancers categorized as ER+ in the METABRIC
dataset (n=1,445). Increased SLC7A5 mRNA expression was similarly
demonstrated to be significantly associated with histopathological
grade in these tumors (Fig. 5).
However, there were also insignificant associations between
increased SLC7A5 mRNA expression and lymph node infiltration, as
well as progesterone receptor (PR) negativity and HER2-positivity.
When investigating differences regarding SLC7A5 mRNA expression in
ER+ tumor subtypes, higher z-values could be observed in
tumors classified as the more aggressive luminal B subtype compared
with luminal A tumors (Fig.
5).
 | Figure 5.Association between SLC7A5 mRNA
expression and clinicopathological variables in
estrogen-receptor-positive tumors (METABRIC, n=1,445). Association
between SLC7A5 expression and (A) tumor stage, (B)
histopathological grade (Elston grade), (C) histological type, (D)
molecular subtype classified via three gene expression profiling,
(E) genetic subtype based on PAM50 gene expression analysis, (F)
lymph node infiltration, (G) PR-status according to genetic
analysis, (H) HER2-status according to genetic analysis. SLC7A5,
solute carrier family 7 member 5; PR, progesterone receptor; HER2,
human epidermal growth factor receptor 2. *p<0.05, **p≤0.01,
***p≤0.001, and ****p≤0.0001 and ns, non-significant (p>0.05)
values. |
SLC7A5 mRNA expression and breast
cancer molecular subtype
By including all primary breast cancer cases in the
METABRIC dataset, regardless of ER status (n=1,903), it became
possible to further evaluate SLC7A5 expression with regards to
clinicopathological factors in the context of molecular breast
cancer profiles. The results showed a higher SLC7A5 mRNA expression
among tumors with higher histopathological grade. In addition,
increased genomic expression of SLC7A5 was more strongly coupled to
ER- molecular subtypes, with higher expression of SLC7A5 mRNA
z-values in HER2-enriched and triple-negative breast cancer cases
compared with tumors classified as ER+ (Fig. 6).
 | Figure 6.Association between SLC7A5 mRNA
expression and clinicopathological variables in all tumors
regardless of ER-status (METABRIC, n=1903). Association between
SLC7A5 and (A) tumor stage, (B) genetic subtype based on PAM50 gene
expression analysis, (C) histological type, (D) molecular subtype
classified via three gene expression profiling, (E)
histopathological grade (Elston grade), (F) lymph node
infiltration, (G) ER-status according to genetic analysis, (H)
PR-status according to genetic analysis and (I) HER2-status
according to genetic analysis. SLC7A5, solute carrier family 7
member 5; PR, progesterone receptor; HER2, human epidermal growth
factor receptor 2; ER, estrogen receptor. **p≤0.01, ***p≤0.001,
****p≤0.0001 and ns, non-significant (p>0.05) values. |
SCL7A5 mRNA expression in correlation
with genes related to breast cancer biology, hypoxia and cell
metabolism
To also evaluate the biological role of SLC7A5, mRNA
expression was assessed in connection with the expression of
several other breast cancer related genes and genes related to
hypoxia and cell metabolism (Table
II). This assessment was performed both on the local Örebro
cohort (Fig. S1) and on METABRIC
data (Fig. S2,Fig. S3,Fig. S4). In the Örebro cohort, positive
correlations were found between SLC7A5 expression and both MKI67
and HIF1A expression (Table II).
However, when testing for correlations between SLC7A5 expression
and the other genes aforementioned, no significant correlations
could be found.
 | Table II.Association between SLC7A5 mRNA
expression and genes related to breast cancer, hypoxia and cell
metabolism in the Örebro cohort and the Molecular Taxonomy of
Breast Cancer International Consortium (METABRIC) dataset. |
Table II.
