Introduction
Hepatocellular carcinoma (HCC) is currently the
fifth most common solid tumor worldwide and the second leading
cause of cancer-related deaths in China (1,2).
Although marked progress has been made in recent decades, the
details of the molecular mechanisms underlying HCC carcinogenesis
remain to be elucidated (3).
Survival of patients with HCC has shown improvement with
advancements in surgical techniques. However, the median 5-year
survival rate remains at ∼50% (4).
Cancer classification using biomarkers may
effectively define risk of recurrence, which allows for the use of
appropriate treatments in order to obtain a better prognosis
(5). However, few measurable
biomarkers for predicting HCC recurrence have been identified thus
far. Recently, efforts to examine gene methylation by utilizing
genome-wide techniques have revealed that a large number of genes
exhibit aberrant DNA methylation profiles in cancer (6). These changes can be used to stratify
various subtypes of cancer and predict cancer outcomes (7,8).
DNA-methyltransferase (DNMT) 3A plays a crucial role in embryonic
development and aberrant DNA methylation in carcinogenesis.
Polymorphisms of the DNMT3A gene may regulate gene
expression, affect its enzymatic activity and potentially
contribute to susceptibility to cancer. In a previous study, we
found a novel DNMT3A functional single-nucleotide polymorphism
(SNP), −448A>G (rs1550117) and demonstrated that a DNMT3A
promoter genetic variant increased its transcription activity and
contributed to the genetic susceptibility to gastric cancer (GC)
and colorectal cancer (CRC) in a Chinese population (9,10).
However, an association between the DNMT3A −448A>G
polymorphism and HCC remains to be elucidated. This study was
performed to assess the association between rs1550117 in the
DNMT3A promoter and the risk for HCC in a Chinese Han
population.
Materials and methods
Study population and clinical
samples
A total of 108 HCC cases and 225 controls were
recruited in this case-control study (Table I). Cases and controls were matched
by age, gender and were selected from the same hospital. Thirteen
fresh HCC tissues and paired adjacent non-cancer tissue samples
were randomly selected from patients undergoing hepatectomy from
the First Affiliated Hospital of Nanjing Medical University. The
samples were obtained following written consent and analyzed
anonymously. This study was performed with the approval of the
Medical Ethics Committee of the Medical School of Southeast
University Key Laboratory of Developmental Genes and Human
Diseases, Ministry of Education (Jiangsu, China). Each sample was
frozen and stored at −80°C. Paired non-cancerous tissues were
isolated from at least 2 cm away from the tumor border.
 | Table I.Characteristics of the study
population. |
Table I.
Characteristics of the study
population.
| HCC cases
(n=108) | Controls (n=225) | P-value |
|---|
| Variable | no. (%) | no. (%) | |
|---|
| Age, years | | | 0.239 |
| <50 | 34 (31.48) | 57 (25.33) | |
| ≥50 | 74 (68.52) | 168 (74.67) | |
| Gender | | | 0.062 |
| Male | 85 (78.70) | 155 (68.89) | |
| Female | 23 (21.30) | 70 (31.11) | |
Real-time quantitive PCR (qPCR)
Total RNA was extracted using TRIzol reagent
(Invitrogen, NY, USA). RNA (1 μg) was used to synthesize
cDNA using the Reverse Transcription System (Promega, Madison, WI,
USA). qPCR was performed using the SYBR®Premix Ex
Taq™ (Perfect Real Time; Takara., Otsu, Japan) in an ABI 7300
detection system (Applied Biosystems, Grand Island, NY, USA)
according to the manufacturer's instructions. The primers used for
the PCR amplifications were: forward: 5′-TATTGATGAGCGCACAAGAGAGC-3′
and reverse: 5′-GGGTGTTCCAGGGTAACATTGAG-3′. For the reactions,
annealing was carried out at 65°C, and 40 cycles of amplification
were performed. Data were normalized using β-actin as a
reference gene (forward primer: 5′-AAAGACCTGTACGCCAACAC-3′ and
reverse primer: 5′-GTCATACTCCTGCTTGCTGAT-3′). The relative levels
of expression of the target gene among the different samples were
calculated accordingly (ABI PRISM 7300 Detection System, USA).
DNA extraction
Venous blood samples (5 ml) were collected in EDTA
vacuum tubes from HCC patients and control subjects. Genomic DNA
was extracted from white blood cells within a week after sample
collection by proteinase K digestion as previously described
(11).
