Introduction
Interferon (IFN) regulatory factors are a family of
transcription factors that are crucial in the regulation of
IFN-stimulated genes (ISGs) and the induction of type I IFNs,
including IFN-α and IFN-β. The human interferon regulatory factor
(IRF) family, which consists of 9 members (IRF1-9) (1), is defined by a highly conserved
amino-terminal DNA-binding domain characterized by a 5-tryptophan
residue repeat that allows the binding of IRFs, as homodimers or
heterodimers, to consensus GAA and AANNNGAA motifs found in
IFN-stimulated response elements (ISREs), including promoters of
type I IFNs and ISGs (2). Each IRF
also contains a unique C-terminal domain, referred to as the IRF
association domain (IAD). This region was shown to be important in
mediating IRF protein-protein interaction. The different IADs
promote the interaction of IRFs with distinct transcription factors
and contribute to the regulation of target genes (3,4).
Alternative splicing is a general mechanism for
regulating gene expression that affects the RNA products of >90%
of human genes (5). It is
associated with numerous diseases, such as Kawasaki disease
(6), systemic lupus erythematosus
(7) and myasthenia gravis (8). IRF-3 includes a variety of spliced
variants which may regulate the wild-type of IRF-3. IRF-3 isoforms
produced by alternative splicing have been reported in humans
(IRF-3a) and mice (mIRF-3a). The expression of IRF-3a results in
potent and specific negative regulation of IRF-3 transcriptional
activity (9). Additionally, Li
et al(10) reported that the
spliced variants IRF-3b, -3c, -3d, -3e and -3f were expressed in
the majority of human cells and tissues and their expression was
more frequent in tumor tissues compared to that in normal
counterparts. It was reported that the ectopic expression of these
spliced variants may inhibit the transactivation capacity of IRF-3
to varying degrees.
A previous study identified two novel spliced
variants of IRF-3, starting from intron 2 of the wild-type of
IRF-3, Int2V1 and Int2V2 (11).
Their translation initiation ATG codons were found to be located
718 and 162 bp, respectively, upstream of the third exon. We
reported that the transcription factor Sp1 upregulates the spliced
variant Int2V1 (12), although the
underlying molecular mechanism has not been fully elucidated. In
this study, we demonstrated that exogenous Sp1 expression led to a
significant increase in IRF-3 spliced variant mRNA expression and
promoter activity. We investigated the molecular mechanism through
which Sp1 upregulated the transcription of human IRF-3 gene through
RNAi, electrophoretic gel mobility shift assays and chromatin
immunoprecipitation assays and demonstrated that Sp1 upregulated
IRF-3 spliced variant transcription through directly binding to the
Sp1 consensus binding sequences at positions −130 to −120 bp
relative to the transcriptional initiation site in the spliced
variant of human IRF-3.
Materials and methods
Cell culture
Human embryonic kidney (HEK)-293T cells (purchased
from the Shanghai Chinese Academy of Sciences Institute for Cell
Biology preserved by the State Key Laboratory of Reproductive
Medicine, the First Affiliated Hospital of Nanjing Medical
University, Nanjing, China) were maintained in Dulbecco’s modified
Eagle’s medium (Thermo Fisher Scientific, Rockford, IL, USA)
containing 10% heat-inactivated fetal bovine serum (Zhejiang
Tianhang Biological Technology Co., Ltd, Hangzhou, China)
supplemented with penicillin (100 U/ml) and streptomycin (100
μg/ml). The cells were incubated at 37ºC with 100% humidity in 5%
CO2 and passaged using standard cell culture
techniques.
Plasmids, transfection and RNAi
The cloning of the spliced variant of human IRF-3
gene promoter region was performed as previously described
(12). The expression plasmids pN3,
pN3-Sp1, (donated by Dr Guntram Suske) (13) were cotransfected by using
Lipofectamine™ 2000 (Invitrogen, Carlsbad, CA, USA) and then
incubated for 24 h. For the RNAi assay, the HEK-293T cells were
transfected with Sp1 siRNA or control vector (primer of Sp1 siRNA:
F, 5′-AUCACUCCAUGGAUGAAAUGATT-3′ and R,
5′-UCSUUUCSUCCSUGGSGUGAUTT-3′; and primer of control: F,
5′-UUCUCCGAACGUGUCACGUTT-3′ and R, 5′-ACGUGACACGUUCGGAGAATT-3′) and
cultured for 48 h. The cells were then harvested to assess the
effectiveness of RNA interference.