Association between SLC7A5 mRNA
expression and genes related to breast cancer, hypoxia and cell
metabolism in the Örebro cohort and the Molecular Taxonomy of
Breast Cancer International Consortium (METABRIC) dataset.
|
| Örebro cohort
(n=80) | METABRIC ER+
(n=1,445) | METABRIC
(n=1,903) |
|---|
|
|
|
|
|
|---|
| Gene name | Spearman r | 95% CI | P-value | Spearman r | 95% CI | P-value | Spearman r | 95% CI | P-value |
|---|
| ESR1 | −0.023 | −0.248 to
0.204 | 0.8399 | −0.136 | −0.188 to
−0.083 | <0.0001 | −0.448 | −0.484 to
−0.410 | <0.0001 |
| ERBB2 | −0.043 | −0.266 to
0.185 | 0.7071 | 0.078 | 0.025 to 0.130 | 0.0031 | 0.010 | −0.036 to
0.056 | 0.668 |
| MKI67 | 0.410 | 0.202 to 0.582 | 0.0002 | 0.348 | 0.301 to 0.394 | <0.0001 | 0.480 | 0.444 to 0.515 | <0.0001 |
| MYC | 0.150 | −0.079 to
0.363 | 0.1851 | 0.046 | −0.008 to
0.098 | 0.084 | 0.137 | 0.091 to 0.182 | <0.0001 |
| MTOR | −0.042 | −0.266 to
0.185 | 0.7089 | 0.013 | −0.040 to
0.066 | 0.6139 | 0.084 | 0.038 to 0.129 | 0.0003 |
| CCND1 | −0.117 | −0.334 to
0.112 | 0.3007 | 0.040 | −0.013 to
0.093 | 0.1297 | −0.186 | −0.231 to
−0.141 | <0.0001 |
| HIF1A | 0.263 | 0.039 to 0.462 | 0.0184 | 0.097 | 0.044 to 0.150 | 0.0002 | 0.192 | 0.147 to 0.236 | <0.0001 |
| VEGFA | 0.062 | −0.166 to
0.284 | 0.585 | 0.243 | 0.192 to 0.292 | <0.0001 | 0.355 | 0.314 to 0.395 | <0.0001 |
| EPAS1 | 0.191 | −0.036 to
0.400 | 0.089 | −0.031 | −0.084 to
0.022 | 0.239 | −0.039 | −0.085 to
0.008 | 0.0929 |
| LDHA | 0.013 | −0.214 to
0.238 | 0.9083 | 0.066 | 0.014 to 0.119 | 0.0118 | 0.149 | 0.103 to 0.194 | <0.0001 |
| SLC2A1 | −0.213 | −0.419 to
0.014 | 0.0579 | 0.261 | 0.211 to 0.310 | <0.0001 | 0.381 | 0.341 to 0.420 | <0.0001 |
| CA9 | −0.010 | −0.235 to
0.217 | 0.9304 | 0.136 | 0.084 to 0.188 | <0.0001 | 0.340 | 0.298 to 0.380 | <0.0001 |
| NDUFA4L2 | −0.037 | −0.261 to
0.190 | 0.7416 | 0.026 | −0.028 to
0.079 | 0.3314 | 0.121 | 0.075 to 0.166 | <0.0001 |
ER+ cancers in the METABRIC dataset
(n=1,445) showed concordant statistically significant correlations
between SLC7A5 expression and increased expression of MKI67 and
HIF1A (Table II). In this
dataset, SLC7A5 was also positively correlated with expression of
VEGFA, MYC, ERBB2, and of genes linked to metabolism (LDHA, SLC2A1
and CA9), and negatively correlated with ESR1 expression. When
including all tumors in the METABRIC dataset (n=1,903), positive
correlations were similarly found between SLC7A5 expression and
MKI67, HIF1A, VEGFA, MYC, ERBB2, LDHA, SLC2A1 and CA9 expression.
Positive correlations in this dataset were also found between
SLC7A5 expression and both MTOR and NDUFA4L2 expression. Negative
correlations were demonstrated between SLC7A5 expression and ESR1
and CCND1 expression (Table II).
