Introduction
Gastric cancer is the fourth most common human
malignant disease and the second most frequent cause of
cancer-related mortality worldwide, causing ~800,000 deaths
annually. Approximately two-thirds of gastric cancer cases occur in
developing countries, with 42% occurring in China alone (1). Although the optimal combination of
surgical and non-surgical approaches has been used to treat gastric
cancer, a considerable number of patients develop metastases to
other sites due to the lack of reliable diagnostic techniques for
early-stage detection (2).
Therefore, diagnosis in the early stages is crucial for the
selection of the most effective treatment for gastric cancer
patients.
MicroRNAs (miRNAs) are small, non-coding RNAs, 19–24
nucleotides in length, which were first described by Lee et
al(3). The mature
single-stranded microRNAs bind to the 3′ untranslated region of
potentially hundreds of target genes of imperfect complementarity,
resulting in degradation of target mRNAs and inhibition of
translation (4). Over the past 10
years, accumulating evidence strongly supports the role of miRNAs
in crucial cellular processes, including development,
differentiation, stress response, apoptosis and proliferation
(5). Particularly in tumors, miRNAs
may function as oncogenes and/or tumor suppressor genes (6).
Thus far, apart from traditional methods, the
molecular technique is considered the optimal method for early
diagnosis and prognosis prediction in cancer. Recent studies
demonstrated that miRNAs are frequently dysregulated in human
malignancies (7), including
increased expression [i.e., miR-221 (8), -223 (9) and -135a (10)] and decreased expression [i.e.,
miRNA-200c (11), -451 (12) and -34b (13)]. This altered miRNA expression
pattern provided novel opportunities for the use of biomarkers in
cancer diagnosis (14). Several
miRNA analyses were reported in human malignancies and the
differences in expression between tumor tissues and their benign
counterparts may be useful for cancer diagnosis (15). However, more ideal biomarkers or
co-markers for gastric cancer are required for optimizing treatment
selection for patients.
In this study, we compared the expression of 8
miRNAs (miR-21, -103, -106a, -126, -143, -195, -221 and -222)
between gastric cancer and normal gastric tissue and investigated
the association between their expression and clinical significance
in order to identify potential diagnostic markers for gastric
cancer.
Materials and methods
Sample collection
Gastrectomy samples were obtained from gastric
cancer patients who underwent total gastrectomy at the Shaanxi
Provincial People’s Hospital and the First Affiliated Hospital of
Xi’an Jiaotong University between 2009 and 2011. A total of 20
paired samples (gastric cancer and normal tissues, at a distance
>5 cm from the tumor) were randomly selected and independently
examined. The pathological characteristics of the patients are
summarized in Table I.
 | Table ISummary of pathological
characteristics of samples. |
Table I
Summary of pathological
characteristics of samples.
| Pathological
characteristics | Patient no.
(n=20) |
|---|
| Age (years) |
| <60 | 7 |
| >60 | 13 |
| Depth of invasion
(T) |
| T1 | 2 |
| T2 | 4 |
| T3 | 3 |
| T4 | 11 |
| Lymph node metastasis
(N) |
| Negative (N0) | 2 |
| Positive
(N1–N3) | 18 |
| Haematogenous
metastasis (M) |
| Negative (M0) | 17 |
| Positive (M1) | 3 |
| Stage |
| I | 2 |
| II | 5 |
| III | 7 |
| IV | 6 |
|
Differentiation |
| Poor | 12 |
| Moderate | 6 |
| High | 2 |
Isolation of total RNA from
formaldenhyde-fixed, paraffin-embedded (FFPE) tissue
Total RNA was isolated from FFPE tissue sections
with RecoverAll™ Total Nucleic Acid Isolation kit (Ambion, Austin,
TX, USA) according to the manufacturer’s instructions. Briefly, the
sections were deparaffinized with xylene. Following addition of
digestion buffer and protease, the samples were incubated at 50°C
for 15 min and at 80°C for 15 min. Subsequently, isolation additive
and 100% ethanol were added. After washing 3 times, RNA was eluted
in 60 μl of elution solution and stored at −80°C.
