Introduction
The Wilms' tumor 1 (WT1) gene has been extensively
used in monitoring minimal residual disease (MRD) in acute myeloid
leukemia (AML) as a panleukemic marker, due to its overexpression
(1–3), particularly in cases lacking known
fusion genes (4). The WT1 gene,
which is considered to be an oncogene, is mainly involved in
promoting stem cell proliferation and hampering cell
differentiation in AML (5,6). However, the number of studies
investigating WT1 mutations has been on the increase, although
their prognostic significance in predicting the outcome in AML has
been controversial (7–11). WT1 mutations are encountered in ~10%
of AML patients and the resulting defect affects the interaction of
WT1 with other transcription factors, which may contribute to
leukemogenesis and resistance to chemotherapy (7). The hotspots of WT1 mutations are
mainly located in exon 7 (85.41–87.06%) (9,10),
with frameshift mutations leading to the formation of a premature
stop codon and a truncated protein lacking the C-terminal zinc
fingers. Mutations in exon 9 (7.06–8.33%) (9,10) are
rare and predominantly of the missense type, which interrupt DNA
binding capacity by affecting the amino acid residues directly
involved in DNA binding or essential to the structure of the zinc
finger motif (12).
In this study, we screened the rate of WT1 gene
mutations in exon 7 and analyzed their effect on pediatric AML.
Furthermore, we used the software [ExPASy Translate Tool
(http://www.expasy.ch/tools/dna.html)]
for homology modeling and optimization of molecular dynamics to
evaluate the spatial configuration of WT1 with frameshift mutations
in exon 7.
Homology modeling, also referred to as comparative
or knowledge-based modeling, develops a three-dimensional model
from a protein sequence based on the structures of homologous
proteins. Evolutionarily related proteins have similar sequences
and naturally occurring homologous proteins have similar
structures. It has been demonstrated that the three-dimensional
protein structure is evolutionarily more conserved than would be
expected due to sequence conservation (13). By utilizing this method, we
demonstrated that WT1 frameshift mutations in exon 7 affected the
spatial configuration of WT1.
Materials and methods
Patients and samples
Cryopreserved bone marrow samples collected at
diagnosis from 60 patients with newly diagnosed AML were provided
by the Children's Hospital of Soochow University. The diagnosis and
classification of AML were based on morphological, cytogenetic and
immunophenotypic criteria according to the WHO classification.
Children with AML were treated according to the protocol for
Chinese AML children, as determined by the Subspecialty Group of
Hematology, Society of Pediatrics, Chinese Medical Association
(14). More detailed information is
provided in Table I.
 | Table IClinical and genetic characteristics
of the 60 children with newly diagnostic acute myeloid
leukemia. |
Table I
Clinical and genetic characteristics
of the 60 children with newly diagnostic acute myeloid
leukemia.
| FAB subtype | Cases | Gender (%
female) | Median age
(years) | Median WBC
(×109/l) | Karyotype (%
normal) |
|---|
| M1 | 2 | 100 | 4 | 19.9 | 50 |
| M2 | 12 | 56 | 6 | 8.7 | 33 |
| M3 | 21 | 53 | 8 | 11.7 | 10 |
| M4 | 13 | 22 | 8.5 | 31.2 | 25 |
| M5 | 12 | 55 | 11 | 38.6 | 38 |
The study was approved by the ethics committee of
the hospital and informed consent was provided by the parents or
the legal guardians of the patients. The procedures were approved
by the hospital's Institutional Review Board.
Polymerase chain reaction (PCR)
For mutation analysis of the exon 7 of the WT1 gene,
PCR amplification was performed with the use of specific primers
(15): 7F: 5′-CTCCAG
TGCTCACTCTCCCTC-3′; 7R: 5′-CCTTAGCAGTGTGAG AGCCTG-3′.