When categorizing the data by ER status, as based on IHC staining,
ER+ tumors tended to follow a general pattern of lower
relative expression of all assessed genes compared with SLC7A5 mRNA
expression (Fig. S4).
Discussion
ER+ breast cancers present with varying
clinical characteristics and prognosis. Current therapy targets
aberrant estrogen-signaling, in most cases with good effect;
however, a high number of patients see a progression of the
disease, possibly due to endocrine resistance. To overcome therapy
resistance, finding novel diagnostic markers and therapeutic
targets has become increasingly important (19). The amino acid transporter SLC7A5
(or LAT1) was attributed some pro-oncogenic properties and is
overexpressed in various types of cancer, including breast cancer,
and may therefore constitute a potential new marker and treatment
target.
The results from the present study demonstrated that
SLC7A5 protein expression was associated with histopathological
grade in ER+ breast cancers, which may suggest the role
of the amino acid exchanger in breast cancer biology and tumor
progression. We were not able to find statistically significant
associations between SLC7A5 protein expression and other prognostic
markers (e.g. lymph node infiltration or tumor size) or patient
outcome (i.e. distant recurrence) in the Örebro cohort. However,
Mihály et al (20) reported
that increased SLC7A5 expression is associated with shorter
recurrence free survival in a larger study population of tamoxifen
treated patients (n=1,044). Considering the small size of the
Örebro cohort and the risk of failing to achieve statistical
significance due to inadequate study power, the results from the
present study were in concordance with those from El Ansari et
al (21). El Ansari et
al (21) also reported that
SLC7A5 protein expression is an independent risk factor for
worsened patient outcome in highly proliferative ER+
tumors and HER2-enriched breast cancer subtypes. SLC7A5 protein
expression has been evaluated and reported in different ways, which
complicates comparison between studies. In our present cohort, 15%
of the tumors were defined as SLC7A5 positive, which was similar to
the 17% positivity from the 2,664 breast cancer cases of El Ansari
et al Bodoor et al (22) demonstrated that 84.4 and 64.3% of
luminal A (n=32) and luminal B/triple positive (n=42) tumors,
respectively, were SLC7A5 positive, and that SLC7A5 expression was
overall negatively correlated with HER2 expression. However, this
study used a polyclonal antibody, which also stains cytoplasmic
structures, as well as a scoring system developed by the authors.
El Ansari et al used an H-score of >15 as cut-off for
positivity, whereas we applied the HER2 evaluation guideline
identifying score 2+ and 3+ as SLC7A5 positivity in the present
study. The HER2 evaluation guideline was established for
pathological assessment of HER2 status and could be adapted for
future clinical examination of SLC7A5, which is also a
membrane-bound protein.
Comparing SLC7A5 protein expression and mRNA
expression in the same patients can be seen as arbitrary. This
issue needs to be addressed, as protein expression scoring is based
upon tumor cell membrane expression while mRNA expression is based
upon analysis of tumor tissue samples, which also contain stromal
cells and parts of normal tissue. In the present study, being
graded as SLC7A5-negative in protein expression (0 or 1+) meant
that <10% of all tumor cells in the specimen were positively
stained. Furthermore, there was a variability in SLC7A5 expression,
as some specimens with low protein expression still showed
substantial mRNA expression levels (Fig. 2). A spatial expression of SLC7A5
could possibly explain this phenomenon and must be taken into
consideration.
In the present study, high SLC7A5 protein expression
was found to correspond to high mRNA expression, thus providing a
rationale for validating our findings in the METABRIC cohort.
