Introduction
Bladder cancer (BCa) is one of the most common
urological cancers. The most common histologic type of BCa is
uroepithelial carcinoma, which accounts for ~95% of cases (1,2). Since
the cancer cells originate from the metastatic epithelium of the
bladder, it is also known as metastatic cell carcinoma. Based on
the depth of tumor infiltration, BCa can be classified as
non-muscle-invasive BCa (NMIBC, ~75% of cases) and muscle-invasive
BCa (MIBC; ~25% of cases) (3).
NMIBC is usually treated conservatively, which preserves bladder
structure and function. Progression or recurrence can be prevented
by transurethral resection followed by intravesical instillation
(4). However, recurrence can occur
in >50% of patients with NMIBC; of these cases, 10 to 30%
progress to MIBC or even metastasis (5). MIBC is highly malignant, has a rapid
disease progression, and is characterized by metastasis and
recurrence (6). The main treatment
options for MIBC are radical surgery and radiotherapy; 25% of
patients with MIBC have a poor prognosis (7,8). In
addition, the existing diagnostic methods for BCa, such as
cystoscopy and urine cytology, are invasive procedures that can
lead to patient discomfort or are costly and have a poor
sensitivity (9). Therefore,
clarifying the underlying pathogenesis of BCa and identifying
biomarkers that accurately identify early cases are urgent research
goals.
A number of studies have verified that
monomethylated (m) modifications on the sixth nitrogen atom of
adenine (A) of RNA (designated as m6A) are essential in tumor
development. m6A modifications usually occur in a variety of RNAs,
including messenger RNAs (mRNAs), long non-coding RNAs (lncRNAs),
circular RNAs, small nuclear RNAs, transfer RNAs and ribosomal RNAs
(10,11). m6A modification sites are highly
conserved and tend to appear on the common sequence RR m6ACH
(R=G/A, H=A/C/U), which are mainly located near the 3′ untranslated
repeat, stop codon, or long exon of RNA. The sites are crucial for
the splicing of RNA precursors, 3′ end processing, extranuclear
transport, degradation and translation (12). The m6A modification is reversible.
Its regulation is dynamic and consists of methylesterase (writer),
demethylase (eraser) and methyl recognition protein (reader), which
perform different functions (13,14).
Methylation enzymes catalyze the methylation of the target. The
enzymes are m(6)A methyltransferase
(METTL)3, METTL14 and Wilms' tumor 1-associating protein (WTAP),
which form the METTL3/METTL14/WTAP complex (15). Demethylases catalyze demethylation.
With the involvement of ferrous ion (Fe2+) and
α-ketoglutarate, m6A methylation can be cleared by fat mass and
obesity-associated protein and AlkB homolog 5, RNA demethylase
(ALKBH5) (16). The demethylase
readers, including YTH N6-methyladenosine RNA binding
protein 3 (YTHDF1-3), YTHDC1-2, eukaryotic initiation factor 3,
heterogeneous nuclear ribonucleoproteins, HUR and insulin-like
growth factor-2 mRNA-binding proteins (IGF2BPs) can recognize
methylation information (17). As a
result, m6A is involved in a closed-loop, reversible process that
regulates its own methylation level and the expression of upstream
and downstream related genes.
METTL3 is the most common and most extensively
studied methyltransferase of m6A. m6A modifications significantly
modulate RNA stability, localization, transport, splicing and
translation (18). METTL3 also
plays an integral role in the development and progression of
various of tumors. METTL3 promotes homologous recombination repair
through the regulation of the EGF/RAD51 signaling axis, thereby
regulating the chemotherapeutic response in breast cancer (19). METTL3 mediates the m6A modification
of six-transmembrane epithelial antigen of the prostate-2 (STEAP2)
and promotes the malignant progression of papillary thyroid cancer
by inhibiting the Hedgehog signaling pathway and
epithelial-mesenchymal transition (20). METTL3 can promote poly(ADP-ribose)
polymerase 1 mRNA stability and thus resistance to oxaliplatin in
CD133+ stem cells of gastric cancer (21). In addition, METTL3 is also involved
in the carcinogenesis and progression of BCa. c-Jun N-terminal
kinase (JNK) signaling promotes immune escape from BCa by
modulating the METTL3-mediated m6A modification of programmed
death-ligand 1 (PD-L1) (22).
METTL3 can promote the malignant progression of BCa by regulating
the ALF transcription elongation factor 4/nuclear factor-κB/MYC
signaling network (23). METTL3 can
accelerate the maturation of pri-microRNA (miRNA/miR)-221/miR-222
and can thus promote the proliferation of BCa cells in an
m6A-dependent manner (22).
However, the specific biological effects and molecular mechanisms
of METTL3 in BCa need to be further explored.
The present study detected the expression level of
METTL3 in BCa tissues and cell lines, and validated the biological
role of METTL3 in BCa. In addition, the role of the METTL3/RAS
related (RRAS)/YTHDF2 regulatory axis in the tumorigenesis and
development of BCa was examined. The collective findings may
provide a theoretical basis for the comprehensive clinical
diagnosis and treatment of BCa.
Materials and methods
Bioinformatics analysis
The expression levels of METTL3 in BCa were analyzed
by utilizing the UANCAL database (http://ualcan.path.uab.edu/). In The Cancer Genome
Atlas (TCGA), urothelial bladder carcinoma (BLCA) data were
selected, METTL3 was entered, and the Expression function was
selected to analyze the association of METTL3 expression levels
with sample types, individual cancer stages and nodal metastasis
status. The Gene Expression Profiling Interactive Analysis 2
(GEPIA2) visual network analysis tool (http://gepia2.cancer-pku.cn/) was utilized for the
analysis. Survival analysis utilized BLCA data. METTL3 was entered,
the Group Cut-off was median, and Axis Units were months. The high
and low group was depicted in red and blue, respectively, and
survival curves were plotted.
Specimen collection
A total of 40 BCa tissues and para-cancerous tissues
were obtained from specimens surgically resected from bladder
cancer at the Department of Urology, Sir Run Run Hospital, Nanjing
Medical University from January, 2019 to December, 2021. The
inclusion and exclusion criteria for the selection of patients with
BCa are listed in Table SI. None
of the patients had been treated with radiotherapy and chemotherapy
prior to surgery. All tissue specimens were identified by two or
more pathologists. Ethics approval was provided by the Ethics
Committee of Sir Run Run Hospital, Nanjing Medical University
(2019-SR-S019). All patients provided signed informed consent and
agreed to the publication of their data.
Cells, cell culture and
transfection
Normal control (NC) cells (SV-HUC-1) (cat. no.
TCHu169) and the BCa cell lines, UM-UC-3 (cat. no. TCHu217), T24
(cat. no. TCHu 55), J82 (cat. no. TCHu218), TCCSUP (cat. no.