DNMT3A genotyping
The DNMT3A −448A>G SNP was analyzed by
polymerase chain reaction-restriction fragment length polymorphisms
(PCR-RFLPs). PCR was performed in a 25 μl volume containing
100 ng of genomic DNA, 2.5 μl of 10X PCR buffer, 2.0 mM
MgCl2, 0.1 mM dNTPs (mixture of dATP, dTTP, dCTP and
dGTP), 10 pmol of each primer (forward: 5′-ACACACCGCCCTCACCCCTT-3′
and reverse: 5′-TCCAGCAATCCCTGCCCACA-3′), 1 unit of Taq DNA
polymerase (Biocolor BioScience and Technology Co., Shanghai,
China). Amplification was performed as follows: an initial
denaturation at 94°C for 5 min followed by 32 cycles at 94°C for 30
sec, 66°C for 30 sec, 72°C for 30 sec, and a final extension at
72°C for 10 min. A 10 μl aliquot of PCR product was then
digested with TaaI (Fermentas Co., Glen Burnie, MD, USA) at
65°C for 5 min. Restriction fragments were then separated on a 3.0%
agarose gel stained with ethidium bromide (EB) and visualized under
UV light. The wild-type G allele had a TaaI restriction site
that resulted in 3 bands (153, 94 and 87 bp), while the variant A
allele resulted in 4 bands (247, 153, 94 and 87 bp). Genotyping
quality was evaluated by the repeated genotyping of 10% randomly
selected samples. To confirm the genotyping results, PCR-amplified
DNA samples were selected and examined by DNA sequencing.
Statistical analysis
Data were analyzed with SPSS version 13.0 (SPSS
Inc., Chicago, IL, USA). Patients and controls were compared using
the Student's t-test for continuous variables and the Chi-square
(χ2) test for categorical variables. Allele and genotype
frequencies between control and HCC subjects were obtained using
the Chi-square (χ2) test, and the standard
goodness-of-fit test was used to test the Hardy-Weinberg
equilibrium. P<0.05 was considered statistically
significant.
Results
Previously, we revealed the depletion of DNMT3A
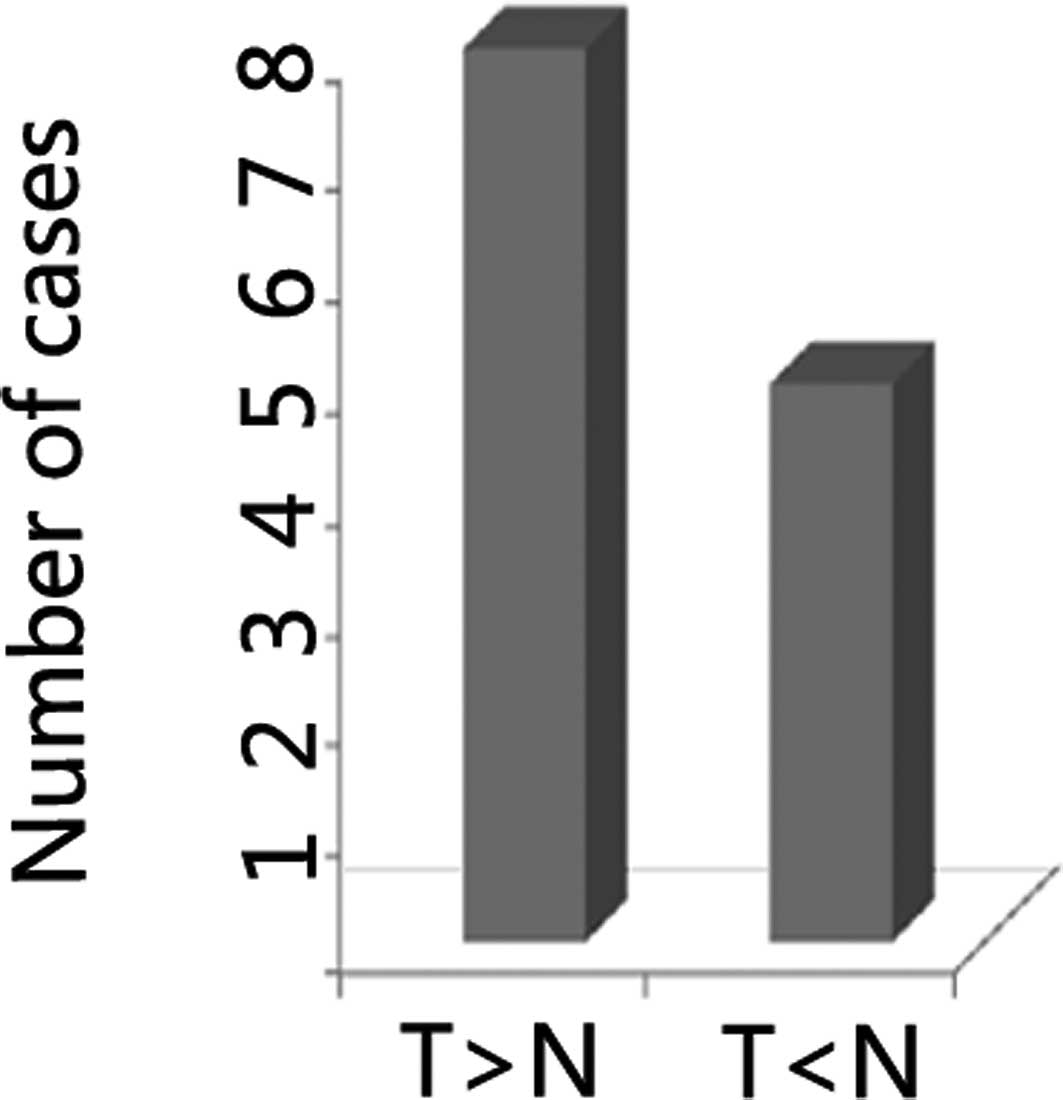
suppressed cell proliferation in HCC cells (1). In the present study, we found that the
frequency of overexpression of DNMT3A was 62% in HCC, which
was higher than that in the non-cancer cases (Fig. 1). These data suggested that DNMT3A
is involved in the development of carcinogenesis. Recently, we
found that a functional SNP of DNMT3A −448A>G polymorphism
affected the promoter activity of DNMT3A. Thus, we hypothesized