Furthermore, associations between SLC7A5 gene expression and
clinical variables, such as histopathological grade, was connected
to poor outcome, which has previously been described in the
METABRIC data set (21). In
METABRIC ER+ breast cancers from the present study,
SLC7A5 expression was associated with HER2-positivity and PR
negativity. Furthermore, a negative correlation between SLC7A5
expression and ESR1 expression was demonstrated. Among the luminal
tumors, it is well known that these observations correspond to a
lower likelihood of endocrine treatment response as they influence
estrogen signaling. Expression of SLC7A5 seems to vary in luminal
cancers and is of great interest in terms of endocrine response
(23). Guan et al (23) compared differentially expressed
genes in tamoxifen-sensitive and tamoxifen-resistant cell lines as
well as in clinical breast cancer samples, and developed a two-gene
pair signature (TOP2A, SLC7A5; NMU, PDSS1) that could predict
tamoxifen therapy outcome. However, in the Örebro cohort, which
consisted of tamoxifen-treated patients only, there was no
significant difference in distant recurrence between
SLC7A5-positive and SLC7A5-negative cases; however, there was a
trend for SLC7A5-positive cases to have lower survival rates.
SLC7A5 has previously been featured as one of the
five immunohistochemical markers constituting the
Mammostrat® risk assessment tool, which is intended to
predict the risk of recurrent disease in cases of tamoxifen treated
ER+ cancer (24). Using
this tool, Bartlett et al (24) demonstrated that increased SLC7A5
expression is on its own significantly associated with shorter
recurrence-free survival. However, previous studies reported that
the analysis of individual expression of SLC7A5 is insufficient to
predict the prognosis and response to endocrine therapy in these
patients (25–27). These studies suggested that SLC7A5
should be assessed together with SLC3A2, which is a key helper of
SLC7A5 membrane transportation, to predict patient outcome in
ER+ breast cancer. The role of SLC7A5 as a marker in
endocrine disease is therefore disputed, and the present findings
further highlight this.
To determine the biological role of SLC7A5, the
present study evaluated the correlation between SLC7A5 expression
and the expression of genes involved in cell proliferation,
metabolism, and hypoxia. The results demonstrated that SLC7A5 gene
expression was positively correlated with HIF1A and MKI67
expression in both the Örebro cohort and the METABRIC dataset,
indicating the importance of SLC7A5 expression regarding the
proliferative capabilities of breast cancer cells. Our findings
also suggested a connection between SLC7A5 expression and tissue
hypoxia in breast cancer, which has been previously reported in
renal cell carcinoma (9). HIF1A
overexpression is known to be positively correlated with numerous
unfavorable carcinogenic characteristics, such as increased
angiogenesis, cell invasion and metastasis (28). Regarding SLC7A5, increased
expression together with co-expression of proliferative and hypoxic
genes, in primary and metastatic tumor sites could indicate adverse
disease (29). SLC7A5 expression
has previously been positively associated with expression of
hypoxia related genes such as HIF2A in breast cancer, although it
was only seen in luminal B tumors (21). The present study provided further
insight into the possible connection between amino acid
transporters such as SLC7A5 and hypoxia-inducible factors in
ER+ disease. In the future, validation of protein
expressions in breast cancer will be required, alongside functional
in vitro investigation. HIF1A signaling has been reported to
stimulate the expression of several glycolytic enzymes as well as
pH-regulating factors in cancer, including LDHA, SLC2A1 and CA9
(30–32). In the present study, we also
observed positive correlation between the expression of SLC7A5 and
LDHA, which encodes the enzyme lactate dehydrogenase, and of
SLC2A1, which encodes the glucose transporter GLUT1. Both are
crucial to accelerate glycolysis and stimulate the Warburg effect
(33–35). SLC7A5 expression was also
significantly correlated with CA9 expression, which encodes the
enzyme carbonic anhydrase. This enzyme is involved in cellular pH
buffering and has been demonstrated to provide a more favorable
tumor microenvironment in breast cancer (36,37).
These results suggested that SLC7A5 may either be part of or be
crucial to the metabolic reprogramming of breast cancer cells.
Tumors may upregulate SLC7A5 together with other factors such as
LDHA, SLC2A1 and CA9, as a way to counteract the negative effects
of oxygen and substrate starvation that arises as the tumor
progressively grows.