SCSP-571) and 5637 (cat. no. TCHu 1), were obtained from The Cell
Bank of Type Culture Collection of the Chinese Academy of Sciences.
The UM-UC-3 and SV-HUC-1 cells were cultured in Dulbecco's modified
Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented
with 10% fetal bovine serum and 2% double antibiotics (Gibco;
Thermo Fisher Scientific, Inc.). The T24, J82, TCCSUP and 5637
cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum and 2%
double antibiotics (Gibco; Thermo Fisher Scientific, Inc.) at 37°C
in a 5% CO2 atmosphere. Both the T24 and J82 cells were
cultured to 80% confluency, and transfection was performed with a
concentration of 2.5 µg overexpression plasmids (OE)-Vector (or
OE-METTL3, OE-RRAS and OE-YTHDF2) and 30 nM small interfering RNA
(siRNA)-NC (or si-METTL3 and si-YTHDF2) in six-well plates at
1×105 cells/well using LipofectamineTM
2000® Transfection Reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions at
37°C for 48 h. The time interval between transfection and
subsequent experimentation was 48 h. After the transfection
efficiency was verified using reverse transcription-quantitative
PCR (RT-qPCR) and western blot analysis, the experiments were
performed. All the siRNAs and overexpression plasmids were
purchased from Shanghai GeneChem Co., Ltd. The siRNA sequences are
presented in Table SII.
Lentiviruses and infection
The lentiviral vectors (LV)-small hairpin RNA
(shRNA) based on a third generation lentiviral system targeting
METTL3 (LV-shMETTL3) and negative control (LV-shNC) were purchased
from Shanghai GeneChem Co., Ltd. Briefly, 2.0 µg shRNA vector and
2.0 µg packaging mix plasmids were transfected into 293T cells
(cat. no. C6008; Beyotime Institute of Biotechnology) using
Lipofectamine 3000® reagent (Thermo Fisher Scientific,
Inc.) for 24 h at 37°C, and the medium was then harvested and the
virus was extracted by 85,000 × g ultracentrifugation at 4°C for 2
h in a centrifuge (Beckman Coulter, Inc.). For lentiviral
transduction, the cells were seeded in six-well plates at a density
of 50,000 cells/well. The lentiviral vector was added at a
multiplicity of infection of 20, with 8 µg polybrene
(Sigma-Aldrich; Merck KGaA) per well. After 72 h, the cells
infected with the lentiviral vectors were selected using puromycin
(Beyotime Institute of Biotechnology). The concentration of
puromycin used for selection was 2 µg/ml, and the concentration
used for maintenance was 1 µg/ml. The knockdown efficiency was
evaluated using RT-qPCR.
RNA extraction and RT-qPCR
Total RNA was extracted from the BCa tissues and
cells using TRIzol® reagent (Life; Thermo Fisher
Scientific, Inc.) and the RNA concentration was detected.
Complimentary DNA (cDNA) was synthesized by the reverse
transcription of RNA in accordance with the instructions provided
with the kit (Takara Bio, Inc.). The cDNA was diluted 20 times for
RT-qPCR. qPCR was performed according to the instructions provided
with the SYBR Green mix kit (Takara Bio, Inc.). The reaction
conditions were 95°C for 2 min and 30 cycles of 95°C for 30 sec,
58°C for 30 sec, and 72°C for 30 sec. glyceraldehyde 3-phosphate
dehydrogenase (GAPDH) were used as internal references. The qPCR
fluorescence values were calculated using the 2−ΔΔCq
method (24). The primers used are
listed in Table SIII.
Cell counting kit-8 (CCK-8) assay
The transfected BCa cells were inoculated in wells
of 96-well plates (3,000 cells per well). Following 24 h of cell
culture, 10 µl CCK-8 reagent (Dojindo Laboratories, Inc.) was added
to each well and incubation was continued at 37°C in a 5%
CO2 incubator protected from light. After 2 h, the
absorbance of each well was measured at 450 nm. The results were
compared using an unpaired t-test with GraphPad 7.0 software
(GraphPad Software Inc.).
5-ethynyl-2′-deoxyuridine (EdU)
assay
Following transfection for 48 h, single cell
suspensions were prepared from each group of cells in good
condition at the logarithmic growth stage. Wells of 96-well plates
were inoculated with 5×103 cells/well. Cell
proliferation was assessed using the EdU kit (Guangzhou RiboBio
Co., Ltd.). A total of 50 mM EdU solution was added to the BCa
cells and incubated for at 37°C for 2 h. Following incubation, the
cells were fixed with 4% paraformaldehyde at 20°C for 15 min,
treated with 0.1% Triton X-100 at 20°C for 5 min, closed with 5%
bovine serum albumin (Invitrogen; Thermo Fisher Scientific, Inc.)
for 30 min at 20°C, removed from the cell crawl and washed.
Rhodamine (Merck Co., Ltd.) was added and the cells were incubated
with the EdU reaction mixture for 20 min at 20°C. The nuclei were
re-fixed with Hoechst for 15 min at 20°C. Images were captured
under a fluorescent microscope at ×200 magnification (Olympus
Corporation) to calculate cell proliferation using ImageJ software
(Version 1.45s; National Institutes of Health).
Transwell assay
The transfected BCa cells (~2×105
cells/ml) were inoculated in the upper chamber (200 µl/well) of a
Transwell device (Corning, Inc.). The lower chamber was incubated
with culture medium containing 10% fetal bovine serum (60 µl/well)
and incubated for 24 h at 37°C. The cells were washed with PBS,
fixed by the addition of paraformaldehyde for 20 min at 20°C, and
stained with 0.1% crystal violet (Beyotime Institute of
Biotechnology) for 10 min at 20°C. The number of migrating cells
was observed under a fluorescent microscope (Olympus Corporation)
for 10 min. For the Transwell invasion assay, the pre-cooled
culture medium was used to dilute the Matrigel matrix (0.8 µm;
Corning, Inc.) gel, which was added to the upper chamber and
incubated for 5 h at 37°C. The BCa cell suspension was then added,
and the subsequent experimental steps were the same as those for
the cell migration assay. The number of invading cells was
determined under a fluorescent microscope (Olympus Corporation).
Images were captured under a fluorescent microscope at ×200
magnification (Olympus Corporation) to calculate cell migration and
invasion using ImageJ software (Version 1.45s; National Institutes
of Health).
