Introduction
Glioblastoma multiforme (GBM) is the most common
type of malignant brain tumor in adults, with 3.6 cases per 100,000
patients diagnosed annually in Europe (1). Median survival for patients with GBM
is 12–15 months following diagnosis, despite surgical resection,
chemo- and radiotherapy (2).
Genetic heterogeneity is characteristic of GBM
(3). Among the high mutational
background, mutations in TP53 have been detected in 31% of
cases of primary GBM (4).
TP53 status affects transcriptional profiles in tumor cell
lines (5,6), including GBM cells exposed to
irradiation (7,8).
The existence of cancer stem cells (CSCs) in tumors
may be associated with chemo- and radioresistance (9–11).
In brain tumors, cluster of differentiation (CD) 133 expression is
considered a marker of stem cells (12,13)
and is increased in GBM cell lines grown as spheres, compared with
adherent cells (14). There is
evidence that GBM tumors may originate from CSCs that are present
in neurospheres, and these cells often reflect the
histopathological features of the tumor, indicating their
suitability to reproduce the cellular heterogeneity of human GBM
(13). Transcription factor 12
(TCF12; HEB) is a transcription factor (TF) involved
in the proliferation of neuronal stem cells via the maintenance of
their undifferentiated state during embryonic and adult
neurogenesis (15).
Novel therapies are required to improve overall
survival, and therapies targeting molecules involved in
proliferation, survival and invasiveness of GBM cells have been
evaluated (16). Increased cell
death was reported using LY294002 (a phosphoinositide 3-kinase
inhibitor) in association with cisplatin (17), and using methoxyamine (a base
excision repair blocker) (18) or
apurinic apyrimidinic endonuclease redox effector factor-1
silencing (19) in combination
with temozolomide in GBM cell lines.
TFs have been suggested as potential therapeutic
targets for cancer treatment (20), and they have previously been
evaluated in prostate (21) and
breast cancer (22). Our
laboratory has identified certain TFs associated with genes
modulated in GBM compared with healthy brain samples (unpublished
data), following treatment with cisplatin associated with LY294002
(17), and in response to
gamma-rays, where HEB was associated with upregulated genes
in the U87MG cell line (23).
HEB is a member of the helix-loop-helix
protein family (24), and together
with TCF3 (E12, E47) and TCF4
(E2-2) is known as an E-protein with DNA binding properties
(25). HEB functions
include T-cell development (26,27),
myogenesis (28,29), dedifferentiation of renal tubular
epithelial cells (30), and
proliferation of neural and progenitor cells (15). Recently, dysregulated HEB
expression has been reported in leukemia (31,32),
oligodendroglioma (33) and
colorectal cancer (CRC) (34,35).
In glioma, HEB was demonstrated to be highly expressed
(36).
HEB is involved in neural and stem cell
proliferation, is overexpressed in certain tumor types, and is
associated with upregulated genes in irradiated U87MG cells.
Therefore, the present study aimed to test the hypothesis that
HEB knockdown may sensitize GBM cells by affecting cell
proliferation, cell death and cell cycle kinetics, as well as
affecting the formation of neurospheres and differentiation in
irradiated GBM cells. In addition, the effects of HEB
silencing combined with irradiation were evaluated.
Materials and methods
Cell lines and monolayer cultures
The U87MG human GBM cell line, [American Type
Culture Collection (ATCC)® HTB-14™] was obtained from
the ATCC (Manassas, VA, USA) and the ACBRI-371 human primary
astrocyte cell line was obtained from Cell Systems Corporation
(Kirkland, WA, USA). Cells were cultured in Dulbecco's modified
Eagle's medium (DMEM) Ham's F-10 (Sigma-Aldrich; Merck Millipore,
Darmstadt, Germany), supplemented with 10% fetal bovine serum (FBS;
Cultilab, Campinas, Brazil) and 1% penicillin (10.000 units)
/streptomycin (10 mg) (Sigma-Aldrich; Merck Millipore). Cells were
incubated at 37°C in an atmosphere containing 5% CO2 until they
reached semi-confluence, when they were used for experiments.
ACBRI-371 was used to compare HEB protein expression by western
blotting and immunocytochemistry.
Neurosphere cell culture
U87MG and ACBRI-371 cells were initially cultured in
monolayers, dissociated with Accutase® (EMD Millipore,
Billerica, MA, USA), and plated in neurosphere culture medium,
containing DMEM/F12 (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), 20 ng/ml basic fibroblast growth factor
(PeproTech EC Ltd., London, UK), 20 ng/ml epidermal growth factor
(PeproTech EC Ltd.), 10 ng/ml leukemia inhibitory factor (Merck
Millipore), 20 µl/ml B27 supplement (1:50; Thermo Fisher
Scientific, Inc.) and 1% penicillin (10.000 units)/streptomycin (10
mg). The neurosphere medium was replaced every 2–3 days. This
protocol was based on previous studies of neurosphere culture
(12,14,37).
For neurosphere formation, ACBRI-371 cells were cultured for 14
days, whereas for U87MG cells, the culture time was 3 days for
western blotting and 7 days for neurosphere assays. The
neurospheres derived from ACBRI-371 were used for western blotting
experiments.
Western blotting
Cells were lysed in ProteoJET™ Mammalian Cell Lysis
reagent (Fermentas; Thermo Fisher Scientific, Inc.), supplemented
with Halt™ Protease Inhibitor Cocktail kit (Thermo Fisher
Scientific, Inc.). Protein concentrations were determined using a
Bicinchoninic Acid Protein Assay (Thermo Fisher Scientific Inc.),
according to the manufacturer's protocol. Proteins (30 µg) were
separated by electrophoresis on a NuPAGE 4–12% Bis-Tris gel
(Invitrogen; Thermo Fisher Scientific, Inc.) and were transferred
onto a polyvinylidene difluoride membrane (Invitrogen; Thermo
Fisher Scientific, Inc.) using the XCell SureLock™ Mini-Cell system
(Invitrogen; Thermo Fisher Scientific, Inc.).