Regarding the spatial expression of SLC7A5, its
expression was demonstrated to differ between and within individual
tumors, suggesting that SLC7A5 expression may vary with certain
tumor growth conditions. A previous study on basal cell carcinoma
reported that SLC7A5 tends to be confined to centrally located
tumor cells rather than surrounding cells in the tumor margins
(15). In this context, SLC7A5
protein expression patterns and SLC7A5 mRNA association with MKI67,
HIF1A, LDHA, SLC2A1 and CA9 could further suggest that SLC7A5
upregulation may be crucial for cell proliferation or survival in
certain unfavorable settings, such as for example, in rapidly
growing tumor masses where blood flow successively becomes
inadequate towards the tumor core or as a response to anti-tumor
agents. Sato et al (38)
reported that SLC7A5 protein expression is higher following
chemotherapy, suggesting an enhanced amino acid metabolism when
glucose metabolism is impaired from treatment. Saito et al
(39) also demonstrated that
endocrine resistance in breast cancer is associated with adaption
to nutrient stress, and that SLC7A5 is an important factor and a
potential target to overcome treatment resistance. The presence of
SLC7A5 could be a sub-marker for endocrine resistance, suggesting a
possible benefit from using SLC7A5 inhibitors such as JPH203, which
is currently undergoing a phase II drug trial (UMIN000034080). It
is reasonable to think that the expression of amino acid
transporters might vary with changing cellular growth conditions.
According to our hypothesis, SLC7A5 expression may be induced as a
result of unfavorable tumor microenvironment.
In summary, the results from the present study
suggested that SLC7A5 expression in ER+ tumors may be of
importance for tumor cell proliferation and survival. However, the
biological role as well as the prognostic and predictive values of
SLC7A5 expression require further investigation in different
subtypes of breast cancer.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was funded by the Lions Cancer Research Fund
for Region Uppsala Örebro (2020), the research committee of Region
Örebro County (grant no. OLL-939134) and ALF grants from Region
Örebro County (grant no. OLL-555621, OLL-935730).
Availability of data and materials
Data from the Örebro cohort are available from the
corresponding author on reasonable request. The Molecular Taxonomy
of Breast Cancer International Consortium (METABRIC) dataset is
accessible via cBioPortal for Cancer Genomics.
Authors' contributions
RT wrote the original draft and contributed to the
methodology and data analysis. AGE and ET contributed to the
methodology, data analysis, reviewing and editing of the
manuscript, and were responsible for the project supervision and
acquisition of funding. All authors confirm the authenticity of all
raw data and have read and approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the regional Ethics
Committee of Uppsala, Sweden (approval no. 2011/070) and patients
provided written consent for the use of their clinical
material.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer Statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Goldhirsch A, Winer EP, Coates AS, Gelber
RD, Piccart-Gebhart M, Thürlimann B, Senn HJ, Albain KS, André F,
Bergh J, et al Panel members, : Personalizing the treatment of
women with early breast cancer: Highlights of the St Gallen
International Expert Consensus on the Primary Therapy of Early
Breast Cancer 2013. Ann Oncol. 24:2206–2223. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Harvey JM, Clark GM, Osborne CK and Allred
DC: Estrogen receptor status by immunohistochemistry is superior to
the ligand-binding assay for predicting response to adjuvant
endocrine therapy in breast cancer. J Clin Oncol. 17:1474–1481.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Duffy MJ, Harbeck N, Nap M, Molina R,
Nicolini A, Senkus E and Cardoso F: Clinical use of biomarkers in
breast cancer: Updated guidelines from the European Group on Tumor
Markers (EGTM). Eur J Cancer. 75:284–298. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
El Sayed R, El Jamal L, El Iskandarani S,
Kort J, Abdel Salam M and Assi H: Endocrine and targeted therapy
for hormone-receptor-positive, HER2-negative advanced breast
cancer: Insights to sequencing treatment and overcoming resistance
based on clinical trials. Front Oncol. 9:5102019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Häfliger P and Charles RP: The L-type
amino acid transporter LAT1 - An emerging target in cancer. Int J
Mol Sci. 20:24282019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Scalise M, Galluccio M, Console L, Pochini
L and Indiveri C: The human SLC7A5 (LAT1): The intriguing
histidine/large neutral amino acid transporter and its relevance to
human health. Front Chem. 6:2432018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Elorza A, Soro-Arnáiz I,
Meléndez-Rodríguez F, Rodríguez-Vaello V, Marsboom G, de Cárcer G,
Acosta-Iborra B, Albacete-Albacete L, Ordóñez A, Serrano-Oviedo L,
et al: HIF2α acts as an mTORC1 activator through the amino acid
carrier SLC7A5. Mol Cell. 48:681–691. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gatenby RA, Smallbone K, Maini PK, Rose F,
Averill J, Nagle RB, Worrall L and Gillies RJ: Cellular adaptations
to hypoxia and acidosis during somatic evolution of breast cancer.