Cell cycle analysis
The transfected BCa cells were cultured in six-well
plates for 48 h and then digested and resuspended. Following
centrifugation ×1,500 g at 4°C for 10 min, the supernatant was
discarded and the cells were suspended in 75% ethanol at 4°C and
fixed overnight. Propidium iodide (PI) staining solution (Merck
Co., Ltd.) was added to each tube (0.5 ml) after subsequent
washing, mixed well and incubated in an incubator at 37°C for 30
min protected from light, followed by automated detection at 4°C.
Cell cycle localization at each stage was measured using the
FACSCanto II flow cytometer (BD Biosciences) at a wavelength of 488
nm and the data were analyzed using BD FACSDiva Software version
8.0.2 (BD Biosciences).
Cell apoptosis analysis
The transfected BCa cells were cultured in six-well
plates for 48 h. The cells were then digested, resuspended and
transferred to 15-ml centrifuge tubes. Following centrifugation
×1,500 g at 4°C for 15 min, the supernatant was removed and 195 µl
fluorescein isothiocynate (FITC) conjugate (Merck Co., Ltd.) was
added to each Eppendorf tube (Merck Co., Ltd.). The cells were
resuspended, followed by the sequential addition of 5 µl FITC and
10 µl PI staining (Merck Co., Ltd.) solution. The contents were
mixed well and left static for 30 min at room temperature protected
from light, and then transferred to a refrigerator at 4°C. The
cells were incubated for 30 min at room temperature and then
transferred to a refrigerator at 4°C. Cellular apoptosis was
detected on a FACSCanto II flow cytometer (BD Biosciences), and the
data were analyzed with BD FACSDiva Software version 8.0.2 (BD
Biosciences).
Western blot analysis
Protein extracts were prepared by cell lysis using
radioimmunoprecipitation assay buffer (RIPA, Beyotime Institute of
Biotechnology). The protein concentration was determined using a
BCA protein assay kit (Beyotime Institute of Biotechnology), and 20
mg protein extract were separated by 10% sodium dodecyl
sulfate-poly-acrylamide gel electrophoresis (SDS-PAGE) followed by
transfer onto a polyvinylidene fluoride (PVDF) microporous
membranes (MilliporeSigma). The PVDF membranes were blocked by
placing them in Tris-buffered saline containing 0.1% Tween-20 and
blocked with 5% non-fat milk at room temperature for 2 h and
incubated with rabbit primary antibodies against METTL3 (1:1,000,
cat. no. ab195352), GAPDH (1:3,000, cat. no. ab181602), YTHDF2
(1:1,000, cat. no. ab246514) and RRAS (1:1,000, cat. no. ab154962)
(all from Abcam) at 4°C for 12 h. The PVDF membranes were incubated
with HRP-conjugated Affinipure goat anti-mouse IgG (H+L) (1:10,000;
cat. no. SA00001-1; ProteinTech Group, Inc.) or HRP-conjugated
Affinipure goat anti-rabbit IgG (H+L) (1:10,000; cat. no.
SA00001-2; ProteinTech Group, Inc.) at room temperature for 2 h.
The western blot bands were visualized using an ECL Kit (Vazyme
Biotech Co., Ltd.) and images were captured using the Tanon 4600
system (Tanon Science and Technology Co., Ltd.). Finally, ImageJ
software (version 1.45s; National Institutes of Health) was used to
calculate the relative gray value.
m6A-RNA immunoprecipitation assay
Total RNA (100 µg) was pre-cleared by incubation
with 20 µl recombinant protein-G sepharose bead suspension
(Invitrogen; Thermo Fisher Scientific, Inc.) in Magna Methylated
RNA Immunoprecipitation (MeRIP) buffer (10 mM Tris-HCl, pH 7.5, 150
mM NaCl, 0.1% NP-40, 2 mM ribonucleoside vanadyl complexes and 200
U/ml RNasin) for 1 h at 4°C according to the manufacturer's
instructions. The pre-cleared RNA was incubated with 1 µg anti-m6A
antibody (cat. no. ab286164, Abcam) for 4 h at 4°C. Recombinant
protein-G sepharose beads (50 µl) were blocked using 0.5 mg/ml
bovine serum albumin in meRIP buffer for 1 h at 4°C. The
RNA-antibody mixture was incubated with the pre-cleared beads and
for 2 h at 4°C. The beads were then washed five times with the
meRIP buffer, and the mRNA bound to the beads was eluted using 100
µl elution buffer (meRIP buffer supplemented with 6.7 mM
N6-methyladenosine base; Selleck Chemicals) at 4°C for 1 h. The
fragmented RNA containing m6A was eluted and purified using the RNA
purification kit (Qiagen, Inc.). The enriched fragments were
analyzed using RT-qPCR.
RNA binding protein
immunoprecipitation (RIP) assay
RIP experiments were performed using the Magna RIP
kit (MilliporeSigma) according to the instructions provided by the
manufacturer. Briefly, the cells (1×107 cells per 150
µl) were collected and lysed in a complete radioimmunoprecipitation
assay buffer containing a protease inhibitor cocktail and RNase
inhibitor. Magnetic beads coated with 5 µg specific antibodies
against mouse IgG (SAB5600281, MilliporeSigma, Merck Co., Ltd.) or
YTHDF2 (1/30, ab220163, Abcam) were pre-bound to protein A/G
magnetic beads in immunoprecipitation buffer (20 mM Tris-HCl pH
7.5, 140 mM NaCl, 0.05% Triton X-100) (MilliporeSigma, Merck Co.,
Ltd.) for 2 h and then incubated with 100 µl cell lysate overnight
at 4°C with rotation. RNA was eluted from the beads by incubation
with 400 µl elution buffer for 2 h, precipitated with ethanol and
dissolved in RNase-free water. The enrichment of certain fragments
was determined using RT-qPCR.
Luciferase reporter gene assay
DNA fragments containing the wild-type or mutant CDS
region of RRAS were inserted into the pmirGLO plasmids (GeneChem
Co., Ltd.). Plasmids (2 µg) containing the wild-type or mutant
fragments of the RRAS CDS region were co-transfected into 293T
cells with METTL3 overexpression plasmids (2 µg) or NC using
Lipofectamine 3000® reagent (Thermo Fisher Scientific,
Inc.). Following 48 h of transfection, analysis was performed using
a dual-luciferase reporter gene analysis system (Promega
Corporation). The ratio of Firefly luciferase signal to the
Renilla luciferase signal was calculated to evaluate the
modification effect of METTL3 on m6A of RRAS.
Xenograft experiments
The animal experiment was approved (approval no.