Membranes were incubated in blocking buffer
(WesternBreeze Chromogenic kit; Invitrogen; Thermo Fisher
Scientific, Inc.) for 1 h at room temperature prior to the addition
of a rabbit primary antibody recognizing HEB (1:1,000; catalog no.
sc-357; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), followed
by incubation with an alkaline phosphatase-conjugated goat
anti-rabbit secondary antibody (catalog no. WB7105; WesternBreeze
Chromogenic kit) for 30 min at room temperature. A rabbit
anti-β-actin antibody (ACTB; 1:1,000; catalog no. 4967; Cell
Signaling Technology, Inc., Danvers, MA, USA) served as an
endogenous control. Protein bands were visualized using the Western
Breeze Chromogenic kit. Densitometric analysis of protein bands was
performed using Gel Pro Analyzer software version 4.0 (www.gelanalyzer.com/download.html); HEB
expression was calculated relative to ACTB.
Immunocytochemistry
To assess HEB expression in monolayer cultures,
50,000 cells were seeded in 6-well plates containing one
coverslip/well. After 24 h, cells were fixed with 3%
paraformaldehyde and 2% sucrose in phosphate-buffered saline (PBS)
and were permeabilized with 0.5% Triton X-100. Cells were incubated
with the primary antibody [anti-HEB; 1:100 in PBS/2% bovine serum
albumin (BSA)] for 30 min at 37°C, and subsequently with the
secondary antibody [Alexa Fluor 488-conjugated anti-rabbit
immunoglobulin (Ig)G; 1:100 in PBS/2%BSA; catalog no. A21441;
Thermo Fisher Scientific, Inc.] for 30 min at 37°C. Nuclei were
counterstained with 4′,6-diamidino-2-phenylindole (DAPI; 90 mM in
PBS; Sigma-Aldrich; Merck Millipore) for 10 min. Coverslips were
then mounted using Vectashield (Vector Laboratories, Inc.,
Burlingame, CA, USA), sealed and stored at 4°C in the dark until
analysis. Subsequently, the slides were analyzed under a
fluorescence microscope (Carl Zeiss AG, Oberkochen, Germany)
equipped with filters for DAPI and Alexa Fluor 488. Images were
captured at magnification, ×20.
Gene silencing by small interfering
(si) RNA
U87MG cells (1×106) were seeded in 25
cm2 culture flasks containing 3 ml DMEM Ham's F-10
supplemented with 10% FBS without antibiotics. Cells were incubated
until they reached 60–80% confluence (18–24 h). Specific oligomers
for HEB siRNA were used (catalog no. sc-35552; Santa Cruz
Biotechnology, Inc.), which includes three sequences of 20–25
nucleotides for HEB silencing. Oligomer transfections into
U87MG cells were performed using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Cells were transfected for 6 h, followed
by a 24 h incubation and irradiation. Cells were then analyzed for
cell death, proliferation and cell cycle kinetics (at 24, 48 and 72
h following irradiation). To assess the efficiency of transfection,
1×106 cells were seeded and harvested after 72 h for
western blotting. The scrambled sequence (SCR; catalog no.
sc-37007; Santa Cruz Biotechnology, Inc.) served as a negative
control and was used at an identical concentration to HEB
siRNA. For neurosphere culture, 24 h following transfection in
monolayers, 500 cells were seeded in 96-well plates in triplicate,
in 200 µl specific medium for neurosphere formation, as described
earlier. Cells were irradiated or sham-irradiated 24 h later and
cultured for 6 days at 37°C and 5% CO2, at which point they were
harvested for neurosphere counting.
Cell irradiation
U87MG cells cultured as monolayers or neurospheres
were irradiated or sham-irradiated 24 h following transfection with
HEB siRNA or SCR. Cells were irradiated with 4 Gy of
gamma-rays (60Co source; dose rate, 0.5 Gy/min;
Gammatron S-80; Siemens AG, Munich, Germany).
Proliferation, cell death and cell
cycle analyses
Following HEB knockdown by siRNA, U87MG cells
were incubated for 24 h, irradiated with 4 Gy, and harvested at 24,
48 and 72 h subsequent to irradiation. Cells were detached with
Accutase® followed by measurements of cell viability and
cell death, using the Guava ViaCount kit (Merck Millipore),
according to the manufacturer's protocol. Samples were analyzed
using the Guava cytometer EasyCyte Mini system (Merck Millipore),
and GuavaCytoSoft software version 4.2.1 (Merck Millipore); ≥1,000
events were analyzed for each sample.
Following the removal of an aliquot of cells for the
proliferation assay, the remaining cells were fixed in 70% ethanol
and frozen at −20°C for ≥24 h, prior to cell cycle analysis. Cells
were incubated with Guava Cell Cycle reagent (Merck Millipore),
according to the manufacturer's protocol, and analyzed using the
Guava cytometer EasyCyte Mini system and GuavaCytoSoft software
version 4.2.1; ≥5,000 events were analyzed for each sample.
Neurosphere formation, determination
of cell number and cell death
Neurospheres >60 microns were counted using a
Nikon inverted microscope (TS100; Nikon Corporation, Tokyo, Japan)
at magnification, ×100. The following day, the neurospheres were
dissociated with Accutase® in order to determine cell
number and death using the Guava ViaCount kit as described in the
previous section.