Br J Cancer. 97:646–653. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Salisbury TB and Arthur S: The regulation
and function of the L-type amino acid transporter 1 (LAT1) in
cancer. Int J Mol Sci. 19:23732018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Adamaki M: Cancer and the cellular
response to hypoxia. Pediatr Ther. S1:0022012.
|
|
13
|
Cormerais Y, Giuliano S, LeFloch R, Front
B, Durivault J, Tambutté E, Massard PA, de la Ballina LR, Endou H,
Wempe MF, et al: Genetic disruption of the multifunctional
CD98/LAT1 complex demonstrates the key role of essential amino acid
transport in the control of mTORC1 and tumor growth. Cancer Res.
76:4481–4492. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Göthlin Eremo A, Lagergren K, Othman L,
Montgomery S, Andersson G and Tina E: Evaluation of
SPP1/osteopontin expression as predictor of recurrence in tamoxifen
treated breast cancer. Sci Rep. 10:14512020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tina E, Prosén S, Lennholm S, Gasparyan G,
Lindberg M and Göthlin Eremo A: Expression profile of the amino
acid transporters SLC7A5, SLC7A7, SLC7A8 and the enzyme TDO2 in
basal cell carcinoma. Br J Dermatol. 180:130–140. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl1. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Curtis C, Shah SP, Chin S-F, Turashvili G,
Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et
al METABRIC Group, : The genomic and transcriptomic architecture of
2,000 breast tumours reveals novel subgroups. Nature. 486:346–352.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rondón-Lagos M, Villegas VE, Rangel N,
Sánchez MC and Zaphiropoulos PG: Tamoxifen resistance: Emerging
molecular targets. Int J Mol Sci. 17:13572016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mihály Z, Kormos M, Lánczky A, Dank M,
Budczies J, Szász MA and Győrffy B: A meta-analysis of gene
expression-based biomarkers predicting outcome after tamoxifen
treatment in breast cancer. Breast Cancer Res Treat. 140:219–232.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
El Ansari R, Craze ML, Miligy I,
Diez-Rodriguez M, Nolan CC, Ellis IO, Rakha EA and Green AR: The
amino acid transporter SLC7A5 confers a poor prognosis in the
highly proliferative breast cancer subtypes and is a key
therapeutic target in luminal B tumours. Breast Cancer Res.
20:212018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bodoor K, Almomani R, Alqudah M, Haddad Y
and Samouri W: LAT1 (SLC7A5) overexpression in negative Her2 group
of breast cancer: A potential therapy target. Asian Pac J Cancer
Prev. 21:1453–1458. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guan Q, Song X, Zhang Z, Zhang Y, Chen Y
and Li J: Identification of tamoxifen-resistant breast cancer cell
lines and drug response signature. Front Mol Biosci. 7:5640052020.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bartlett JM, Thomas J, Ross DT, Seitz RS,
Ring BZ, Beck RA, Pedersen HC, Munro A, Kunkler IH, Campbell FM, et
al: Mammostrat as a tool to stratify breast cancer patients at risk
of recurrence during endocrine therapy. Breast Cancer Res.