Dual-luciferase reporter assays
The HEK-293T cells were seeded in 96-well plates 24
h prior to transfection. Sp1 expression plasmid or empty vector was
individually cotransfected into the cells, together with the
appropriate IRF-3 spliced variant promoter reporter plasmids, using
Lipofectamine 2000 (Invitrogen). For the RNAi assay, Sp1 siRNA or
control vector was individually cotransfected into HEK-293T cells,
with the appropriate IRF-3 spliced variant promoter reporter
plasmids. The pRL-TK plasmid (2 ng/sample; Promega, Madison, WI,
USA) containing the Renilla luciferase gene driven by the
herpes simplex virus thymidine kinase promoter was cotransfected
with the constructs and the luciferase activity was normalized. The
preparation of cell lysates and measurements of luciferase activity
were performed using the Dual Reporter Assay system (Promega) and
the TD-20/20 luminometer (Turner Designs, Inc., Sunnyvale, CA, USA)
according to the manufacturer’s instructions.
Nuclear extract preparation and
electromobility shift assay
Nuclear extracts from HEK-293T cells were prepared.
Oligonucleotides (probes) were labeled at the 5′ end with ATP using
T4 polynucleotide kinase (Fermentas, Vilnius, Lithuania). The
proteins Sp1 (Promega) or nuclear extracts from the HEK-293T cells
(100–300 ng) were pre-incubated in binding buffer (5X binding
buffer, Promega) for 20 min at 25ºC in a volume of 19 μl, with and
without an excess of unlabeled oligonucleotide competitors.
Following the addition of 1 μl of labeled DNA, the mixture was
incubated for 60–90 min at 4ºC. Each reaction mixture was then
loaded into the well of a 4% non-denaturing polyacrylamide gel
(Beyotime Institute of Biotechnology, Shanghai, China) and
electrophoresed at 100 V in 0.5X TBE buffer at 4ºC for 1–2 h. For
competition experiments and antibody supershift, the competing
unlabeled probes and antibodies were pre-incubated for 20 min at
room temperature with the nuclear extracts prior to the addition of
the radiolabeled probe. The DNA-protein complexes were resolved on
4% non-denaturing polyacrylamide gels for 3 h at 100 V in 0.5X TBE
buffer. After electrophoresis, the gel was dried and exposed to
autoradiography film at −80ºC.
Chromatin immunoprecipitation assay
(ChIP)
The chromatin immunoprecipitation assay was
performed using the ChIP-IT kit (Active Motif, Carlsbad, CA, USA)
following the manufacturer’s instructions. Briefly, three
100-cm2 dishes of 80–90% confluent HEK-293T cells were
treated with 1% formaldehyde in phosphate-buffered saline (PBS) for
10 min at room temperature. The formaldehyde was inactivated by the
addition of 0.125 M glycine in PBS to the cells for 5 min at room
temperature. The cells were then washed in ice-cold PBS and lysed
with lysis buffer containing 1% sodium dodecyl sulfate. Sonication
of cross-linked chromatin was performed at 200 W with five rounds
of 20-sec pulses, so that the chromatin fragments thus obtained
ranged from 500 to 1,000 bp in size. Soluble chromatin was
subjected to overnight immunoprecipitation with anti-IgG or
anti-Sp1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA). A portion
of the chromatin solution was kept to measure the amount of input
DNA in different samples prior to immunoprecipitation. For each
immunoprecipitation, 2 μg of the appropriate antibody was incubated
with a precleared chromatin aliquot overnight at 4ºC. Following
immunoprecipitation and elution, the eluent was heated to 65ºC for
6 h to reverse the cross-link and DNA then was purified using the
minicolumns provided with the kit. The purified DNA was amplified
by the promoter-specific primers (ChIP-F, 5′-CACCCCTCGTCAACACCC-3′
and ChIP-R, 5′-CGCGGGAAAGTTGAACTAATA-3′] and polymerase chain
reaction (PCR) was performed under the following conditions: 1
cycle at 94ºC for 5 min; 36 cycles of 30 sec at 94ºC, 30 sec at
59°C and 30 sec at 72ºC; and a final extension step for 10 min at
72ºC. The PCR products were analyzed by electrophoresis on a 2%
agarose gel.