The following PCR conditions were used: 2 min at
50°C, 10 min at 95°C, 35 cycles for 10 sec at 95°C and 30 sec at
60°C, with a final extension step for 30 sec at 72°C. The wild-type
amplicon comprised 309 base pairs. The purified PCR products were
directly sequenced from the two strands using the described primers
and analyzed on the Applied Biosystems 3730 Genetic Analyzer
(Applied Biosystems, Carlsbad, CA, USA).
Statistical analysis
The sequence data were analyzed using Chromas
software version 2.31. According to the structure of the Wilms'
tumor suppressor protein zinc finger domain bound to DNA (16), homology modeling and optimization of
molecular dynamics was performed with the ExPASy Translate Tool to
investigate the spatial configuration of the WT1 gene with
frameshift mutations in exon 7.
Results
Study population
A total of 60 newly diagnosed AML samples collected
from The Children's Hospital of Soochow University, were screened
for WT1 gene mutations. The patient characteristics are provided in
Table I.
Mutation analysis of the WT1 exon 7
We analyzed the samples for mutations in the exon 7
of the WT1 gene. Of the 60 cases of patients with AML, only three
cases harboured a frameshift mutation, which accounts for 5%. The
three cases are described in detail below.
Case 1 had antecedent myelodysplastic
syndrome (MDS)
Case 1, a 3-year-old female was diagnosed as AML-M1
with a white blood cell (WBC) count of 19.9×109/l and an
abnormal karyotype 46,XX,t(2;11)(q31;p15),del(12)(p12) (15). The patient had a history of MDS for
6 months prior to the diagnosis of AML-M1. Complete remission (CR)
was achieved after the first induction therapy. The patient has
been on consolidation therapy since CR. The c.[1319delG] was
detected, which caused a frameshift mutation compared to the
wild-type sequence (Table II).
Alanine (Ala) coded by GCC was replaced by proline (Pro) coded by
CCC, since 1319 G was deleted and the 1320 C behind G moved forward
to form a new codon with 1317 C and 1318 C. Accordingly, all the
amino acids behind Ala were changed due to the recombined codons.