Detection of CD133 by
immunofluorescence
Neurospheres formed from U87MG adherent cells were
dissociated 7 days following treatment. Cells were re-incubated in
cellular differentiation medium (DMEM/F12 containing 10% FBS and 1%
penicillin/streptomycin) and harvested following an additional
period of 8 days. Cells were detached using Accutase®,
washed in 1X PBS and fixed in 4% paraformaldehyde for 15 min at
room temperature. Subsequent to washing in 1X PBS, cells were
incubated in blocking solution (0.5% BSA in PBS), followed by
incubation with a rabbit primary anti-CD133 antibody (1:100 in
PBS/2%BSA; catalog no. 3663; Cell Signaling Technology, Inc.).
Cells were subsequently incubated with an Alexa Fluor
488-conjugated anti-rabbit IgG secondary antibody (1:100 in
PBS/2%BSA). The samples were analyzed using the Guava cytometer
EasyCyte Mini system and GuavaCytoSoft software version 4.2.1.
The presence of CD133 protein is characteristic of
certain types of tumor stem cells and is associated with their
differentiation capacity. Therefore, neurospheres cultured for 8
days in neurosphere culture medium were used as a positive
control.
Statistical analysis
In general, at least three independent experiments
were performed, except for HEB protein expression analysis by
western blotting and immunofluorescence, for which one experiment
was conducted. The results were analyzed by Student's t-test if
less than three groups were compared, or by one-way analysis of
variance followed by Holm-Sidak pairwise multiple comparison test
if more than two groups were compared. P<0.05 was considered to
indicate a statistically significant difference. Statistical
analyses were performed using SigmaStat for Windows software
version 3.5 (Systat Software, Inc., San Jose, CA, USA), and the
graphs were plotted in Microsoft Excel version 2010 (Microsoft
Corporation, Redmond, WA, USA). Data are expressed as the mean ±
standard deviation.
Results
HEB protein expression
HEB protein expression was assessed in U87MG and
ACBRI-371 cells in adherent monolayer cultures in the absence of
radiation treatment. Using immunofluorescence, nuclear localization
of HEB protein in U87MG (Fig. 1A)
and ACBRI-371 (Fig. 1B) was
observed.
 | Figure 1.HEB expression and silencing. HEB
protein expression was visualized by immunocytochemistry in (A)
U87MG and (B) ACBRI-371 cell lines. HEB was localized to the nuclei
of the two cell lines. Magnification, ×20. (C) HEB expression was
analyzed by western blotting in U87MG monolayers and neurospheres
and ACBRI-371 astrocyte monolayers and neurospheres. (D) Relative
protein expression levels of HEB calculated by densitometric
analysis relative to ACTB. HEB protein expression levels were
increased in U87MG neurospheres compared with ACBRI-371
neurospheres. *P<0.05. (E) HEB protein expression in U87MG cells
transfected with 25 nM HEB siRNA or SCR, analyzed 72 h following
transfection. (F) HEB protein expression was decreased by 97.8% in
cells transfected with HEB siRNA compared with SCR. HEB,
transcription factor 12; U87, U87MG monolayers; U87N, U87MG
neurospheres; ACBRI, ACBRI-371 monolayers; ACBRI N, ACBRI-371