Reverse transcription and quantitative
polymerase chain reaction (qPCR)
RNA was reverse-transcribed to cDNA by priming with
a mixture of looped primers and preamplified according to the
manufacturer’s instructions. The primers for miRNA were designed by
our team (Table II) and
commercially obtained from AuGCT Biotechnology Corporation (AuGCT,
Beijing, China). The PrimeScript® RT reagent kit
(Perfect Real Time; Takara Bio Inc., Shiga, Japan) was used. The
reverse transcriptase reactions contained 20 ng of RNA samples, 2
μl of 5X PrimeScript® Buffer (for Real Time), 0.5 μl of
PrimeScript® RT Enzyme Mix I, 4.5 μl of RNase-free water
and 1 μl of stem-loop RT primers. The 10-μl reactions were
incubated for 15 min at 37°C and for 5 sec at 85°C and maintained
at 4°C.
 | Table IIPrimers for reverse transcription
(RT) quantitative polymerase chain reaction of 8 microRNAs and
U6. |
Table II
Primers for reverse transcription
(RT) quantitative polymerase chain reaction of 8 microRNAs and
U6.
| miRNAs | Primer
sequences |
|---|
| miR-103 |
| Forward: |
ATCCAGTGCGTGTCGTG |
| Reverse: |
TGCTAGCAGCATTGTACAGG |
| RT: |
GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACTCATAGC |
| miR-106a |
| Forward: |
ATCCAGTGCGTGTCGTG |
| Reverse: |
TGCTAAAAGTGCTTACAGTG |
| RT: |
GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACCTACCTG |
| miR-143 |
| Forward: |
CAGTGCGTGTCGTGGAG |
| Reverse: |
GCGGTGAGATGAAGCACT |
| RT: |
GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACGAGCTAC |
| miR-145 |
| Forward: |
CAGTGCGTGTCGTGGAGT |
| Reverse: |
AGGTCCAGTTTTCCCAGG |
| RT: |
GGAGTCGGCAATTGCACTGGATACGACAGGGATT |
| miR-195 |
| Forward: |
CAGTGCGTGTCGTGGAGT |
| Reverse: |
ACGGTAGCAGCACAGAAATA |
| RT: |
GGAGTCGGCAATTGCACTGGATACGACGCCAATA |
| miR-21 |
| Forward: |
ATCCAGTGCGTGTCGTG |
| Reverse: |
TGCTTAGCTTATCAGACTG |
| RT: |
TGGAGTCGGCAATTGCACTGGATACGACTCAACAT |
| miR-221 |
| Forward: |
ATCCAGTGCGTGTCGTG |
| Reverse: |
TGCTAGCTACATTGTCTGCT |
| RT: |
GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACGAAACCC |
| miR-222 |
| Forward: |
ATCCAGTGCGTGTCGTG |
| Reverse: |
TGCTAGCTACATCTGGCT |
| RT: |
GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACACCCAGT |
| U6 |
| Forward: |
GCTTCGGCAGCACATATACTAAAAT |
| Reverse: |
CGCTTCACGAATTTGCGTGTCAT |
| RT: |
CGCTTCACGAATTTGCGTGTCAT |
qPCR was performed using SYBR®-Green I
with the SYBR® Premix Ex Taq™ II (Perfect Real Time).
The qPCR reactions included 1 μl of RT product dilution (100 ng),
10 μl of 2X SYBR Premix Ex Taq II, 7 μl of RNase-free water, 1 μl
of forward primer and 1 μl of reverse primer. U6 was used as an
internal control to normalize the expression levels of the target
genes. The qPCR was initiated with an initial denaturation step at
95°C for 30 sec, 40 cycles of 5 sec at 95°C and 30 sec at 60°C. All
the reactions were run in triplicate using the IQ-5 Real-Time PCR
system (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
The ΔCt and 2−ΔΔCt methods were used for
analysis. The ΔCt value was calculated by the difference between
the Ct values of the specific miRNA and U6: ΔCt =
CtmiRNA − CtU6.ΔΔCt = ΔCtcancer
tissues − ΔCtnormal tissues. The value of
2−ΔCt represented the miRNA expression of each sample
and the value of 2−ΔΔCt represented the relative
quotient (RQ) of the expression of the target gene to that of the
control. In the present study, RQ represented the value of the
ratio of miRNA expression in cancer tissue to that in normal
tissue. An RQ<1 indicated that the miRNA expression levels in
cancer tissue were lower compared to those in normal tissue.