that DNMT3A promoter SNP (rs1550117) variants may have an effect on
DNMT3A expression in HCC.
We genotyped the DNMT3A promoter −448A>G
polymorphisms in 108 HCC patients and 225 controls by PCR-RFLP. The
genotyping was confirmed by DNA sequencing, and the results of the
PCR-RFLP genotyping and sequencing analysis were 100% concordant.
The genotypic and allelic frequencies of DNMT3A −448A>G
are summarized in Tables I and
II. The distributions of −448A>G
genotypes in the HCC were AA, 3.70%; AG, 40.74%; GG, 55.56%. The A
allele frequency was 24.07%, while the frequency for the controls
was 5.33, 37.78, 56.89%, respectively, and 24.22% for the A allele.
No significant differences were observed in the genotypic and
allelic frequencies in the two groups. HCC was then stratified by
age and gender. No significantly different frequencies of
−448A>G were observed in HCC patients (Table III).
 | Table II.DNMT3A −448A>G genotype and allele
frequency among cases and controls and association with HCC. |
Table II.
DNMT3A −448A>G genotype and allele
frequency among cases and controls and association with HCC.
| Genotype/allele | HCC (n=108)
No. (%) | Control subjects
(n=225)
No. (%) | Crude OR (95%
CI) | P-valuea |
|---|
| Genotype | | | | |
| AA | 4 (3.70) | 12 (5.33) | 1 | |
| AG | 44 (40.74) | 85 (37.78) | 1.553
(0.473–5.098) | 0.465 |
| GG | 60 (55.56) | 128 (56.89) | 1.406
(0.435–4.542) | 0.567 |
| Allele | | | | |
| A | 52 (24.07) | 109 (24.22) | 1 | |
| G | 164 (75.93) | 341 (75.78) | 1.008
(0.690–1.473) | 0.967 |
 | Table III.DNMT3A −448 A>G genotypes and
allele frequencies in HCC cases. |
Table III.
DNMT3A −448 A>G genotypes and
allele frequencies in HCC cases.
| Group | Genotype
| Allele
| P-valuea |
|---|
AA
No. (%) | AG
No. (%) | GG
No. (%) | G
% |
|---|
| Total | 4 (3.70) | 44 (40.74) | 60 (55.56) | 75.93 | |
| Age, years | | | | | 0.248b |
| <50 | 2 (1.85) | 9 (8.33) | 23 (21.30) | 80.88 | |
| ≥50 | 2 (1.85) | 35 (32.41) | 37 (34.26) | 73.65 | |
| Gender | | | | | 0.420c |
| Male | 4 (3.70) | 35 (32.41) | 46 (42.60) | 74.71 | |
| Female | 0 (0.00) | 9 (8.33) | 14 (12.96) | 80.43 | |
The DNMT3A −448A>G polymorphism was
evaluated in relation to the risk of HCC in this case-control
study. The HCC patients and control subjects were first grouped by
gender (male and female), and then by age (<50 and ≥50 years).