2021-AR-008) and monitored by the Animal Ethics Committee of Sir
Run Run Hospital, Nanjing Medical University. A total of 16 male,
4- to 6-week-old BALB/c nude mice weighing 18–26 g, were purchased
from the Institute of Model Animals, Nanjing University and raised
in a specific pathogen-free (SPF) environment for the xenograft
model. The mice were raised in animal individually ventilated cage
(IVC cages) with a room temperature of 24°C and relative humidity
of 70% and the air exchange rate was 15 times/h. The mice could
drink filtered tap water and commercial feed ad libitum
under a strict 12-h light/dark cycle. The animal laboratory was
cleaned twice one day and sterilized with ultraviolet light for 1 h
each week. Each mouse was injected with 100 µl PBS containing
1×106 T24 cell lines expressing either LV-NC or
LV-shMETTL3 subcutaneously under the right armpit. Tumor volumes
were recorded on day 7 after the injection and then measured each
week. At day 28, the mice were sacrificed by cervical dislocation
following the inhalation of 3% isoflurane anesthesia and the death
of the mice was confirmed by respiratory arrest. Tumor weights were
also compared. The data from each group of mice are expressed as
the mean ± standard deviation (SD).
Statistical analysis
Each experiment was independently repeated in
triplicate. GraphPad Prism 8.0 (GraphPad Software Inc.) and SPSS
22.0 (IBM Corp.) were used for data analysis. The data are
presented as mean ± SD. The data from two groups were compared
using an unpaired t-test. Differences between three or more groups
were evaluated using a one-way ANOVA multiple comparison test,
followed by Tukey's post hoc test. Pearson's correlation analysis
was performed to assess the correlations between genes. A P-value
<0.05 was considered to indicate a statistically significant
difference.
Results
METTL3 is significantly overexpressed
in BCa
To verify the expression level of METTL3 in BCa, the
GEPIA database was first used for analysis. The expression level of
METTL3 was significantly higher in BCa tissues than in normal
control tissues (Fig. 1A). The
association between the METTL3 expression levels and BCa cancer
stages and nodal metastasis status was also analyzed using TCGA
data. The results revealed that a high expression level of METTL3
was positively associated with BCa cancer stages and nodal
metastasis (Fig. 1B and C).
Subsequently, the association between the METTL3 expression levels
and the prognosis of patients with BCa was analyzed. No significant
association was observed between the METTL3 expression levels and
the overall and disease-free survival of patients with BCa
(Fig. 1D and E). The METTL3
expression levels were also verified in BCa and paraneoplastic
tissues collected from Sir Run Run Hospital, Nanjing Medical
University. As shown in Fig. 1F and
G, the mRNA expression level of METTL3 was markedly higher in
BCa tissues than in paraneoplastic tissues. Finally, the expression
patterns of METTL3 were examined in BCa cells. The expression level
of METTL3 was markedly higher in BCa cells. The most significant
increase was observed in the T24 and J82 cells (Fig. 1H and I). These cell lines were thus
used in subsequent assays.
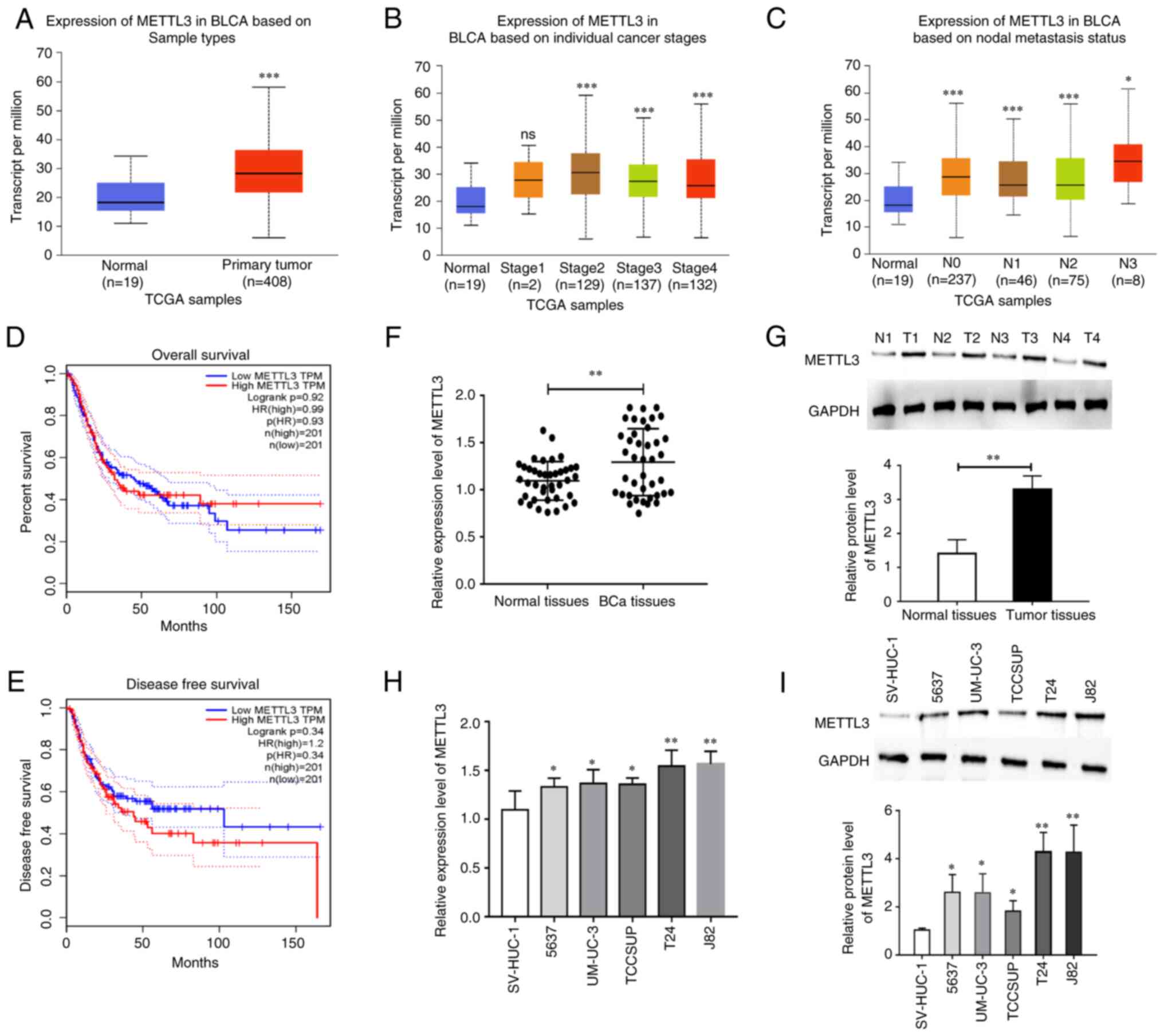 | Figure 1.METTL3 is significantly overexpressed
in BCa. (A) The expression of METTL3 was found to be significantly
higher in BCa tumor tissues than in normal control tissues using
TCGA database analysis. (B) The association of METTL3 expression
levels with BCa cancer stages examined using TCGA database
analysis. (C) The association of METTL3 expression levels with BCa
nodal metastasis status. (D) No significant association was found
between the METTL3 expression level and overall survival rate of
patients with BCa using GEPIA database analysis. (E) No significant
association was found between the METTL3 expression level and the
disease-free survival rate of patients with BCa using GEPIA
database analysis. (F) The mRNA expression of METTL3 in BCa tumor
tissues was significantly higher than that in normal control
tissues, as shown using RT-qPCR. (G) The protein expression of
METTL3 in BCa tumor tissues was significantly higher than that in
normal control tissues, as shown using western blot analysis. (H)
The mRNA expression of METTL3 in BCa cells was significantly higher
than that in normal control tissues, as shown using RT-qPCR. (I)
The expression of METTL3 in BCa cells was significantly higher than
that in normal control cells, as shown using western blot analysis.