This mutation was previously reported by Gaidzik et
al(15), although not in
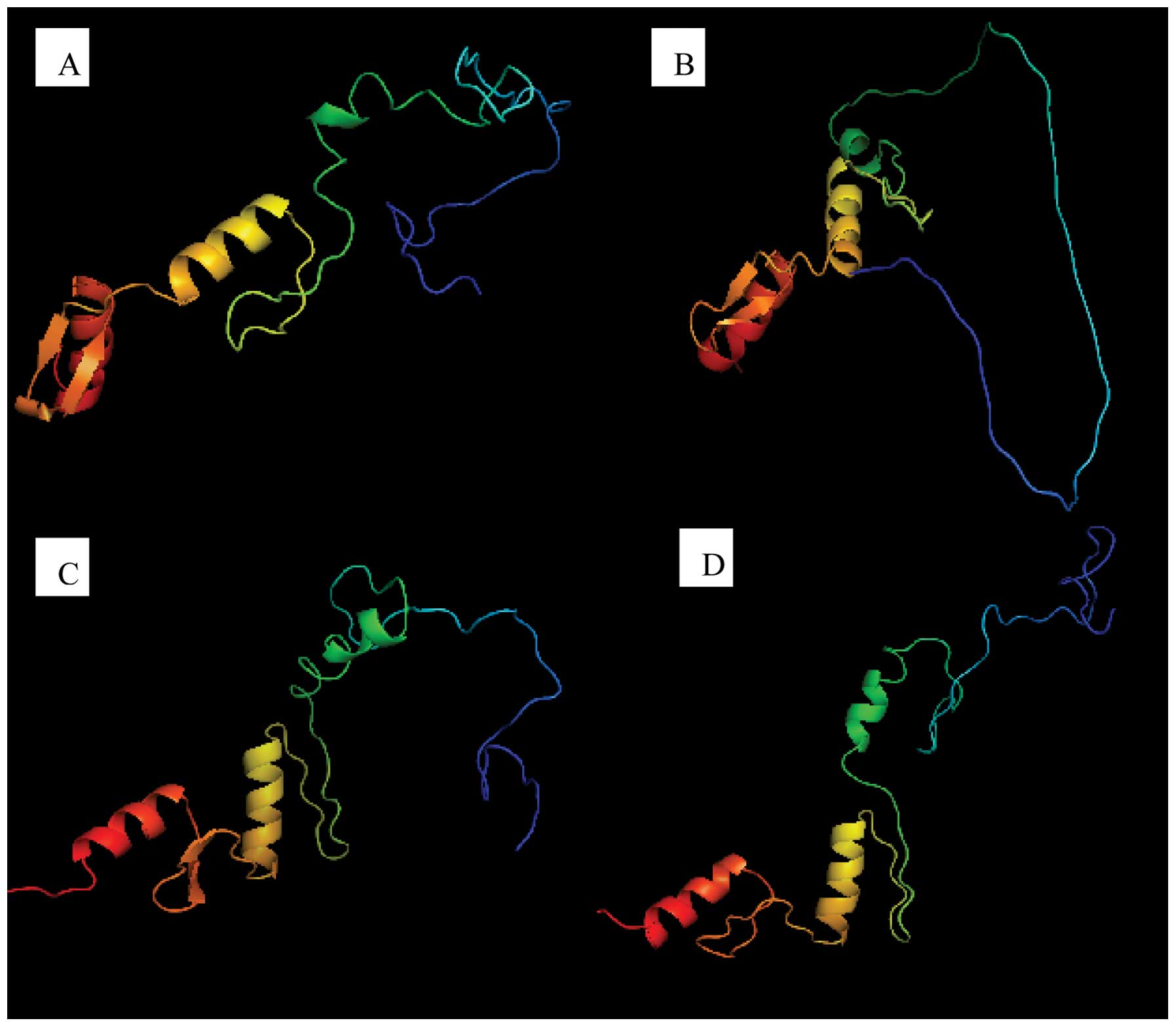
association with patient characteristics (16). The spatial configuration alterations
due to the mutation compared to the wild-type may be visualized
based on homology modeling and optimization of molecular dynamics
(Fig. 1A and B).
 | Table IISequence of exon 7 of the WT1 gene
(wild-type and cases with frameshift mutations). |
Table II
Sequence of exon 7 of the WT1 gene
(wild-type and cases with frameshift mutations).
| Exon 7 | Sequence |
|---|
| Wild-type | 1295-GAT GTG CGA CGT
GTG CCT GGA GTA GCC CCG ACT CTT GTA CGG TCG GCA
TCT GAG ACC AGT GAG AAA CGC CCC TTC ATG TGT GCT TAC CCA
GGC TGC AAT AAG AGA TAT TTT AAG CTG TCC CAC TTA CAG ATG
CAC AGC AGG AAG CAC ACT G-1445 |
| Case 1 | 1295-GAT GTG CGG CGT
GTG CCT GGA GTA (del G)CC CCG ACT CTT GTA CGG TCG
GCA TCT GAG ACC AGT GAG AAA CGC CCC TTC ATG TGT GCT TAC
CCA GGC TGC AAT AAG AGA TAT TTT AAG CTG TCC CAC TTA CAG
ATG CAC AGC AGG AAG CAC ACT G-1445 |
| Case 2 | 1295-GAT GTG CGA CGT
GTG CCT GGA GTA GCC CCG ACT CTT GTA CGG TCG GC(del
A)
TCT GAG A(ins A)CC A (del G)T G(ins G)AG AAA
CGC CCC TTC
ATG TGT GCT TAC CCA GGC T(del G)C AAT AAG AGA T(ins
T)AT
TT(del T) AAG CTG T(ins C) CC CAC TTA CAG AT(del
G) CA (ins A)C
AGC AGG AAG CAC ACT G-1445 |
| Case 3 | 1295-GAT GTG CGG CGT
GTG CCT GGA GTA GCC CCG ACT CTT GTA CGG CCG GCT
TC(del T) GAG A(ins A)CC A(del G)T G(ins
G)AG AAA CGC CCC TTC ATG
TGT GCT TAC CCA GGC TGC A(del A)T AAG AGA TAT TTT AAG CTG
TCC
CAC TTA CAG ATG CAC AGC AGG AAG CAC ACT G-1445 |
Cases 2 and 3 exhibited primary
resistance to chemotherapy
Case 2, a 2-year-old male, was diagnosed as AML-M3
with a WBC count of 53.3×109/l and a normal karyotype
(46,XY). PML-RARα was negative. CR was achieved after 4 cycles of
chemotherapy and the patient relapsed 6 months later. The patient
had a mutation in c.[1342delA; 1349–1350insA; 1353delG;
1355–1356insG; 1392delG; 1400–1401insT;
1405delT];c.[1415–1416insC];c.[1431delG];c.[1433–1434insA]
(Table II), which had not been
reported before.