The genotype and allele frequencies were evaluated, the OR and
their 95% confidence intervals (CIs) were calculated using the more
common homozygous variant genotype as the reference group. No
differences between the HCC patients and control subjects were
observed (Table IV).
 | Table IV.Distribution of 448 A >G DNMT3A
genotypes and associated OR in relation to age and gender in HCC
cases. |
Table IV.
Distribution of 448 A >G DNMT3A
genotypes and associated OR in relation to age and gender in HCC
cases.
|
Characteristics | Genotype | HCC cases
No. (%) | Controls
No. (%) | OR (95% CI) | P-value |
|---|
| Male | AA | 4 (3.70) | 11 (4.89) | 1 | |
| AG + GG | 81 (75.00) | 144 (64.00) | 1.547
(0.477–5.016) | 0.464 |
| Age, years | | | | | |
| <50 | AA | 2 (1.85) | 5 (2.22) | 1 | |
| AG + GG | 26 (24.07) | 41 (18.22) | 1.585
(0.286–8.782) | 0.595 |
| ≥50 | AA | 2 (1.85) | 6 (2.67) | 1 | |
| AG + GG | 45 (41.67) | 103 (45.78) | 1.311
(0.255–6.744) | 0.746 |
| Female | AA | 0 (0.00) | 1 (0.44) | 1 | |
| AG + GG | 23 (21.30) | 69 (30.67) | 0.750
(0.667–0.844) | 1 |
| Age, years | | | | | |
| <50 | AA | 0 (0.00) | 0 (0.00) | | |
| AG + GG | 6 (5.56) | 11 (4.89) | - | - |
| ≥50 | AA | 0 (0.00) | 1 (0.44) | | |
| AG + GG | 17 (15.74) | 58 (25.78) | 0.773
(0.684–0.874) | 1 |
Discussion
HCC is one of the most common malignant tumors, with
a poor survival rate. It is particularly prevalent in China and
Asia. While surgery is the most effective treatment for liver
tumor, ∼80% of HCC patients are inoperable at presentation and
succumb to the disease early due to late diagnosis (12). The development of biomarkers for the
early diagnosis and accurate prognosis of HCC is crucial for
improving patient survival. Various tissue and serum potential
biomarkers including AFP-L3, DCP, AFP, and hepatocyte growth factor
(HGF) are involved in HCC (13).
In mammals, DNA methylation is essential for
embryonic development and is involved in gene expression, genomic
imprinting, X chromosome inactivation, and maintenance of genome
integrity. Aberrant changes of genomic methylation patterns or
abnormal interpretation of the DNA methylation signals are
associated with several human disorders, most notably
immunodeficiency, centromeric instability, facial anomalies
syndrome, Rett syndrome and cancer (14). The expression of these DNMTs is
significantly elevated in various types of cancer including breast,
colon, endometrium, prostate, stomach and in uterine leiomyomata
(15). In a previous study, DNMT3A
expression was increased in 3/6 HCC cell lines and 16/25 (64%) HCC
tissues, suggesting that DNMT3A is involved in hepatocellular
carcinogenesis (16). Abnormal DNA
methylation can lead to tumor suppressor gene silencing by DNA
methylation on CpG islands in their promoter regions in cancer
cells, and thus the overall level of DNA methylation is higher in
cancer cells compared with normal cells (17,18).
SNPs of DNMTs are important indicators of genetic susceptibility to
cancer development. Therefore, genetic polymorphism assays have
been used to investigate the aetiology of malignant diseases
(19).
Little is known about the association of DNMTs and
the genetic susceptibility to HCC. Therefore, we investigated the
effect of DNMT3A polymorphisms −448A>G and risk of HCC in
a hospital-based case-control study in a Chinese population.
Overexpression of DNMT3A was detected in HCC cases at the
transcription level. The functional SNP −448A>G of DNMT3A
was evaluated in HCC patients and healthy controls as a
case-control study. The results suggest that the DNMT3A
448A>G polymorphism was not associated with the risk of HCC in
the study population. This finding was different from those of
previous studies (11) possibly due
to the genetic background of HCC being different from that of GC
and CRC. In addition, DNMT3A has four alternatively spliced
variants, and different isoforms of the same enzyme may have
altered catalytic activity or target-site specificity that may play
different roles in carcinogenesis (20). However, additional studies with
larger sample sizes and different populations are required to
confirm findings of the present study.