*P<0.05, **P<0.01 and ***P<0.001, vs. normal tissues or
cells. ns, no significant difference; METTL3,
methyltransferase-like 3; BCa, bladder cancer; TCGA, The Cancer
Genome Atlas; RT-qPCR, reverse transcription-quantitative PCR. |
Inhibition of METTL3 significantly
suppresses BCa cell proliferation, migration and invasion
To investigate the biological effects of METTL3 in
BCa, the expression level of METTL3 in BCa cells we decreased or
increased by siRNA and an overexpression plasmid, respectively, and
the transfection efficiency was verified using RT-qPCR and western
blot analysis (Fig. S1). Cell
proliferation was then examined using CCK-8 and EdU assays
following the silencing of METTL3 expression in BCa cells. The
results revealed that METTL3 siRNA significantly inhibited cell
proliferation (Fig. 2A and B).
Furthermore, METTL3 siRNA also inhibited cell migration and
invasion, as shown by Transwell assay (Fig. 2C and D). In addition, the results of
flow cytometry revealed that the silencing METTL3 repressed the
cell cycle, while no significant changes in the cell apoptotic
ratio were observed after the silencing of METTL3 expression in BCa
cells (Fig. 2E and F). In addition,
cell phenotype changes were examined in BCa cells transfected with
METTL3 overexpression plasmid using an in vitro assay, and
it was found that the overexpression of METTL3 produced opposite
biological effects (Fig. S2).
These results indicated that METTL3 may function as an oncogene in
BCa.
RRAS is a potential downstream target
of METTL3
To explore the potential downstream targets of
METTL3, potential genes that were highly correlated with METTL3 in
the GEPIA database were screened (Table SIV). The expression of the
potential genes was then analyzed in BCa tissues and only three
genes exhibited a differential expression [low density lipoprotein
receptor class A domain containing 2 (LDLRAD2), RRAS and proteasome
20S subunit beta 10 (PSMB10)]. LDLRAD2 has been reported to
function as an oncogene in previous studies (25,26),
while PSMB10 was upregulated in BCa tissues according to the
results of the online database (GEPIA, http://gepia.cancer-pku.cn/ and UALCAN, http://ualcan.path.uab.edu/), which was inconsistent
with the correlation with METTL3. Hence, RRAS was selected for
further analysis. RRAS negatively correlated with METTL3 in BCa
tissues (Fig. 3A). The analysis of
the GEPIA database revealed that the expression of RRAS was
significantly lower in BCa tissues than in normal control tissues
(Fig. 3B). These findings suggest
that METTL3 may negatively regulate the expression of RRAS. The
METTL3 methyltransferase can regulate gene expression by regulating
the level of m6A modification of genes. To verify this mechanism of
the METTL3-regulated expression of RRAS, m6A modification sites in
the coding sequence region of RRAS were identified through database
analysis (Fig. 3C). The MeRIP assay
revealed that the overexpression of METTL3 increased the m6A
modification level of RRAS, while the silencing of METTL3
significantly decreased the m6A modification level of RRAS
(Fig. 3D), which also confirmed the
initial hypothesis. Further detection using RT-qPCR and western
blot analysis revealed that the overexpression of METTL3 suppressed
the mRNA and protein expression levels of RRAS, while the silencing
of METTL3 produced the opposite result (Fig. 3E and F). The results of the
dual-luciferase reporter gene assay indicated that the
overexpression of METTL3 suppressed the transcriptional activity of
RRAS in an m6A-dependent manner, while the silencing of METTL3
resulted in a significant increase in the transcriptional activity
of RRAS (Fig. 3G and H). These
experimental results indicate that METTL3 may inhibit RRAS
expression by elevating the level of m6A modification of RRAS.
 | Figure 3.RRAS is a potential downstream target
of METTL3. (A) A significant negative correlation between METTL3
and RRAS was found using GEPIA database analysis. (B) The
expression of RRAS in BCa tumor tissues was found to be
significantly lower than that in normal control tissues using TCGA
database analysis. (C) The presence of m6A modification sites in
the coding sequence region of RRAS was found using database
analysis. (D) The level of m6A modification of RRAS was found to be
significantly lower by MeRIP assay. MeRIP assay revealed that the
overexpression of METTL3 increased the m6A modification level of
RRAS, while the silencing of METTL3 significantly decreased the m6A
modification level of RRAS. (E and F) Reverse
transcription-quantitative PCR and western blot assays revealed
that the overexpression of METTL3 inhibited the mRNA and protein
expression levels of RRAS. (G and H) Dual luciferase reporter gene
assays revealed that the overexpression of METTL3 inhibited the
transcriptional activity of RRAS, while the silencing of METTL3
significantly increased the transcriptional activity of RRAS.
*P<0.05 and **P<0.01. ns, no significant difference; METTL3,
methyltransferase-like 3; BCa, bladder cancer; GEPIA, Gene
Expression Profiling Interactive Analysis; TCGA, The Cancer Genome
Atlas; RRAS, RAS related; m6A, N6-methyladenosine; MeRIP, Magna
methylated RNA immunoprecipitation. |
Silencing of METTL3 significantly
inhibits tumor growth
To confirm the effects of METTL3 on tumor growth, a
T24 cell line (LV-shMETTL-T24) with a stable low expression of
METTL3 was constructed and injected into nude mice. The tumorigenic
potential in mice in the LV-shMETTL3 group was significantly
diminished compared with that of mice in the LV-NC group (Fig. 4A-C). This finding suggested that
METTL3 functions as a pro-oncogenic factor in BCa. Furthermore, the
expression levels of METTL3 and RRAS were examined using RT-qPCR
and western blot analysis. The results revealed that LV-shMETTL3
suppressed the expression level of METTL3, while it promoted RRAS
expression at both the mRNA and protein level (Fig. 4D-F).