Conversely, an RQ>1 indicated higher miRNA expression in cancer
compared to normal tissues.
Statistical analysis
Data were analyzed with SPSS software, version 12.0
(SPSS Inc., Chicago, IL, USA) and Excel software (Microsoft,
Redmond, USA). The paired samples t-test was used to compare the
expression of miRNAs between cancer and normal tissues. One-way
ANOVA was used to investigate the association between cancer and
normal tissues. P<0.05 was considered to indicate a
statistically significant difference.
Results
Dysregulated miRNAs in cancer and normal
tissues of gastric cancer patients
Among the 20 paired samples, 19 cases (95%)
exhibited a higher expression of miR-21, 15 cases (75%) exhibited a
higher expression of miR-103 and -106a and 13 cases (65%) exhibited
a higher expression of miR-221 and -222 in gastric cancer tissues
compared to that in normal tissues (Fig. 1), whereas miR-126 exhibited no
identical or significant differences between gastric cancer and
normal tissues (higher in 10 and lower in the remaining 10 cases).
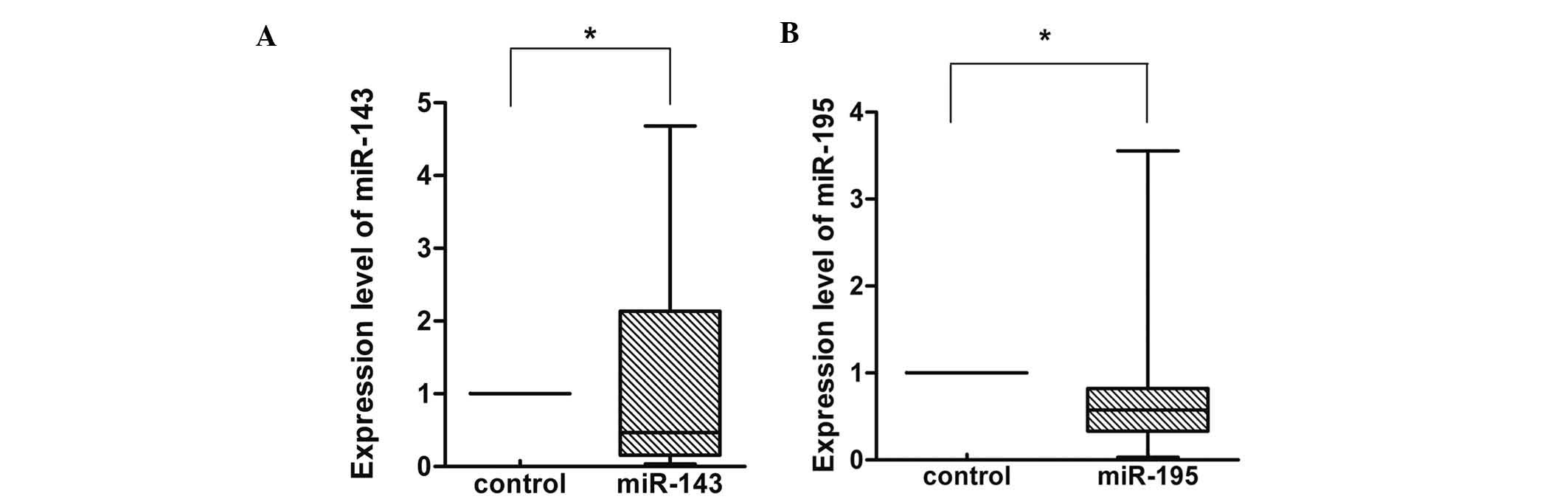
Furthermore, miR-143 was shown to be decreased in 15 of the 20
pairs (75%) (Fig. 2A). The average
fold change of miR-143 between gastric cancer and normal tissues
was 1.121 (±1.589). miR-195 was also decreased in 16 of the 20
pairs (80%) (Fig. 2B). The average
fold change of miR-195 between gastric cancer and normal tissues
was 0.799 (±0814).