neurospheres; ACTB, β-actin; siRNA, small interfering RNA; SCR,
scrambled sequence. |
Western blotting revealed a relative HEB expression
of 1.35±0.16 and 1.31±0.01 (neurospheres) in U87MG cells, and
1.14±0.24 and 0.76±0.11 (neurospheres) in ACBRI-371 cells (Fig. 1C and D). No significant differences
were observed between each cell line and their respective
neurospheres. Although U87MG cells have greater HEB protein
expression levels compared with astrocytes, the difference was not
significant. Notably, U87MG neurospheres have significantly greater
HEB protein expression levels compared with ACBRI-371 neurospheres
(P=0.001). HEB silencing resulted in a 97.8% reduction in
HEB protein expression levels 72 h following transfection (Fig. 1E and F). These results indicated
that HEB protein is a potential target for therapeutic strategies
based on molecular inhibition.
Effects of HEB silencing on U87MG
monolayer cells
To determine whether HEB knockdown affects
the proliferation and cell death of U87MG cells, cell cultures were
irradiated or sham-irradiated 24 h following transfection with
HEB siRNA; cells were analyzed 24, 48 and 72 h later.
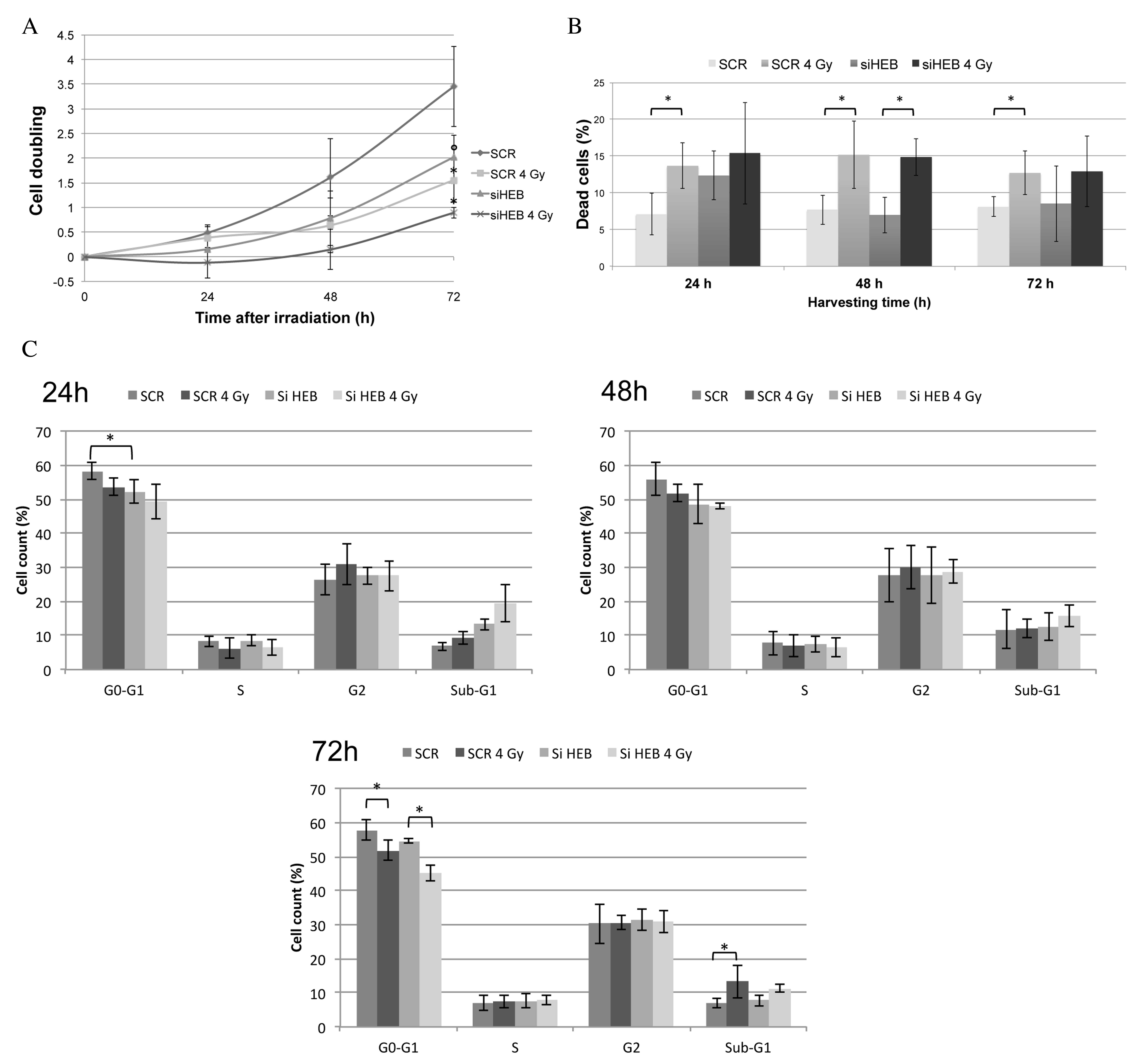
Inhibiting HEB decreased cell proliferation at all time
points, compared with the SCR group (Fig. 2A). This decrease was significant at
72 h (siHEB, 2.0±0.4; SCR, 3.4±0.8; P=0.008). In addition,
HEB silencing in combination with irradiation reduced U87MG
cell doubling (0.9±0.1) compared with the non-irradiated siHEB
group (2.0±0.4; P=0.013) at 72 h. An increase in cell death was
detected in irradiated compared with non-irradiated siHEB cells
(P=0.007); however, this increase was proportional to SCR
irradiated cells (P=0.005), at 48 h (Fig. 2B).
Cell cycle kinetics and sub-G1 cells were analyzed
by flow cytometry (Fig. 2C). A
significantly decreased proportion of HEB-silenced cells
were in G0/G1 phase (P=0.042) and a non-significantly increased
proportion was detected in the sub-G1 fraction at 24 h compared
with the SCR group. Irradiation of HEB siRNA-transfected
cells further decreased the percentage of cells in G0/G1 phase
(P=0.001) and non-significantly increased the percentage of sub-G1
cells at 72 h compared with sham-irradiated HEB-silenced
cells. In addition, a reduction in the proportion of G0/G1 cells
(P=0.01) and an increase in the sub-G1 population (P=0.007) were
observed in the irradiated SCR group at 72 h compared with the
non-irradiated SCR group.
Effects of HEB silencing on
neurosphere formation
HEB silencing significantly reduced the
neurosphere number compared with the SCR group (P=0.002; Fig. 3A); however, no significant
differences were observed in the number of cells (Fig. 3B) or the percentage of cell death
(Fig. 3C). Irradiation
significantly decreased cell number (P=0.006) and increased cell
death (P=0.005) independent of HEB silencing. The irradiated
HEB-silenced group demonstrated a non-significantly
decreased number of neurosphere cells compared with the irradiated
SCR group, but without a concurrent decrease in neurosphere number,
which was however reduced in the SCR irradiated group compared with
the respective control (P=0.003).