Case 3, a 6-year-old female, was diagnosed as
AML-M2a with a WBC count of 43.6×109/l and a complex
karyotype 46,XX,t(8;12;21)/45,idm,-X (1,8). No
positive fusion genes were detected. The ratio of blast cells in
the bone marrow was 22% following administration of daunorubicin,
cytarabine and etoposide as induction therapy for 7 days, and even
increased to 57% after a prolonged 3 days of cytarabine. The
patient had a mutation in c.[1345delT;1349–1350insA; 1353delG;
1355–1356insG; 1392delA], which was first reported by our group
(Table II).
The mutations in these two cases affected the
spatial configuration of exon 7 (Fig.
1C and D), although to a lesser extent compared to case 1
(Fig. 1B).
Discussion
The WT1 gene is located on 11p13, encoding a protein
of 429 amino acids (17). The
protein is constituted of the C-terminal of the DNA-binding region
and the N-terminal of the transcriptional regulatory region. A
previous study by King-Underwood et al(1) was the first to report WT1 gene
mutations in AML and their potential effect on AML. Since then, WT1
mutations have been sporadically reported until recently, when the
results on WT1 mutations from cohort studies (8–11,15)
have begun to attract attention again. However, the effect of WT1
gene mutations on the prognosis of AML varies widely among
different groups, even leading to opposite conclusions (8–11).
This inconsistency may be attributed to differences in the
populations investigated, such as age (children vs. adults),
karyotype (normal vs. abnormal), combination with other mutations
such as FLT3-ITD, or different ethnicities (17). The mutation rate in exon 7 in our
study was 5% (3/60), which was lower compared to that reported by
previous studies on pediatric AML (9,12).
This may be due to our limited patient sample or due to the fact
that M3 was included in this cohort. Our data demonstrated that the
three cases with frameshift mutations had a short term survival
which indicated that frameshift mutations may be related to their
initial characterics (antecedent MDS or resistance to
chemotherapy). Hollink et al(10) reported similar results, according to
which WT1 gene mutations conferred an independent poor prognostic
significance. By contrast, Ho et al(9) expanded the population to 842 patients
and observed that WT1 mutations alone are of no independent
prognostic significance in predicting the outcome in pediatric AML.