RNA purification and quantitative reverse
trancription (qRT-PCR)
Total RNA extraction was performed using TRIzol
reagent (Invitrogen) followed by chloroform-isopropanol extraction
and ethanol precipitation. Subsequently, duplicate samples of 1 μl
of each cDNA were used as a template. The quantification of gene
transcripts was performed by qPCR using SYBR-Green I dye
(Invitrogen) and the ABI PRISM 7700 Sequence Detection system (PE
Applied Biosystems, Wellesley, MA, USA). The specificity of
amplification was assessed for each sample by melting curve
analysis. The expression values were normalized with control GAPDH.
The primers used were as follows: spliced variant of IRF-3: sense
primer, 5′-ACGGGATTAGACACCAAGTT-3′ and antisense primer,
5′-TGGGATTACAGGCATGAGCT-3′; and GAPDH: sense primer,
5′-AGGTCGGAGTCAACGGAT-3′ and antisense primer,
5′-TCCTGGAAGATGGTGATG-3′.
Statistical analysis
The results were analyzed by the paired two-tailed
Student’s-t test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Overexpression of Sp1 leads to the
upregulation of IRF-3 spliced variant promoter activity
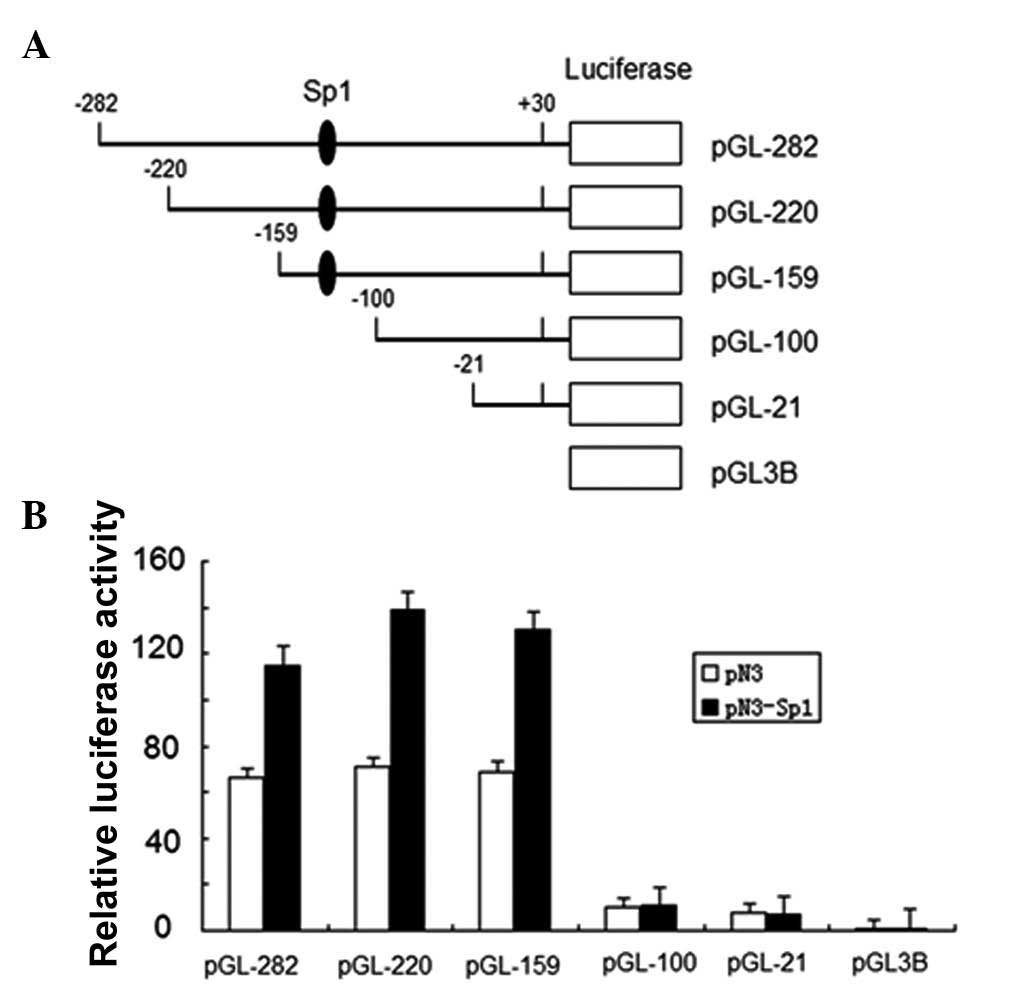
To confirm the effect of Sp1 on IRF-3 spliced