Overexpression of RRAS suppresses BCa
cell proliferation, migration and invasion
To explore the biological role of RRAS in BCa, the
expression level of RRAS and METTL3 was increased in BCa cells
using an overexpression plasmid and the cell proliferative,
migratory and invasive abilities were examined using EdU and
Transwell assays. The results demonstrated that the overexpression
of RRAS significantly inhibited cell proliferation, migration and
invasion, while the co-overexpression of METTL3 partially
attenuated the inhibitory effects of RRAS overexpression (Fig. 5). These results demonstrate that
RRAS may function as a tumor suppressor in BCa tissues.
YTHDF2 preferentially binds to the m6A
sites of RRAS
In order to examine the potential m6A recognition
processes that can identify RRAS, the correlation between the
expression levels of all m6A recognition proteins and RRAS was
analyzed in BCa tissues. The results revealed that IGF2BP2,
IGF2BP3, YTHDC1, YTHDC2, YTHDF1 and YTHDF2 exhibited a significant
correlation with RRAS (Fig. S3).
The expression levels of these m6A recognition proteins in BCa were
then analyzed. The expression of levels IGF2BP3, YTHDF1 and YTHDF2
were significantly increased, while the expression of YTHDC1 was
significantly decreased in BCa tissues (Fig. S4). IGF2BP3 can recruit RNA
stabilizers (27), YTHDF1 can
facilitate mRNA translation efficiency (28), and YTHDC1 can recruit the RNA
splicing and control the nuclear export (29). These biological functions are
inconsistent with their expression levels and the reduced
expression level of RRAS. YTHDF2 can promote mRNA degradation and
decrease gene expression (30–32),
which may be the reason for the decrease in RRAS expression.
Therefore, YTHDF2 was selected for further analysis. The expression
levels of YTHDF2 negatively correlated with RRAS expression
(Fig. 6A). Further analysis
revealed that the m6A modification site of RRAS was located within
the YTHDF2 protein binding region (Fig.
6B). The RIP assay revealed the binding of the YTHDF2 protein
to RRAS in BCa cells, indicating that YTHDF2 can bind the m6A
modification site of RRAS (Fig.
6C). The overexpression of YTHDF2, as verified using RT-qPCR
and western blot analysis, inhibited the mRNA and protein
expression levels of RRAS, while the silencing of YTHDF2 produced
the opposite results (Fig. 6D and
E). These results suggest that YTHDF2 may be able to bind the
m6A modification site of RRAS and lead to the degradation of RRAS
mRNA.
Discussion
The main reason for the high mortality rate of
patients with BCa is the recurrence and metastasis that occurs in
some patients, even following treatment with conventional regimens
(33,34). The mechanisms that are involved are
complex. The molecular staging of BCa is still exploratory. Several
staging schemes have been proposed based on different research
directions. These schemes are currently mainly used in the
prognostic assessment of the therapeutic sensitivity of targeted
chemotherapy and immune drugs (35,36).
The lack of in-depth research on pathogenesis and difficulties in
the development of new drugs and technology have prevented
significant progress being made in research on systemic treatment
and early diagnostic methods for BCa in the past decades. There is
an urgent need to identify new therapeutic targets for bladder
muscle cancer. Further in-depth studies on the molecular mechanisms
associated with BCa progression and metastasis are of utmost
importance.
m6A is the most common and abundant mRNA
modification in eukaryotic mRNAs and lncRNAs (14). Numerous in vitro data have
established the involvement of m6A modifications in multiple
aspects affecting mRNA metabolism, including RNA splicing,
processing, translocation, stability and translation (16). m6A modifications are involved in
regulating mRNA splicing, nuclear export, mRNA stability,
translation and miRNA processing (37,38).
The wide range of m6A modifications has led to their involvement in
a variety of physiological and pathological processes, such as
cancer and immune responses (39,40).
Previous studies have also demonstrated that m6A modifications are
closely associated with the development and progression of BCa.
Fine particulate matter induces the METTL3-mediated m6A
modification of BIRC5 mRNA in BCa, leading to malignant tumor
progression (41). The JNK
signaling axis promotes the BCa immune escape by regulating
METTL3-mediated m6A modification of PD-L1 mRNA (42). circ0008399 promotes m6A modification
by interacting with ALKBH5, which inhibits cell proliferation and
sensitizes BCa cells to cisplatin through m6A-CK2α-mediated
glycolysis (43). However, the
molecular mechanisms and biological effects of m6A modification in
BCa warrant further investigation.
The present study, to the best of our knowledge,
provides the first evidence from bioinformatics analysis and assay
validation that METTL3 expression is significantly increased in BCa
tissues and demonstrates a strong association between high METTL3
expression and BCa staging and staging. However, no significant
association was found between the METTL3 expression levels and the
prognosis of patients with BCa. The patients with BCa were not
followed-up for 5 years and thus the data from Sir Run Run
Hospital, Nanjing Medical University were not analyzed. The
long-term importance of the findings of the present study will be
explored in future studies. Furthermore, the ex vivo
experimental results indicated that METTL3 may play a role as a
pro-cancer factor in BCa by promoting cell proliferation, migration
and invasion. The bioinformatics analysis suggested RRAS as a
potential downstream target of METTL3. RRAS has important
biological roles in breast cancer (44), gastric cancer (45), colorectal cancer (46) and melanoma (47). However, its role in BCa remains
unclear. Mechanistic analyses in the present study indicated that
METTL3 may inhibit the expression of RRAS by increasing its m6A
modification level. YTHDF2 was the first specific m6A recognition
protein identified to regulate mRNA degradation (48,49).
YTHDF2 also plays an integral role in tumorigenesis and malignant
progression (50,51). Evidence provided in the present
study indicates that YTHDF2 may bind to the m6A modification site
of RRAS and lead to the degradation of RRAS mRNA.
There are several limitations to the present study.
First, the present study did not include a sufficient number of
clinical specimens and detailed clinical information to further
explore the diagnostic efficacy of METTL3. The authors aim to
address this limitation in future research. Second, while the
results confirmed that METTL3 promotes tumor cell migration and
invasion, these findings were not validated in vivo. Of
note, the possible downstream targets of METTL3 were not screened
by m6A sequencing and high-throughput sequencing, which limits the
innovation of the study renders the findings unconvincing.
Moreover, the underlying mechanism of METTL3 upregulation was not
investigated in the present study. Ni et al (42) demonstrated that JNK signaling was
associated with an increased METTL3 expression in BCa. In addition,
Liu et al (41) identified
that PM2.5 may enhance the expression of METTL3 by inducing the
promoter hypomethylation of its promoter and increasing the binding
affinity of the transcription factor HIF1A. Due to the limitations
regarding time and resources, the present study not explore and
verify the mechanisms for the increase of METTL3. In the future,
the authors aim to explore the potential mechanism for the increase
of METTL3 in BCa cells.