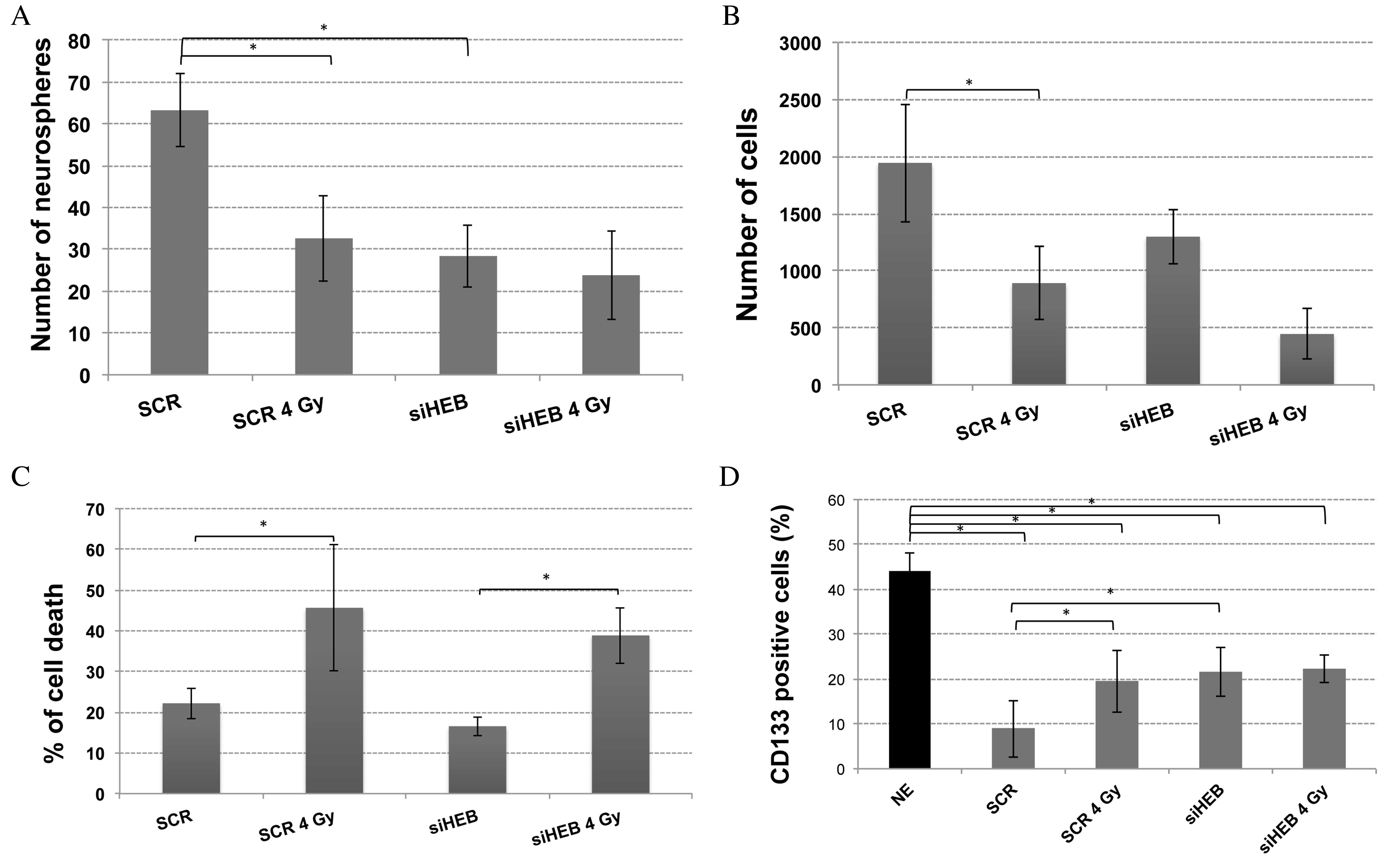 | Figure 3.Effects of HEB silencing on
neurospheres. U87MG neurospheres were transfected with SCR or
HEB siRNA and irradiated or sham-irradiated 24 h following
transfection. (A) Neurosphere number formed from 500 cells, at 6
days post-irradiation. The neurosphere number was decreased in
irradiated SCR cells, and irradiated and non-irradiated siHEB
cells, compared with non-irradiated SCR cells. (B) Number of cells
present in neurospheres, at 7 days post-irradiation. The number of
cells within neurospheres was decreased by irradiation. (C)
Percentage of dead cells, at 7 days post-irradiation. The
proportion of dead cells was increased by irradiation. (D)
Percentage of CD133+ cells, measured following 8 days
culture in differentiation medium, initiated 7 days following
irradiation, and compared with cells maintained in neurosphere
formation medium. The proportion of CD133+ cells was
decreased in all differentiation medium-cultured cells compared
with neurosphere medium-cultured cells, and further decreased in
SCR non-irradiated cells. *P<0.05. HEB, transcription factor 12;
siRNA, small interfering RNA; SCR, scrambled sequence; CD133,
cluster of differentiation 133. |
A high percentage of neurospheres cultured for 8
days expressed CD133 (44.1±4.1%; Fig.
3D). Neurospheres dissociated and cultured under
differentiation conditions demonstrated a significant reduction
(P<0.0001) in the percentage of CD133+ cells (SCR
transfected group, 8.9±6.4%). These results confirm the presence of
CD133+ stem cells in neurospheres, and indicate that the
differentiation medium reduced the proportion of CD133+
cells, which may be directly correlated with the differentiation of
stem cells.
Irradiation did not affect the CD133+
population in HEB-silenced cells, whereas a greater
percentage of irradiated SCR transfected cells were
CD133+ compared with non-irradiated SCR cells. A greater
proportion of siHEB-transfected cells were CD133+
(21.6±5.5%; P=0.0010) compared with the SCR group; however, this
remained almost 3 times lower than cells cultured under neurosphere
growing conditions (P<0.0001). These results indicate that
irradiation or HEB silencing inhibits or delays GBM cell
differentiation; however when combined, the effect is not additive
or synergistic.
Discussion
The present study investigated the effects of
HEB knockdown on the proliferation of GBM cells, and whether
HEB-silenced cells may be sensitized to irradiation. HEB
protein expression was analyzed in the nuclei of U87MG and
ACBRI-371 monolayers and neurosphere cells. Although significant
differences were not observed between the two cell lines, U87MG
cells demonstrated greater HEB protein expression levels compared
with astrocytes when cultured in monolayers. Consistent with these
results, HEB has been revealed to be transcriptionally
induced in oligodendroglioma and astrocytic glioma compared with
healthy brain tissue (36). A
previous study of 120 patients with CRC associated HEB
overexpression with metastasis and poorer survival, using
microarray data and validation (34). In addition, the same study observed
high levels of HEB in certain CRC cell lines.
In the present study, HEB silencing was
confirmed by western blotting 72 h following transfection, and
reached 97.8% inhibition compared with the SCR group. HEB
silencing induced a significant reduction in cell proliferation at
72 h, and a decrease of cells in the G0/G1 phase at 24 h. This
transient effect of HEB silencing at 24 h may be due to the
effect of different quantities of HEB protein at this time, as the
inhibition increased over time, at least at 48 and 72 h following
transfection. HEB protein may act via threshold levels, as for the
E2 factor family of transcription factors, in which one threshold
is associated with apoptosis, and a lower threshold is associated
with proliferation (38).