From the data of that study it was indicated that the
FLT3-ITD status affected the evaluation of WT1
mutations. We investigated FLT3-ITD,
FLT3-TKD, NPM1, CEBPA, C-kit,
DNMT3A 882AA and WT1 exon 7 and 9 mutation in 16
cases of AML and did not observe WT1 mutation overlapping with any
other mutations (Table III).
 | Table IIIMutation panel of AML patients. |
Table III
Mutation panel of AML patients.
| ID | Gender | Age | FAB | NPM1 | FLT3a | C-kit | DNMT3A 882AA | WT1E7 | WT1E9 | CEBPA |
|---|
| 1 | F | 12 | M3v | P | N | N | N | N | N | N |
| 2 | M | 8 | M2 | N | N | N | N | N | N | P |
| 3 | M | 1 | M4Eo | N | N | N | N | N | N | N |
| 4 | F | 3 | M4 | N | P | N | N | N | N | N |
| 5 | F | 6 | M2-relapse | N | N | N | N | N | N | N |
| 6 | F | 9 | AML | N | P | N | N | N | N | N |
| 7 | M | 11 | M3 | N | N | N | N | N | N | N |
| 8 | F | 1 | M4Eo | N | N | P | N | N | N | N |
| 9 | M | 13 | M2a | N | P | N | N | N | N | N |
| 10 | F | 3 | M5b | N | N | N | N | N | N | N |
| 11 | F | 3 | M5 | N | N | N | N | N | N | N |
| 12 | F | 2 | M5 | N | N | N | N | N | N | N |
| 13 | M | 8 | M4 | N | P | N | N | N | N | N |
| 14 | M | 9 | M2 | N | N | N | N | N | N | P |
| 15 | F | 11 | M5 | N | P | N | N | N | N | N |
| 16 | F | 8 | M2 | N | N | P | N | N | N | N |
Gene expression regulated by transcriptional factors
is one of the important regulatory mechanisms in the proliferation
and differentiation of hematopoietic cells (18). The WT1 gene is a transcription
factor which is crucial in the early differentiation of
hematopoietic cells. WT1 may inhibit blood-related gene
transcription, such as (Bcl-2, c-Myc and CSF-1) and is closely
associated with hematological disorders (19,20).
Stoll et al(16)
demonstrated that exon 7 (zinc finger 1) played an important role
in enhancing the WT1 binding activity with its target DNA in a
non-specific manner. Zinc finger structure is a supersecondary
structure that is able to regulate transcription by specifically
combining with nucleic acid binding sites or by forming connections
between the zinc finger proteins. In our study, three cases with
WT1 frameshift mutations in exon 7 were demonstrated to exhibit
spatial configuration alterations, which may disturb the
interaction with other transcription factors, conferring
transformation of MDS into AML or leukemia cell resistance to
chemotherapy. Further investigations of the effect of WT1 exon 7
mutations on the mechanism of AML are required, with the use of
bioinformatics technology.
Acknowledgements
This study was supported by 81170513, 81100371,
81170468 from the National Natural Science Foundation of China,
BK2009127 from the Natural Science Foundation of Jiangsu Province,
H200921 from the Department of Public Health of Jiangsu Province,
333 Project of Jiangsu Province. The authors would like to thank
Professor Chien-Shing Chen, Loma Linda University Medical Center,
for his critical review of the manuscript.
References
|
1
|
King-Underwood L, Renshaw J and
Pritchard-Jones K: Mutations in the Wilms' tumor gene WT1 in
leukemias. Blood. 87:2171–2179. 1996.
|
|
2
|
Inoue K, Ogawa H, Yamagami T, et al:
Long-term follow-up of minimal residual disease in leukemia
patients by monitoring WT1 (Wilms tumor gene) expression levels.
Blood. 88:2267–2278. 1996.PubMed/NCBI
|
|
3
|
Inoue K, Sugiyama H, Ogawa H, et al: WT1
as a new prognostic factor and a new marker for the detection of
minimal residual disease in acute leukemia. Blood. 84:3071–3079.
1994.PubMed/NCBI
|
|
4
|
Inoue K, Ogawa H, Sonoda Y, et al:
Aberrant overexpression of the Wilms tumor gene (WT1) in human
leukemia. Blood. 89:1405–1412. 1997.PubMed/NCBI
|
|
5
|
Yang L, Han Y, Suarez Saiz F and Minden
MD: A tumor suppressor and oncogene: the WT1 story. Leukemia.