variant promoter activity, we investigated the effects of Sp1
overexpression on IRF-3 promoter activity and performed luciferase
reporter assays with a series of IRF-3 spliced variant promoter
deletion mutants (12). The
schematic representation of the constructs used in this assay is
presented in Fig. 1. Sp1 expression
plasmid and empty vector were transfected into HEK-293T cells
together with each IRF-3 promoter reporter plasmid indicated in
Fig. 1B. Equal concentrations of
Renilla reporter plasmid pRL-TK were also cotransfected as
an internal control. Luciferase activity was measured and the
relative fold activation of each IRF-3 spliced variant promoter
normalized to the pRL-TK internal standard was presented in the
histogram with arbitrary units. All the IRF-3 spliced variant
promoter fragments, except pGL3-100 and pGL3-21, which do not have
Sp1 binding sites, exhibited a significant upregulation under
exogenous Sp1 expression (Fig. 1A).
This result indicated that human IRF-3 spliced variant promoter was
positively regulated by Sp1 in HEK-293T cells.
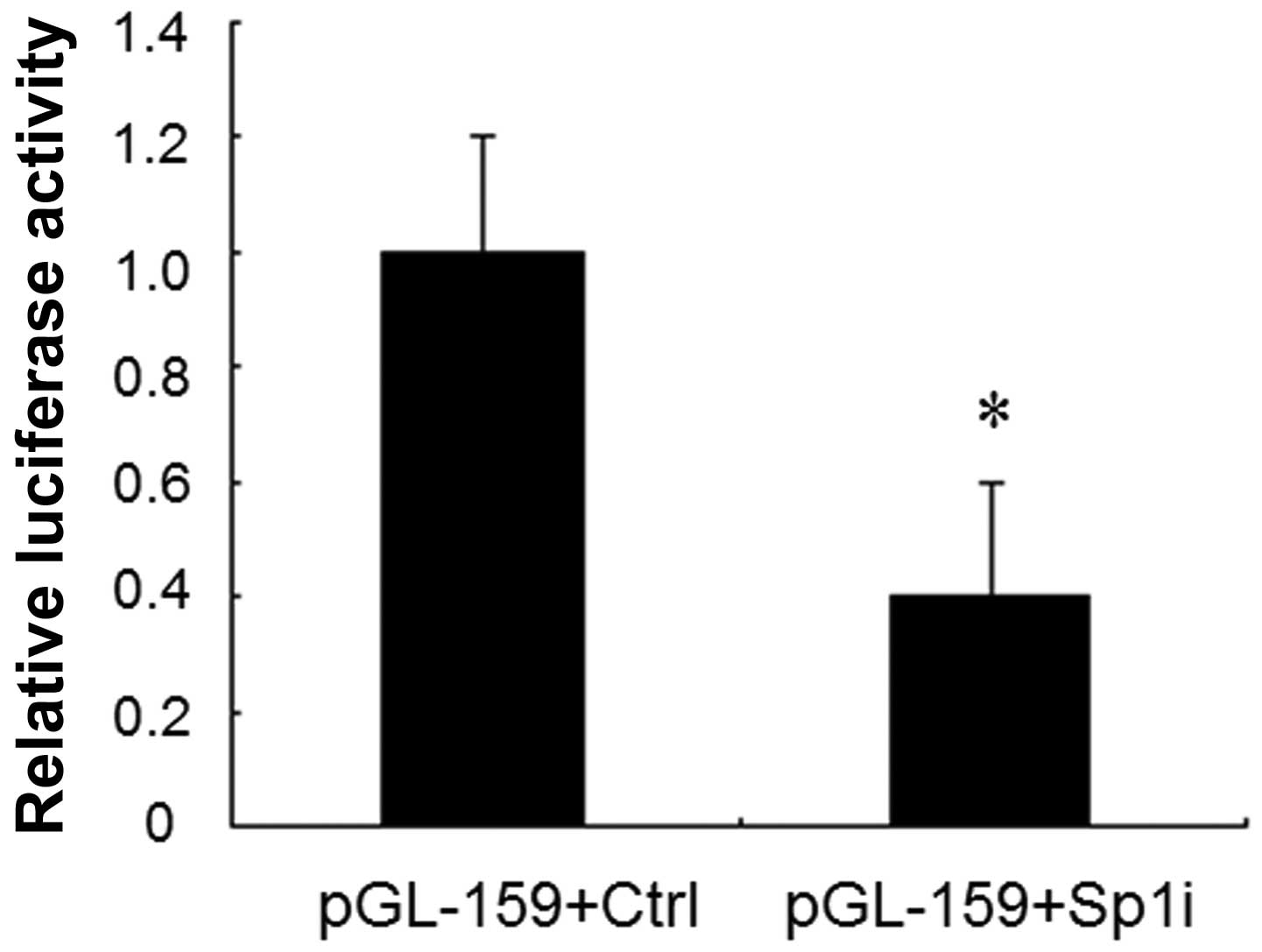
Knockdown of Sp1 expression by siRNA
decreases IRF-3 spliced variant promoter activity
Our previous study demonstrated that a Sp1 binding