Association between miRNA level and
clinicopathological factors in patients with gastric cancer
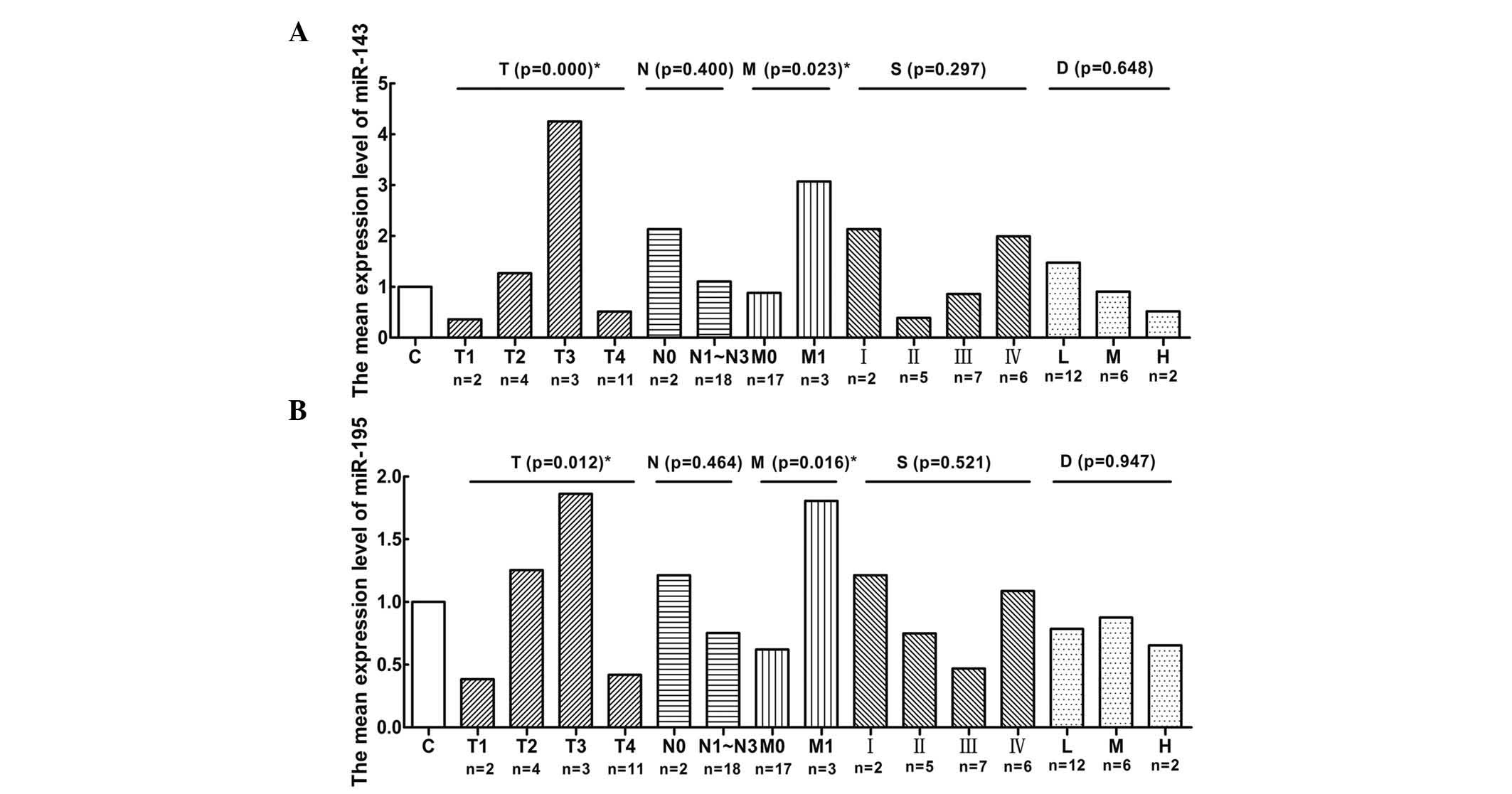
To further investigate the roles of miR-143 and -195
expression in gastric cancer progression, the association between
the expression of miR-143 (Fig. 3A)
and -195 (Fig. 3B) was analyzed by
one-way ANOVA. A decreased expression of miR-143 and -195 in
gastric cancer was not associated with age (>60 vs. <60
years), lymph node metastasis (negative vs. positive), stage and
differentiation (poor vs. moderate vs. high). However, miR-143 and
-195 underexpression was found to be significantly associated with
depth of invasion (P=0.000 and P=0.012, respectively) and
haematogenous metastasis (P=0.023 and P=0.016, respectively).