In conclusion, the findings of the present study
indicate that METTL3 expression is significantly increased in BCa.
METTL3 may contribute to the malignant progression of BCa by
regulating the RRAS/YTHDF2 signaling axis, thus promoting cell
proliferation, migration and invasion.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Science and Technology
Development Fund of Nanjing Medical University (grant no.
NMUB2019084).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SZ, DMC, JXC and XLX were involved in the
conceptualization of the study, as well as in the study
methodology, and in the writing, reviewing and editing of the
manuscript. DW and YX were involved in the investigative aspects of
the study, as well as in data curation, and in the writing and
preparation of the original draft. SZ, JXC and XLX were involved in
visualization, data validation, supervision and in the provision of
software. JXC and XLX confirm the authenticity of all the raw data.
All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The research protocol conformed to the principles
outlined in the Declaration of Helsinki. All patients provided
written informed consent and the protocol of the study was approved
by the Research Ethics Committee of Sir Run Run Hospital, Nanjing
Medical University. The animal experiment was approved (approval
no. 2021-AR-008) and monitored by the Animal Ethics Committee of
Sir Run Run Hospital, Nanjing Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Tran L, Xiao JF, Agarwal N, Duex JE and
Theodorescu D: Advances in bladder cancer biology and therapy. Nat
Rev Cancer. 21:104–121. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lenis AT, Lec PM, Chamie K and Mshs MD:
Bladder cancer: A review. JAMA. 324:1980–1991. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Patel VG, Oh WK and Galsky MD: Treatment
of muscle-invasive and advanced bladder cancer in 2020. CA Cancer J
Clin. 70:404–423. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Joensen UN, Maibom SL and Poulsen AM:
Surgical management of muscle invasive bladder cancer: A review of
current recommendations. Semin Oncol Nurs. 37:1511042021.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Witjes JA, Bruins HM, Cathomas R, Compérat
EM, Cowan NC, Gakis G, Hernández V, Linares Espinós E, Lorch A,
Neuzillet Y, et al: European association of urology guidelines on
muscle-invasive and metastatic bladder cancer: Summary of the 2020
guidelines. Eur Urol. 79:82–104. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nason GJ, Ajib K, Tan GH and Kulkarni GS:
Bladder-sparing treatment options in localized muscle-invasive
bladder cancer. Expert Rev Anticancer Ther. 20:179–188. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nadal R and Bellmunt J: Management of
metastatic bladder cancer. Cancer Treat Rev. 76:10–21. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Alifrangis C, McGovern U, Freeman A,
Powles T and Linch M: Molecular and histopathology directed therapy
for advanced bladder cancer. Nat Rev Urol. 16:465–483. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jordan B and Meeks JJ: T1 bladder cancer:
Current considerations for diagnosis and management. Nat Rev Urol.
16:23–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jia J, Wu S, Jia Z, Wang C, Ju C, Sheng J,
He F, Zhou M and He J: Novel insights into m6A
modification of coding and non-coding RNAs in tumor biology: From
molecular mechanisms to therapeutic significance. Int J Biol Sci.
18:4432–4451. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen J, Fang Y, Xu Y and Sun H: Role of
m6A modification in female infertility and reproductive system
diseases. Int J Biol Sci. 18:3592–3604. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Qu X, Zhang Y, Sang X, Ren D, Zhao H and
Wong STC: Methyladenosine modification in RNAs: From regulatory
roles to therapeutic implications in cancer. Cancers (Basel).
14:31952022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang F, Liu H, Duan M, Wang G, Zhang Z,
Wang Y, Qian Y, Yang Z and Jiang X: Crosstalk among m6A
RNA methylation, hypoxia and metabolic reprogramming in TME: From
immunosuppressive microenvironment to clinical application. J
Hematol Oncol. 15:842022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Murakami S and Jaffrey SR: Hidden codes in
mRNA: Control of gene expression by m6A. Mol Cell.
82:2236–2251. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shi B, Liu WW, Yang K, Jiang GM and Wang
H: The role, mechanism, and application of RNA methyltransferase
METTL14 in gastrointestinal cancer. Mol Cancer. 21:1632022.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shen D, Wang B, Gao Y, Zhao L, Bi Y, Zhang
J, Wang N, Kang H, Pang J, Liu Y, et al: Detailed resume of RNA
m6A demethylases. Acta Pharm Sin B. 12:2193–2205. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen D, Cheung H, Lau HC, Yu J and Wong
CC: N6-methyladenosine RNA-binding protein YTHDF1 in
gastrointestinal cancers: Function, molecular mechanism and
clinical implication. Cancers (Basel). 14:34892022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu X, Ye W and Gong Y: The role of RNA
methyltransferase METTL3 in normal and malignant hematopoiesis.
Front Oncol. 12:8739032022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li E, Xia M, Du Y, Long K, Ji F, Pan F, He
L, Hu Z and Guo Z: METTL3 promotes homologous recombination repair
and modulates chemotherapeutic response in breast cancer by
regulating the EGF/RAD51 axis. Elife. 11:e752312022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhu Y, Peng X, Zhou Q, Tan L, Zhang C, Lin
S and Long M: METTL3-mediated m6A modification of STEAP2 mRNA
inhibits papillary thyroid cancer progress by blocking the Hedgehog
signaling pathway and epithelial-to-mesenchymal transition. Cell
Death Dis. 13:3582022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li H, Wang C, Lan L, Yan L, Li W, Evans I,
Ruiz EJ, Su Q, Zhao G, Wu W, et al: METTL3 promotes oxaliplatin
resistance of gastric cancer CD133+ stem cells by promoting PARP1
mRNA stability. Cell Mol Life Sci. 79:1352022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu
HC, Yuan WB, Lu JC, Zhou ZJ, Lu Q, et al: METTL3 promote tumor
proliferation of bladder cancer by accelerating pri-miR221/222
maturation in m6A-dependent manner. Mol Cancer. 18:1102019.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cheng M, Sheng L, Gao Q, Xiong Q, Zhang H,
Wu M, Liang Y, Zhu F, Zhang Y, Zhang X, et al: The m6A
methyltransferase METTL3 promotes bladder cancer progression via
AFF4/NF-κB/MYC signaling network. Oncogene. 38:3667–3680. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wei Y, Zhang F, Zhang T, Zhang Y, Chen H,
Wang F and Li Y: LDLRAD2 overexpression predicts poor prognosis and
promotes metastasis by activating Wnt/β-catenin/EMT signaling
cascade in gastric cancer. Aging (Albany NY). 11:8951–8968. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li J, Huang W, Han Q, Xiong J and Song Z:
LDLRAD2 promotes pancreatic cancer progression through Akt/mTOR
signaling pathway. Med Oncol. 38:22021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wan W, Ao X, Chen Q, Yu Y, Ao L, Xing W,
Guo W, Wu X, Pu C, Hu X, et al: MEMETTL3/IGF2BP3 axis inhibits
tumor immune surveillance by upregulating
N6-methyladenosine modification of PD-L1 mRNA in breast
cancer. Mol Cancer. 21:602022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xia H, Wu Y, Zhao J, Cheng C, Lin J, Yang
Y, Lu L, Xiang Q, Bian T and Liu Q: N6-methyladenosine-modified
circSAV1 triggers ferroptosis in COPD through recruiting YTHDF1 to
facilitate the translation of IREB2. Cell Death Differ. Feb
24–2023.(Epub ahead of print). View Article : Google Scholar
|
|
29
|
Timcheva K, Dufour S, Touat-Todeschini L,
Burnard C, Carpentier MC, Chuffart F, Merret R, Helsmoortel M,
Ferré S, Grézy A, et al: Chromatin-associated YTHDC1 coordinates
heat-induced reprogramming of gene expression. Cell Rep.