site mutation resulted in a 50% decrease in the promoter activity
compared to that of the unmodified promoter of Sp1 (12). To further confirm the role of Sp1 in
the regulation of IRF-3 spliced variant promoter activity, we used
RNA interference to knock down Sp1 expression in HEK-293T cells. We
cotransfected the reporter plasmids pGL3-159 together with Sp1
siRNA or control vector into HEK-293T cells individually. A 60%
decrease of luciferase activity was observed in the presence of Sp1
siRNA compared to that observed with the control vector (Fig. 2). These results further validated
that Sp1 upregulated the transcriptional activity of the IRF-3
spliced variant promoter.
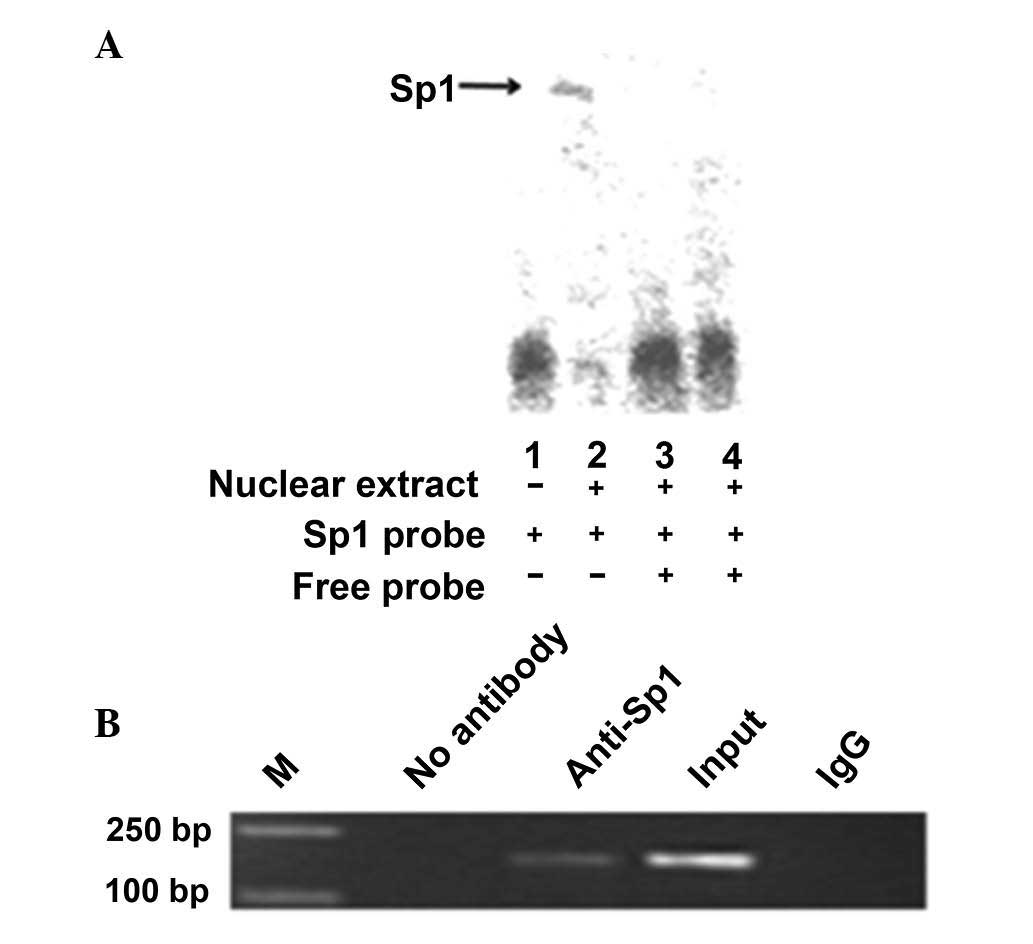
Sp1 binds to the IRF-3 spliced variant
promoter in vitro and in vivo
An electrophoretic mobility shift assay was
performed to investigate the possibility of Sp1 directly binding to
and upregulating the IRF-3 spliced variant promoter. A wild-type
oligonucleotide containing the Sp1 consensus binding site
(GGGGGATGGT) in the context of the IRF-3 promoter was synthesized.
The oligonucleotide was biotin-labeled and incubated with the
nuclear protein from HEK-293T cells. The nuclear protein bound to
the wild-type oligonucleotide, forming a protein-DNA complex
(Fig. 3A, lane 2). By contrast,
there was no protein-DNA complex formation when no nuclear extract
was incubated with the wild-type oligonucleotides (Fig. 3A, lane 1). Competition assays (using
10- and 100-fold excess of cold competitor oligonucleotides)
confirmed the absence of protein-DNA complex formation (Fig. 3A, lanes 3 and 4). The composition of
this protein-DNA complex was investigated by the addition of
antibodies against Sp1. Thus, Sp1 may upregulate the IRF-3 spliced
variant via directly binding to its promoter.
To determine whether Sp1 binds to the IRF-3 promoter
in vivo, we performed a ChIP assay (Fig. 3B), which was used for the detection
of proteins and the specific regions of DNA binding in vivo.
The HEK-293T cells were fixed with formaldehyde, lysed and the
chromatin was cleaved by nuclease digestion. The chromatin was then
immunoprecipitated with anti-IgG and anti-Sp1 antibodies and the
DNA precipitated in the complexes was subjected to PCR
amplification with primers flanking the region containing the
Sp1-binding site. The results indicated that Sp1 may directly
interact with the IRF-3 promoter in vivo.