Discussion
Alterations in miRNA expression have been described
in various cancer types and shown to be significantly correlated
with cancer progression (16).
miRNAs may function as oncogenes in human cancer. Our data
demonstrated that miR-21, -103, -106a, -221 and -222 exhibited a
higher expression in gastric cancer compared to normal gastric
tissues (Fig. 1). The expression
level of miR-21 was reported to be significantly higher in breast
cancer compared to normal breast tissues in 109 patients who
underwent surgery between 2002 and 2004; its overexpression was
associated with mastectomy, larger tumor size, advanced stage,
higher grade, estrogen receptor-negative status, human epidermal
growth factor receptor 2-positive status, higher Ki-67 expression
and mortality and it was significantly associated with lower
overall survival (17). miR-103
expression was shown to be increased in patients with colorectal
cancer (CRC) and was associated with poor prognosis; its actions
are mediated through targeting the known metastasis suppressors
death-associated protein kinase and Kruppel-like factor 4 in CRC
cells, resulting in increased cell motility and cell-matrix
adhesion and decreased cell-cell adhesion and epithelial marker
expression (18). The expression
analysis of miR-106a in the patient samples demonstrated its
overexpression in CRC, leading to downregulation of the
retinoblastoma protein activity, which plays an important role in
cell cycle progression and, therefore, may be involved in malignant
transformation of colonic cells (19). miR-221 and -222 were found to be
overexpressed in several human cancers, including breast (20), prostate (21), lung (22) and liver cancer (23). Although the expression levels of
miR-126 in CRC tissues were significantly lower compared to those
in normal tissues and miR-126 overexpression may inhibit the growth
of cancer cells (24), it was also
found to be overexpressed in acute myeloid leukemia (25). Although the precise mechanisms
underlying the biological functions of miRNAs have not been fully
elucidated, our data demonstrated that miR-21, -103, -106a, -221
and -222 were overexpressed in gastric cancer, which may indicate
their role as oncogenes in cancerous processes.
Furthermore, previous studies reported the
expression alterations and potential target sites of miR-143 and
-195. Motoyama et al(26)
used a miRNA microarray containing 455 human miRNA probes to
determine the miRNA pattern in human CRC and the results
demonstrated that the expression of miR-143 in cancer tissues was
significantly lower compared to that in normal tissues. In
addition, miRNA microarray analysis demonstrated that miR-143 was
significantly downregulated, which may promote apoptosis and
inhibit tumor formation by targeting Bcl-2 in cervical cancer
(27). Ozata et al(28) reported that miR-195 was
underexpressed in adrenocortical carcinoma (ACC) compared to normal
adrenal cortices and benign adenomas. The overexpression of miR-195
may reduce cell proliferation and lead to significant induction of
cell death in human NCI-H295R ACC cells. miR-195 was also found to
be downregulated in CRC compared to normal colorectal tissue
samples, with this downregulation being observed more frequently
among patients with lymph node metastasis and advanced tumor stage
(29). As regards the potential for
a miRNA-based therapy, several target genes of miR-143 and -195
have already been proven experimentally, such as hexokinase 2
isoform and extracellular signal-regulated kinase-5 (30,31)
and WEE1 (32), respectively. Our
data also demonstrated that miR-143 and -195 were downregulated in
gastric cancer and were associated with depth of invasion (T) and
haematogenous metastasis (M), providing a potential target for
research of human cancer.
The potential usefulness of miRNA-based biomarkers
for the diagnosis of numerous human cancers has been established by
several studies. miR-222 expression in the urine was verified by
in situ hybridization, which provided a high-accuracy method
for bladder cancer diagnosis (33).