41:1117842022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hu L, Yu Y, Shen Y, Huang H, Lin D, Wang
K, Yu Y, Li K, Cao Y, Wang Q, et al: Ythdf2 promotes pulmonary
hypertension by suppressing Hmox1-dependent anti-inflammatory and
antioxidant function in alveolar macrophages. Redox Biol.
61:1026382023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang Y, Yan Y, Yin J, Tang N, Wang K,
Huang L, Hu J, Feng Z, Gao Q and Huang A: O-GlcNAcylation of YTHDF2
promotes HBV-related hepatocellular carcinoma progression in an
N6-methyladenosine-dependent manner. Signal Transduct
Target Ther. 8:632023. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhuang M, Geng X, Han P, Che P, Liang F,
Liu C, Yang L, Yu J, Zhang Z, Dong W and Ji SJ: YTHDF2 in dentate
gyrus is the m6A reader mediating m6A
modification in hippocampus-dependent learning and memory. Mol
Psychiatry. Jan 20–2023.(Epub ahead of print). View Article : Google Scholar
|
|
33
|
Martin A, Woolbright BL, Umar S, Ingersoll
MA and Taylor JA III: Bladder cancer, inflammageing and
microbiomes. Nat Rev Urol. 19:495–509. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lee YC, Lam HM, Rosser C, Theodorescu D,
Parks WC and Chan KS: The dynamic roles of the bladder tumour
microenvironment. Nat Rev Urol. 19:515–533. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu S, Chen X and Lin T: Emerging
strategies for the improvement of chemotherapy in bladder cancer:
Current knowledge and future perspectives. J Adv Res. 39:187–202.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ranti D, Bieber C, Wang YS, Sfakianos JP
and Horowitz A: Natural killer cells: Unlocking new treatments for
bladder cancer. Trends Cancer. 8:698–710. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li Y, Meng L and Zhao B: The roles of
N6-methyladenosine methylation in the regulation of bone
development, bone remodeling and osteoporosis. Pharmacol Ther.
238:1081742022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhuo R, Xu M, Wang X, Zhou B, Wu X, Leone
V, Chang EB and Zhong X: The regulatory role of
N6-methyladenosine modification in the interaction
between host and microbes. Wiley Interdiscip Rev RNA. 13:e17252022.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhou H, Mao L, Xu H, Wang S and Tian J:
The functional roles of m6A modification in T lymphocyte
responses and autoimmune diseases. Cytokine Growth Factor Rev.
65:51–60. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li X, Ma S, Deng Y, Yi P and Yu J:
Targeting the RNA m6A modification for cancer
immunotherapy. Mol Cancer. 21:762022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu H, Gu J, Huang Z, Han Z, Xin J, Yuan
L, Du M, Chu H, Wang M and Zhang Z: Fine particulate matter induces
METTL3-mediated m6A modification of BIRC5 mRNA in
bladder cancer. J Hazard Mater. 437:1293102022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ni Z, Sun P, Zheng J, Wu M, Yang C, Cheng
M, Yin M, Cui C, Wang G, Yuan L, et al: JNK signaling promotes
bladder cancer immune escape by regulating METTL3-mediated m6A
modification of PD-L1 mRNA. Cancer Res. 82:1789–1802. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yu H, Yang X, Tang J, Si S, Zhou Z, Lu J,
Han J, Yuan B, Wu Q, Lu Q and Yang H: ALKBH5 inhibited cell
proliferation and sensitized bladder cancer cells to cisplatin by
m6A-CK2α-mediated glycolysis. Mol Ther Nucleic Acids. 23:27–41.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li H, Prever L, Hsu MY, Lo WT, Margaria
JP, De Santis MC, Zanini C, Forni M, Novelli F, Pece S, et al:
Phosphoinositide conversion inactivates R-RAS and drives metastases
in breast cancer. Adv Sci (Weinh). 9:e21032492022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hu Q, Masuda T, Koike K, Sato K, Tobo T,
Kuramitsu S, Kitagawa A, Fujii A, Noda M, Tsuruda Y, et al:
Oxysterol binding protein-like 3 (OSBPL3) is a novel driver gene
that promotes tumor growth in part through R-Ras/Akt signaling in
gastric cancer. Sci Rep. 11:191782021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Raza A, Pandey MS, Jin Q and Mulder KM:
km23-1/DYNLRB1 regulation of MEK/ERK signaling and R-Ras in
invasive human colorectal cancer cells. Cell Biol Int. 44:155–165.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Sung H, Kanchi KL, Wang X, Hill KS,
Messina JL, Lee JH, Kim Y, Dees ND, Ding L, Teer JK, et al:
Inactivation of RASA1 promotes melanoma tumorigenesis via R-Ras
activation. Oncotarget. 7:23885–23896. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Liu R, Jia Y, Kong G and He A: Novel
insights into roles of N6-methyladenosine reader YTHDF2 in cancer
progression. J Cancer Res Clin Oncol. 148:2215–2230. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lee Y, Choe J, Park OH and Kim YK:
Molecular mechanisms driving mRNA degradation by m6A
modification. Trends Genet. 36:177–188. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chen X, Zhou X and Wang X: m6A
binding protein YTHDF2 in cancer. Exp Hematol Oncol. 11:212022.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang JY and Lu AQ: The biological function
of m6A reader YTHDF2 and its role in human disease. Cancer Cell
Int. 21:1092021. View Article : Google Scholar : PubMed/NCBI
|




