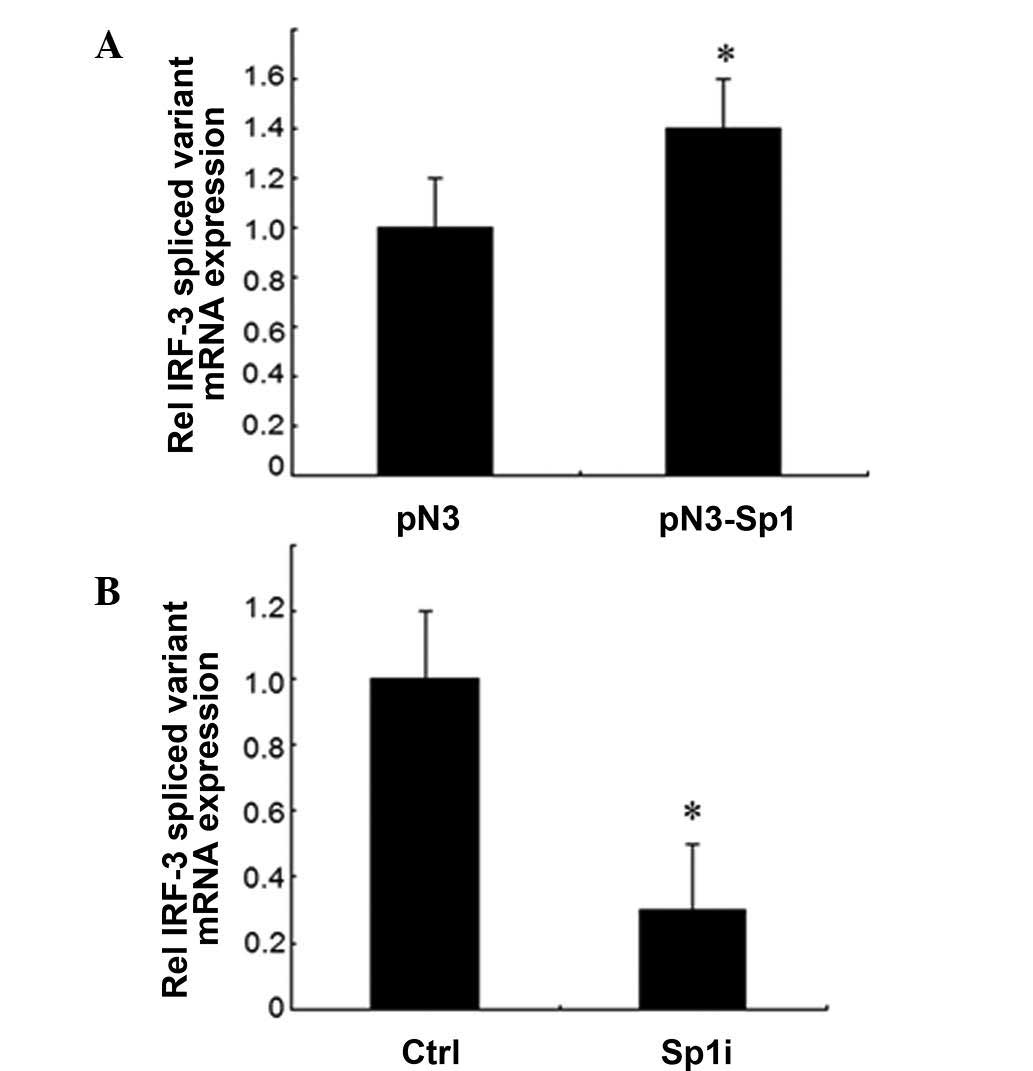
Sp1 leads to the increase of the
expression of IRF-3 spliced variant mRNA
To further investigate whether Sp1 affected the
expression of IRF-3, the mRNA levels of the IRF-3 spliced variant
were determined in HEK-293T cells following transient transfection
with the Sp1 expression vector or siRNA against Sp1 by qRT-PCR. The
transient expression of Sp1 resulted in a 40% increase of IRF-3
spliced variant mRNA expression compared to the control vector
(Fig. 4A). By contrast, the use of
Sp1 siRNA resulted in a 70% reduction of IRF-3 mRNA expression
compared to the control vector (Fig.
4B). These data demonstrated that Sp1 upregulated the
expression of endogenous IRF-3 gene at the level of
transcription.
Discussion
Alternative splicing is a key mechanism for
expanding transcript and protein diversity of mammalian genes.