12:R472010. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yan R, Zhao X, Lei J and Zhou Q: Structure
of the human LAT1-4F2hc heteromeric amino acid transporter complex.
Nature. 568:127–130. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Alfarsi LH, El-Ansari R, Craze ML, Masisi
BK, Mohammed OJ, Ellis IO, Rakha EA and Green AR: Co-expression
effect of SLC7A5/SLC3A2 to predict response to endocrine therapy in
oestrogen-receptor-positive breast cancer. Int J Mol Sci.
21:14072020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
El-Ansari R, Craze ML, Alfarsi L, Soria D,
Diez-Rodriguez M, Nolan CC, Ellis IO, Rakha EA and Green AR: The
combined expression of solute carriers is associated with a poor
prognosis in highly proliferative ER+ breast cancer.
Breast Cancer Res Treat. 175:27–38. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Semenza GL: Targeting HIF-1 for cancer
therapy. Nat Rev Cancer. 3:721–732. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kaira K, Oriuchi N, Imai H, Shimizu K,
Yanagitani N, Sunaga N, Hisada T, Tanaka S, Ishizuka T, Kanai Y, et
al: l-type amino acid transporter 1 and CD98 expression in primary
and metastatic sites of human neoplasms. Cancer Sci. 99:2380–2386.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Semenza GL: HIF-1: Mediator of
physiological and pathophysiological responses to hypoxia. J Appl
Physiol (1985). 88:1474–1480. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rathmell WK and Chen S: VHL inactivation
in renal cell carcinoma: Implications for diagnosis, prognosis and
treatment. Expert Rev Anticancer Ther. 8:63–73. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lock FE, McDonald PC, Lou Y, Serrano I,
Chafe SC, Ostlund C, Aparicio S, Winum JY, Supuran CT and Dedhar S:
Targeting carbonic anhydrase IX depletes breast cancer stem cells
within the hypoxic niche. Oncogene. 32:5210–5219. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Valvona CJ, Fillmore HL, Nunn PB and
Pilkington GJ: The regulation and function of lactate dehydrogenase
A: Therapeutic potential in brain tumor. Brain Pathol. 26:3–17.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Szablewski L: Expression of glucose
transporters in cancers. Biochim Biophys Acta. 1835:164–169.
2013.PubMed/NCBI
|
|
35
|
Vander Heiden MG, Cantley LC and Thompson
CB: Understanding the Warburg effect: The metabolic requirements of
cell proliferation. Science. 324:1029–1033. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Swietach P, Hulikova A, Vaughan-Jones RD
and Harris AL: New insights into the physiological role of carbonic
anhydrase IX in tumour pH regulation. Oncogene. 29:6509–6521. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lou Y, McDonald PC, Oloumi A, Chia S,
Ostlund C, Ahmadi A, Kyle A, Auf dem Keller U, Leung S, Huntsman D,
et al: Targeting tumor hypoxia: Suppression of breast tumor growth
and metastasis by novel carbonic anhydrase IX inhibitors. Cancer
Res. 71:3364–3376. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sato M, Harada-Shoji N, Toyohara T, Soga
T, Itoh M, Miyashita M, Tada H, Amari M, Anzai N, Furumoto S, et
al: L-type amino acid transporter 1 is associated with
chemoresistance in breast cancer via the promotion of amino acid
metabolism. Sci Rep. 11:5892021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Saito Y, Li L, Coyaud E, Luna A, Sander C,
Raught B, Asara JM, Brown M and Muthuswamy SK: LLGL2 rescues
nutrient stress by promoting leucine uptake in ER+
breast cancer. Nature. 569:275–279. 2019. View Article : Google Scholar : PubMed/NCBI
|