21:868–876. 2007.PubMed/NCBI
|
|
6
|
Ariyaratana S and Loeb DM: The role of the
Wilms tumour gene (WT1) in normal and malignant haematopoiesis.
Expert Rev Mol Med. 9:1–17. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
King-Underwood L and Pritchard-Jones K:
Wilms' tumor (WT1) gene mutations occur mainly in acute myeloid
leukemia and may confer drug resistance. Blood. 91:2961–2968.
1998.
|
|
8
|
Hou HA, Huang TC, Lin LI, et al: WT1
mutation in 470 adult patients with acute myeloid leukemia:
stability during disease evolution and implication of its
incorporation into a survival scoring system. Blood. 115:5222–5231.
2010. View Article : Google Scholar
|
|
9
|
Ho PA, Zeng R, Alonzo TA, et al:
Prevalence and prognostic implications of WT1 mutations in
pediatric acute myeloid leukemia (AML): a report from the
Children's Oncology Group. Blood. 116:702–710. 2010. View Article : Google Scholar
|
|
10
|
Hollink IH, van den Heuvel-Eibrink MM,
Zimmermann M, et al: Clinical relevance of Wilms tumor 1 gene
mutations in childhood acute myeloid leukemia. Blood.
113:5951–5960. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Becker H, Marcucci G, Maharry K, et al:
Mutations of the Wilms tumor 1 gene (WT1) in older patients with
primary cytogenetically normal acute myeloid leukemia: a Cancer and
Leukemia Group B study. Blood. 116:788–792. 2010. View Article : Google Scholar
|
|
12
|
Little M and Wells C: A clinical overview
of WT1 gene mutations. Hum Mutat. 9:209–225. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kaczanowski S and Zielenkiewicz P: Why
similar protein sequences encode similar three-dimensional
structures? Theor Chem Acc. 125:543–550. 2010. View Article : Google Scholar
|
|
14
|
Subspecialty Group of Hematology Diseases;
Society of Pediatrics; Chinese Medical Association; and Editorial
Board of Chinese Journal of Pediatrics. Suggestion of diagnosis and
treatment of acute myelocytic leukemia in childhood. Zhonghua Er Ke
Za Zhi. 44:877–878. 2006.(In Chinese).
|
|
15
|
Gaidzik VI, Schlenk RF, Moschny S, et al;
German-Austrian AML Study Group. Prognostic impact of WT1 mutations
in cytogenetically normal acute myeloid leukemia: a study of the
German-Austrian AML Study Group. Blood. 113:4505–4511. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Stoll R, Lee BM, Debler EW, et al:
Structure of the Wilms tumor suppressor protein zinc finger domain
bound to DNA. J Mol Biol. 372:1227–1245. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ho PA, Kuhn J, Gerbing RB, et al: WT1
synonymous single nucleotide polymorphism rs16754 correlates with
higher mRNA expression and predicts significantly improved outcome
in favorable-risk pediatric acute myeloid leukemia: a report from
the Children's Oncology Group. J Clin Oncol. 29:704–711. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Maurer U, Weidmann E, Karakas T, et al:
Wilms tumor gene (wt1) mRNA is equally expressed in blast cells
from acute myeloid leukemia and normal CD34+progenitors.
Blood. 90:4230–4232. 1997.PubMed/NCBI
|
|
19
|
Hewitt SM, Hamada S, McDonnell TJ,
Rauscher FJ III and Saunders GF: Regulation of the proto-oncogenes
bcl-2 and c-myc by the Wilms' tumor suppressor gene WT1. Cancer
Res. 55:5386–5389. 1995.PubMed/NCBI
|
|
20
|
Rossetti S, Van Unen L, Touw IP, et al:
Myeloid maturation block by AML1-MTG16 is associated with Csf1r
epigenetic downregulation. Oncogene. 24:5325–5332. 2005. View Article : Google Scholar : PubMed/NCBI
|