As regards breast cancer, the differential expression of systemic
miRNA-195 provides a possibility for detecting non-invasive and
early-stage disease as a sensitive, specific, non-invasive cancer
biomarker (34). However, the
combination of markers is more reliable compared to a single marker
for cancer diagnosis (35). The
expression level of miR-126 in the patient serum was found to be
associated with soluble mesothelin-related peptides, which are a
specific marker of malignant pleural mesothelioma (MPM). miR-126
expression was correlated with a high risk of developing MPM and
may be used as a marker for the early detection of MPM (36). This study identified miR-143 and
-195 as potentially useful biomarkers in human gastric cancer, due
to their similar performance in the clinicopathological factor
analysis. We also suggest a potential diagnostic biomarker
combination of miR-143 and -195 (co-markers) to improve their
diagnostic sensitivity and specificity.
To validate the performance of biomarkers for the
detection of cancer, further studies are required to verify whether
the miRNAs we selected bear a full potential as either biomarkers
or therapeutic targets in gastric cancer. However, we demonstrated
that miR-143 and -195, alone or in combination, may play a critical
role in cancer progress and may constitute optimal biomarkers for
the diagnosis of gastric cancer.
Acknowledgements
This study was supported by a grant from the Key
Science and Technology Program of Shaanxi (no. 2010ZDKG-50), and
the Program for Changjiang Scholars and Innovative Research Team in
University (PCSIRT: 1171).
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar
|
|
2
|
Hohenberger P and Gretschel S: Gastric
cancer. Lancet. 362:305–315. 2003. View Article : Google Scholar
|
|
3
|
Lee RC, Feinbaum RL and Ambros V: The
C. elegans heterochronic gene lin-4 encodes small RNAs with
antisense complementarity to lin-14. Cell. 75:843–854. 1993.
|
|
4
|
Wang XJ, Reyes JL, Chua NH and Gaasterland
T: Prediction and identification of Arabidopsis thaliana
microRNAs and their mRNA targets. Genome Biol. 5:R652004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Karp X and Ambros V: Developmental
biology. Encountering microRNAs in cell fate signaling. Science.
310:1288–1289. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mraz M, Pospisilova S, Malinova K, Slapak
I and Mayer J: MicroRNAs in chronic lymphocytic leukemia
pathogenesis and disease subtypes. Leuk Lymphoma. 50:506–509. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jiang Q, Wang Y, Hao Y, et al:
miR2Disease: a manually curated database for microRNA deregulation
in human disease. Nucleic Acids Res. 37:D98–D104. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dahiya N, Sherman-Baust CA, Wang TL, et
al: MicroRNA expression and identification of putative miRNA
targets in ovarian cancer. PLoS One. 3:e24362008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li X, Zhang Y, Zhang H, et al: miRNA-223
promotes gastric cancer invasion and metastasis by targeting tumor
suppressor EPB41L3. Mol Cancer Res. 9:824–833. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen Y, Zhang J, Wang H, et al: miRNA-135a
promotes breast cancer cell migration and invasion by targeting
HOXA10. BMC Cancer. 12:1112012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shimono Y, Zabala M, Cho RW, et al:
Downregulation of miRNA-200c links breast cancer stem cells with
normal stem cells. Cell. 138:592–603. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang XC, Tian LL, Jiang XY, et al: The
expression and function of miRNA-451 in non-small cell lung cancer.
Cancer Lett. 311:203–209. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lee YM, Lee JY, Ho CC, et al: miRNA-34b as
a tumor suppressor in estrogen-dependent growth of breast cancer
cells. Breast Cancer Res. 13:R1162011. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang J, Chen J, Chang P, et al: MicroRNAs
in plasma of pancreatic ductal adenocarcinoma patients as novel
blood-based biomarkers of disease. Cancer Prev Res (Phila).
2:807–813. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Paranjape T, Slack FJ and Weidhaas JB:
MicroRNAs: tools for cancer diagnostics. Gut. 58:1546–1554. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
He L, Thomson JM, Hemann MT, et al: A
microRNA polycistron as a potential human oncogene. Nature.