A previous study has reported that HEB
silencing in CRC cell lines resulted in antitumor effects, via
reduced migration, invasion and metastasis, through greater
cell-cell contact and gap-junction activity, and via increased
E-cadherin, connexin 26 and connexin 43, but decreased fibronectin
levels (34). Clinically,
HEB mRNA overexpression has been correlated with E-cadherin
mRNA downregulation in tumor tissues (34). Cadherins are integral membrane
proteins that mediate calcium-dependent cell-cell adhesion, and
they may be involved in the development and maintenance of tissues,
and the invasion and metastasis of malignant tumors (39). Therefore, cadherins are considered
as potential targets to reduce chemo- and radioresistance (40). Our previous study revealed an
upregulation of three cadherins (CDH8, CDH13 and
CD93) and one integrin (ITGA5) in mutated TP53
GBM cell lines compared with wild-type cells (6).
In the present study, the effects of HEB
silencing combined with irradiation were analyzed. Combined
treatment reduced the proliferative capacity (doubling levels) of
U87MG cells when cultured in monolayers; however, cell death was
not increased compared with the SCR irradiated group. It is known
that radiation exposure may not be manifested for several cell
divisions (41). It is therefore
possible that the decrease in proliferation due to HEB silencing
may palliate the effects of irradiation in siHEB cells.
Our previous study identified differentially
expressed genes in irradiated vs. sham-irradiated GBM cell lines,
obtained by microarray (7). By
performing in silico bioinformatics analysis, TFs associated
with those differentially expressed genes were identified; among
these TFs, HEB expression was greater in irradiated U87MG
cells compared with controls (23). Genes controlled by HEB
include neurofibromin 1 (NF1), a disintegrin and
metalloproteinase with thrombospondin type and distal-less homeobox
6 (associated with cell proliferation), and G protein-coupled
receptor 68, mannan binding lectin serine peptidase 2 and
NF1 (involved in wound healing); these were upregulated in
U87MG cells following irradiation. These results are compatible
with those obtained in the present study with HEB silencing,
in which U87MG proliferation was decreased following
irradiation.
The present study revealed greater HEB protein
expression levels in U87MG neurospheres compared with astrocyte
neurospheres. The number of neurospheres formed from
HEB-silenced cells was significantly reduced compared with
the SCR group, whereas there was a non-significant reduction in the
number of cells present within siHEB neurospheres compared with the
SCR group. The reduction of HEB protein expression may alter the
maintenance of U87MG stem cells, possibly via a decrease in
differentiation, thus decreasing the number of neurospheres. HEB
expression in GBM stem cells may have similar proliferative roles
in neural stem cells and progenitor cells (15), supporting the relevance of this TF
in GBM stem cell maintenance.
In the present study, irradiation did not decrease
the number of siHEB neurospheres; however, it did increase cell
death in HEB-silenced cells, and a similar effect was
observed in the irradiated SCR group. Although the number of
neurospheres was unaffected by irradiation, there was a
non-significant reduction in the number of cells within irradiated
HEB-silenced neurospheres, compared with the non-irradiated
siHEB group; in addition, the number of cells was decreased
non-significantly compared with irradiated SCR cells. These results
indicated that siHEB may have a minor influence on the
radioresistance of U87MG GBM cells.
CD133 expression was evaluated 8 days following the
dissociation of neurospheres and was greater in siHEB compared with
SCR cells, indicating a possible decrease in cell differentiation.
These results are in accordance with the literature, as HEB has
been associated with differentiation in diverse cells, including
oligodendrocytes (42). Although
HEB appears to be important in GBM neurosphere
maintenance/formation, siHEB cells exhibited a decrease in
differentiation potential when cells were submitted to a
differentiation stimulus.
Promoting the differentiation of CSCs, thus reducing
tumor growth, is a novel approach to cancer therapy (43). Previous studies have described how
CG500354 (44), a short hairpin
RNA for ubiquitin ligases (45)
and all-trans retinoic acid (46),
may induce stem cell differentiation in GBM. However, in these
studies treatments were performed in neurosphere formation medium,
and stem cell and differentiation markers were analyzed days
following treatment, whereas in the present study, CD133 expression
was assessed following dissociation of neurospheres 7 days
subsequent to treatment, and a further 8 days of culture. The high
proportion of CD133+ cells in the siHEB-silenced group
compared with the SCR group suggested that reduced differentiation
may be a side-effect of HEB silencing. Although under the
differentiation therapy approach, it is desirable to increase
differentiation of tumor stem cells, the primary aim is to reduce
tumor proliferation, which was observed in siHEB cells. These
findings indicated that HEB is a potential target in GBM
treatment. Regarding irradiated cells, the proportion of
CD133+ cells was greater in the HEB-silenced
group compared with the SCR group, independent of irradiation,
indicating that HEB silencing, irradiation or combination
therapy caused similar effects on the differentiation capacity of
cells.
In conclusion, the results of the present study
demonstrated that HEB may be involved in GBM cell
proliferation, as HEB silencing reduced proliferation in
cells cultured as monolayers or neurospheres. Furthermore, the
results suggested a potential role for HEB in the
maintenance of GBM stem cells, as HEB silencing affected the
differentiation capacity of cells. However, only limited effects
were exerted by irradiation, primarily on neurosphere cell number.
HEB may be a potential target to decrease proliferation in
U87MG GBM cells, grown as monolayers or neurospheres, and may
provide important information for the development of novel
strategies for cancer therapy.
Acknowledgements
The present study was supported by the Fundação de
Amparo à Pesquisa do Estado de São Paulo (grant no. 2009/10925-6).