In conclusion, to the best of our knowledge, this is
the first report to investigate the association of an SNP in DNMT3A
with genetic susceptibility to HCC. This finding may provide
valuable insight into hepacellular carcinogenesis, although the
result that DNMT3A −448A>G SNP is associated with the
susceptibility to HCC should be investigated.
Abbreviations:
|
PCR
|
polymerase chain reaction
|
|
RFLP
|
restriction fragment length
polymorphism
|
|
OR
|
odds ratio
|
|
CI
|
confidence interval
|
|
HCC
|
hepatocellular carcinoma
|
|
SNP
|
single-nucleotide polymorphism
|
Acknowledgements
This study was supported by The
National Natural Science Foundation of China, nos. 81171915 and
91229107.
References
|
1.
|
Thomas MB, Jaffe D, Choti MM, et al:
Hepatocellular carcinoma: consensus recommendations of the National
Cancer Institute Clinical Trials Planning Meeting. J Clin Oncol.
28:3994–4005. 2010. View Article : Google Scholar
|
|
2.
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: the next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Takayama T: Surgical treatment for
hepatocellular carcinoma. Jpn J Clin Oncol. 41:447–454. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Fernandez M, Semela D, Bruix J, Colle I,
Pinzani M and Bosch J: Angiogenesis in liver disease. J Hepatol.
50:604–620. 2009. View Article : Google Scholar
|
|
6.
|
Figueroa ME, Lugthart S, Li Y, et al: DNA
methylation signatures identify biologically distinct subtypes in
acute myeloid leukemia. Cancer Cell. 17:13–27. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Noushmehr H, Weisenberger DJ, Diefes K, et
al: Identification of a CpG island methylator phenotype that
defines a distinct subgroup of glioma. Cancer Cell. 17:510–522.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Portela A and Esteller M: Epigenetic
modifications and human disease. Nat Biotechnol. 28:1057–1068.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Fan H, Liu D, Qiu X, et al: A functional
polymorphism in the DNA methyltransferase-3A promoter modifies the
susceptibility in gastric cancer but not in esophageal carcinoma.
BMC Med. 8:122010. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Zhao Z, Li C, Song Y, Wu Q, Qiao F and Fan
H: Association of the DNMT3A −448A>G polymorphism with genetic
susceptibility to colorectal cancer. Oncol Lett. 3:450–454.
2012.
|
|
11.
|
Miller SA, Dykes DD and Polesky HF: A
simple salting out procedure for extracting DNA from human
nucleated cells. Nucleic Acids Res. 16:12151988. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Sun S, Xu MZ, Poon RT, Day PJ and Luk JM:
Circulating Lamin B1 (LMNB1) biomarker detects early stages of
liver cancer in patients. J Proteome Res. 9:70–78. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Behne T and Copur MS: Biomarkers for
hepatocellular carcinoma. Int J Hepatol. 8590762012.
|
|
14.
|
Chen T and Li E: Structure and function of
eukaryotic DNA methyltransferases. Curr Top Dev Biol. 60:55–89.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Kurkjian C, Kummar S and Murgo AJ: DNA
methylation: its role in cancer development and therapy. Curr Probl
Cancer. 32:187–235. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Zhao Z, Wu Q, Cheng J, Qiu X, Zhang J and
Fan H: Depletion of DNMT3A suppressed cell proliferation and
restored PTEN in hepatocellular carcinoma cell. J Biomed
Biotechnol. 7375352010.PubMed/NCBI
|
|
17.
|
Paz MF, Fraga MF, Avila S, et al: A
systematic profile of DNA methylation in human cancer cell lines.
Cancer Res. 63:1114–1121. 2003.PubMed/NCBI
|
|
18.
|
Egger G, Liang G, Aparicio A and Jones PA:
Epigenetics in human disease and prospects for epigenetic therapy.
Nature. 429:457–463. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Wjst M: Target SNP selection in complex
disease association studies. BMC Bioinformatics. 5:922004.
View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Robertson KD, Uzvolgyi E, Liang G, et al:
The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate
mRNA expression in normal tissues and overexpression in tumors.
Nucleic Acids Res. 27:2291–2298. 1999. View Article : Google Scholar : PubMed/NCBI
|