435:828–833. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lee JA, Lee HY, Lee ES, Kim I and Bae JW:
Prognostic implications of microRNA-21 overexpression in invasive
ductal carcinomas of the breast. J Breast Cancer. 14:269–275. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen HY, Lin YM, Chung HC, et al:
miR-103/107 promote metastasis of colorectal cancer by targeting
the metastasis suppressors DAPK and KLF4. Cancer Res. 72:3631–3641.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Catela Ivkovic T, Aralica G, Cacev T,
Loncar B and Kapitanovic S: miR-106a overexpression and pRB
downregulation in sporadic colorectal cancer. Exp Mol Pathol.
94:148–154. 2013.PubMed/NCBI
|
|
20
|
Stinson S, Lackner MR, Adai AT, et al:
TRPS1 targeting by miR-221/222 promotes the
epithelial-to-mesenchymal transition in breast cancer. Sci Signal.
4:ra412011.PubMed/NCBI
|
|
21
|
Sun T, Yang M, Chen S, et al: The altered
expression of MiR-221/-222 and MiR-23b/-27b is associated with the
development of human castration resistant prostate cancer.
Prostate. 72:1093–1103. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang Y, Ma T, Yang S, et al:
High-mobility group A1 proteins enhance the expression of the
oncogenic miR-222 in lung cancer cells. Mol Cell Biochem.
357:363–371. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yoo BK, Santhekadur PK, Gredler R, et al:
Increased RNA-induced silencing complex (RISC) activity contributes
to hepatocellular carcinoma. Hepatology. 53:1538–1548. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li XM, Wang AM, Zhang J and Yi H:
Down-regulation of miR-126 expression in colorectal cancer and its
clinical significance. Med Oncol. 28:1054–1057. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li Z and Chen J: In vitro functional study
of miR-126 in leukemia. Methods Mol Biol. 676:185–195. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Motoyama K, Inoue H, Takatsuno Y, et al:
Over- and under-expressed microRNAs in human colorectal cancer. Int
J Oncol. 34:1069–1075. 2009.PubMed/NCBI
|
|
27
|
Liu L, Yu X, Guo X, et al: miR-143 is
downregulated in cervical cancer and promotes apoptosis and
inhibits tumor formation by targeting Bcl-2. Mol Med Report.
5:753–760. 2012.PubMed/NCBI
|
|
28
|
Ozata DM, Caramuta S, Velazquez-Fernandez
D, et al: The role of microRNA deregulation in the pathogenesis of
adrenocortical carcinoma. Endocr Relat Cancer. 18:643–655. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang X, Wang J, Ma H, Zhang J and Zhou X:
Downregulation of miR-195 correlates with lymph node metastasis and
poor prognosis in colorectal cancer. Med Oncol. 29:919–927. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Peschiaroli A, Giacobbe A, Formosa A, et
al: miR-143 regulates hexokinase 2 expression in cancer cells.
Oncogene. 32:797–802. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Clape C, Fritz V, Henriquet C, et al:
miR-143 interferes with ERK5 signaling, and abrogates prostate
cancer progression in mice. PLoS One. 4:e75422009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bhattacharya A, Schmitz U, Wolkenhauer O,
Schonherr M, Raatz Y and Kunz M: Regulation of cell cycle
checkpoint kinase WEE1 by miR-195 in malignant melanoma. Oncogene.
32:3175–3183. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Puerta-Gil P, Garcia-Baquero R, Jia AY, et
al: miR-143, miR-222, and miR-452 are useful as tumor
stratification and noninvasive diagnostic biomarkers for bladder
cancer. Am J Pathol. 180:1808–1815. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Heneghan HM, Miller N, Kelly R, Newell J
and Kerin MJ: Systemic miRNA-195 differentiates breast cancer from
other malignancies and is a potential biomarker for detecting
noninvasive and early stage disease. Oncologist. 15:673–682. 2010.
View Article : Google Scholar
|
|
35
|
Liang QL, Shi HZ, Qin XJ, Liang XD, Jiang
J and Yang HB: Diagnostic accuracy of tumour markers for malignant
pleural effusion: a meta-analysis. Thorax. 63:35–41. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Santarelli L, Strafella E, Staffolani S,
et al: Association of MiR-126 with soluble mesothelin-related
peptides, a marker for malignant mesothelioma. PLoS One.
6:e182322011. View Article : Google Scholar : PubMed/NCBI
|