P.R.D.V.G. was supported by a fellowship from Coordenação de
Aperfeiçoamento de Pessoal de Nível Superior and Conselho Nacional
de Desenvolvimento Científico e Tecnológico. The authors thank Luiz
A. Costa Junior and Sueli A. Neves (Department of Genetics,
Ribeirão Preto Medical School, University of São Paulo, Ribeirão
Preto, Brazil) for technical assistance, Leonardo L. do Amaral
(Radiotherapy service, Clinical Hospital of Ribeirão Preto,
University of São Paulo) for irradiation procedures and Danilo
Jordão Xavier (Department of Genetics, Ribeirão Preto Medical
School, University of São Paulo) for graphical support.
References
|
1
|
Ohgaki H and Kleihues P: Population-based
studies on incidence, survival rates, and genetic alterations in
astrocytic and oligodendroglial gliomas. J Neuropathol Exp Neurol.
64:479–489. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Stupp R, Mason WP, van den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al: Radiotherapy plus concomitant and adjuvant temozolomide
for glioblastoma. N Engl J Med. 352:987–996. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen J, McKay RM and Parada LF: Malignant
glioma: Lessons from genomics, mouse models, and stem cells. Cell.
149:36–47. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ohgaki H: Genetic pathways to
glioblastomas. Neuropathology. 25:1–7. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
da Silva GN, Evangelista AF, Magalhães DA,
Macedo C, Búfalo MC, Sakamoto-Hojo ET, Passos GA and Salvadori DM:
Expression of genes related to apoptosis, cell cycle and signaling
pathways are independent of TP53 status in urinary bladder cancer
cells. Mol Biol Rep. 38:4159–4170. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Godoy PRDV, Mello SS, Magalhães DAR,
Donaires FS, Montaldi APL, Nicolucci P, Donadi EA, Passos GAS and
Sakamoto-Hojo ET: Portrait of transcriptional expression profiles
displayed by different glioblastoma cell linesMolecular Targets of
CNS Tumors. Garami M: 1. InTech; Rijeka: pp. 265–288. 2011
|
|
7
|
Godoy PR, Mello SS, Magalhães DA, Donaires
FS, Nicolucci P, Donadi EA, Passos GA and Sakamoto-Hojo ET:
Ionizing radiation-induced gene expression changes in TP53
proficient and deficient glioblastoma cell lines. Mutat Res.
756:46–55. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Otomo T, Hishii M, Arai H, Sato K and
Sasai K: Microarray analysis of temporal gene responses to ionizing
radiation in two glioblastoma cell lines: Up-regulation of DNA
repair genes. J Radiat Res. 45:53–60. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Eyler CE and Rich JN: Survival of the
fittest: Cancer stem cells in therapeutic resistance and
angiogenesis. J Clin Oncol. 26:2839–2845. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu Q, Nguyen DH, Dong Q, Shitaku P, Chung
K, Liu OY, Tso JL, Liu JY, Konkankit V, Cloughesy TF, et al:
Molecular properties of CD133+ glioblastoma stem cells derived from
treatment-refractory recurrent brain tumors. J Neurooncol. 94:1–19.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bao S, Wu Q, McLendon RE, Hao Y, Shi Q,
Hjelmeland AB, Dewhirst MW, Bigner DD and Rich JN: Glioma stem
cells promote radioresistance by preferential activation of the DNA
damage response. Nature. 444:756–760. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Singh SK, Clarke ID, Terasaki M, Bonn VE,
Hawkins C, Squire J and Dirks PB: Identification of a cancer stem
cell in human brain tumors. Cancer Res. 63:5821–5828.
2003.PubMed/NCBI
|
|
13
|
Singh SK, Hawkins C, Clarke ID, Squire JA,
Bayani J, Hide T, Henkelman RM, Cusimano MD and Dirks PB:
Identification of human brain tumour initiating cells. Nature.
432:396–401. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rappa G, Mercapide J, Anzanello F,
Prasmickaite L, Xi Y, Ju J, Fodstad O and Lorico A: Growth of
cancer cell lines under stem cell-like conditions has the potential
to unveil therapeutic targets. Exp Cell Res. 314:2110–2122. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Uittenbogaard M and Chiaramello A:
Expression of the bHLH transcription factor Tcf12 (ME1) gene is
linked to the expansion of precursor cell populations during
neurogenesis. Brain Res Gene Expr Patterns. 1:115–121. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu YY, Gao P, Sun Y and Duan YR:
Development of targeted therapies in treatment of glioblastoma.
Cancer Biol Med. 12:223–237. 2015.PubMed/NCBI
|
|
17
|
Carminati PO, Donaires FS, Godoy PRDV,
Montaldi AP, Meador JA, Balajee AS, Passos GA and Sakamoto-Hojo ET:
DNA-PK is a potential molecular therapeutic target for
glioblastomaEvolution of the Molecular Biology of Brain Tumors and
the Therapeutic Implications. Lichtor T: InTech; Rijeka: pp.
459–480. 2013
|
|
18
|
Montaldi AP and Sakamoto-Hojo ET:
Methoxyamine sensitizes the resistant glioblastoma T98G cell line
to the alkylating agent temozolomide. Clin Exp Med. 13:279–288.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Montaldi AP, Godoy PR and Sakamoto-Hojo
ET: APE1/REF-1 down-regulation enhances the cytotoxic effects of
temozolomide in a resistant glioblastoma cell line. Mutat Res Genet
Toxicol Environ Mutagen. 793:19–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mees C, Nemunaitis J and Senzer N:
Transcription factors: Their potential as targets for an
individualized therapeutic approach to cancer. Cancer Gene Ther.
16:103–112. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sankpal UT, Goodison S, Abdelrahim M and
Basha R: Targeting Sp1 transcription factors in prostate cancer
therapy. Med Chem. 7:518–525. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu LY, Chang LY, Kuo WH, Hwa HL, Shyu MK,
Chang KJ and Hsieh FJ: In silico prediction for regulation of
transcription factors ontheir shared target genes indicates
relevant clinical implications in a breast cancer population.
Cancer Inform. 11:113–137. 2012.PubMed/NCBI
|
|
23
|
Godoy PRDV, Mello SS, Donaires FS, Donadi
EA, Passos GAS and Sakamoto-Hojo ET: In silico analysis of
transcription factors associated to differentially expressed genes
in irradiated glioblastoma cell linesEvolution of the Molecular
Biology of Brain Tumors and the Therapeutic Implications. Lichtor
T: InTech; Rijeka: pp. 577–600. 2013
|
|
24
|
Zhang Y, Babin J, Feldhaus AL, Singh H,
Sharp PA and Bina M: HTF4: A new human helix-loop-helix protein.
Nucleic Acids Res. 19:45551991. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Massari ME and Murre C: Helix-loop-helix
proteins: Regulators of transcription in eucaryotic organisms. Mol
Cell Biol. 20:429–440. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sawada S and Littman DR: A heterodimer of
HEB and an E12-related protein interacts with the CD4 enhancer and
regulates its activity in T-cell lines. Mol Cell Biol.
13:5620–5628. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang D, Claus CL, Vaccarelli G, Braunstein
M, Schmitt TM, Zúñiga-Pflücker JC, Rothenberg EV and Anderson MK:
The basic helix-loop-helix transcription factor HEBAlt is expressed
in pro-T cells and enhances the generation of T cell precursors. J
Immunol. 177:109–119. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hu JS, Olson EN and Kingston RE: HEB, a
helix-loop-helix protein related to E2A and ITF2 that can modulate
the DNA-binding ability of myogenic regulatory factors. Mol Cell
Biol. 12:1031–1042. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Conway K, Pin C, Kiernan JA and Merrifield
P: The E protein HEB is preferentially expressed in developing
muscle. Differentiation. 72:327–340. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li Y, Yang J, Luo JH, Dedhar S and Liu Y:
Tubular epithelial cell dedifferentiation is driven by the
helix-loop-helix transcriptional inhibitor Id1. J Am Soc Nephrol.
18:449–460. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
O'Neil J, Shank J, Cusson N, Murre C and
Kelliher M: TAL1/SCL induces leukemia by inhibiting the
transcriptional activity of E47/HEB. Cancer Cell. 5:587–596. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Draheim KM, Hermance N, Yang Y, Arous E,
Calvo J and Kelliher MA: A DNA-binding mutant of TAL1 cooperates
with LMO2 to cause T cell leukemia in mice. Oncogene. 30:1252–1260.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Labreche K, Simeonova I, Kamoun A, Gleize
V, Chubb D, Letouzé E, Riazalhosseini Y, Dobbins SE, Elarouci N,
Ducray F, et al: TCF12 is mutated in anaplastic oligodendroglioma.
Nat Commun. 6:72072015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lee CC, Chen WS, Chen CC, Chen LL, Lin YS,
Fan CS and Huang TS: TCF12 protein functions as transcriptional
repressor of E-cadherin, and its overexpression is correlated with
metastasis of colorectal cancer. J Biol Chem. 287:2798–2809. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen WS, Chen CC, Chen LL, Lee CC and
Huang TS: Secreted heat shock protein 90α (HSP90α) induces nuclear
factor-κB-mediated TCF12 protein expression to down-regulate
E-cadherin and to enhance colorectal cancer cell migration and
invasion. J Biol Chem. 288:9001–9010. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Riemenschneider MJ, Koy TH and
Reifenberger G: Expression of oligodendrocyte lineage genes in
oligodendroglial and astrocytic gliomas. Acta Neuropathol.
107:277–282. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ajeawung N and Deepak Kamnasaran D: An
efficient approach to Enrich Glioma stem cells from Glioma cell
lines in culture. WebmedCentral ONCOLOGY. 1:WMC005572010.
|
|
38
|
Trimarchi JM and Lees JA: Sibling rivalry
in the E2F family. Nat Rev Mol Cell Biol. 3:11–20. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
van Roy F and Berx G: The cell-cell
adhesion molecule E-cadherin. Cell Mol Life Sci. 65:3756–3788.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kraus AC, Ferber I, Bachmann SO, Specht H,
Wimmel A, Gross MW, Schlegel J, Suske G and Schuermann M: In vitro
chemo- and radio-resistance in small cell lung cancer correlates
with cell adhesion and constitutive activation of AKT and MAP
kinase pathways. Oncogene. 21:8683–8695. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hall EJ and Giaccia AJ: Radiobiology for
the Radiologist. 5th. Lippincott Williams & Wilkins;
Philadelphia, PA: pp. 5882000
|
|
42
|
Sussman CR, Davies JE and Miller RH:
Extracellular and intracellular regulation of oligodendrocyte
development: Roles of Sonic hedgehog and expression of E proteins.
Glia. 40:55–64. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Khan IS and Ehtesham M: Targeting
glioblastoma cancer stem cells: The next great hope? Neurosurgical
focus. 37:E72014. View Article : Google Scholar
|
|
44
|
Kang TW, Choi SW, Yang SR, Shin TH, Kim
HS, Yu KR, Hong IS, Ro S, Cho JM and Kang KS: Growth arrest and
forced differentiation of human primary glioblastoma multiforme by
a novel small molecule. Sci Rep. 4:55462014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Low J, Blosser W, Dowless M, Ricci-Vitiani
L, Pallini R, de Maria R and Stancato L: Knockdown of ubiquitin
ligases in glioblastoma cancer stem cells leads to cell death and
differentiation. J Biomol Screen. 17:152–162. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Friedman MD, Jeevan DS, Tobias M, Murali R
and Jhanwar-Uniyal M: Targeting cancer stem cells in glioblastoma
multiforme using mTOR inhibitors and the differentiating agent
all-trans retinoic acid. Oncol Rep. 30:1645–1650. 2013.PubMed/NCBI
|