Several diseases are associated with alternative splicing. Kappova
et al(9) described a second
mRNA that was generated from the IRF-3 gene by alternative splicing
and demonstrated that alternative splicing of the IRF-3
gene-encoded transcript resulted in the production of two isoforms
with antagonistic functions. It was also reported that the relative
value of IRF-3 and IRF-3a exerted an effect on carcinogenesis
(14). We previously identified two
new spliced variants of IRF-3, referred to as Int2V1 and Int2V2. By
generating a series of 5′ deletions, we demonstrated that the
Int2V1 core promoter located within the region −159/−100 bp
relative to the TSS and Sp1 transcription factor positively
regulated the human IRF-3 gene promoter. In this study, we aimed to
investigate the molecular mechanism underlying the upregulation of
promoter activity by Sp1.
Transcription factors may regulate target genes by
direct or indirect interaction with their promoters. The
transcription factor E2F1 represses the expression of IRF-3 by
directly binding to its promoter (15,16).
The CCAAT/enhancer-binding protein (C/EBP) family of transcription
factors augments proximal and distal promoter activation of LMP1 by
binding to a motif in the proximal promoter (17). The Epstein-Barr virus
immediate-early replication and transcription activator (Rta)
protein was shown to regulate the BRLF1 gene by indirect
interaction through the formation of an Sp1-MCAF1-Rta complex at
Sp1 sites (18).
Sp1 is a ubiquitous nuclear factor that plays a key
role in maintaining basal transcription of house-keeping genes. It
was demonstrated that Sp1 is involved in numerous cellular
processes, such as cell growth and differentiation. Sp1 is also
crucial in the growth and metastasis of several tumors by
regulating oncogenes, tumor suppressor genes, cell cycle control
molecules, growth-related signal transduction, angiogenesis-related
factors and apoptosis (19).
The Sp1 transcription factor regulates gene
expression through multiple mechanisms. Sp1 binds to GC-rich motifs
with high affinity and may regulate the expression of
TATA-containing and TATA-deficient genes via protein-protein
interactions or interaction with other transcription factors. It
was reported that Sp1 may be asociated with chromatin remodeling
through interactions with chromatin-modifying factors, such as p300
(20) and histone deacetylases
(21). We recently demonstrated
that the transcriptional factors Sp1 and Sp3 bound to the CD2AP
minimal promoter region and increased CD2AP expression at the mRNA
level in HEK-293 cells (22). In
this study, we demonstrated that the Sp1 protein bound to the Sp1
consensus binding site in the IRF-3 promoter in vitro by
electrophoretic gel mobility shift assays and antibody competition
assays. Chromatin immunoprecipitation assays also demonstrated that
Sp1 interacted with the IRF-3 promoter in vivo.
In summary, our studies identified a
cis-regulatory element within the spliced variant of the
human IRF-3 gene promoter and demonstrated that Sp1 directly bound
to this cis-regulatory element and the exogenous expression
of Sp1 significantly upregulated the transcription of the IRF-3
spliced variant. Characterizing the molecular regulation of human
IRF-3 and the transcription of its spliced variants may be an
important step towards elucidating the key role of IRF-3 in host
defense against viral and bacterial infection and cell growth
regulation.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation of China (nos. 30570863 and 30872804),
the Natural Science Foundation of Jiangsu Province, China (no.
BK2007244) and the Medical Academic Key Talent Program of Jiangsu
Province, China (no. RC2007050).
References
|
1
|
Taniguchi T, Ogasawara K, Takaoka A and
Tanaka N: IRF family of transcription factors as regulators of host
defense. Annu Rev Immunol. 19:623–655. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Paun A and Pitha PM: The IRF family,
revisited. Biochimie. 89:744–753. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mamane Y, Heylbroeck C, Genin P, et al:
Interferon regulatory factors: the next generation. Gene. 237:1–14.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Takaoka A, Tamura T and Taniguchi T:
Interferon regulatory factor family of transcription factors and
regulation of oncogenesis. Cancer Sci. 99:467–478. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Evsyukova I, Somarelli JA, Gregory SG and
Garcia-Blanco MA: Alternative splicing in multiple sclerosis and
other autoimmune diseases. RNA Biol. 7:462–473. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Onouchi Y, Gunji T, Burns JC, et al: ITPKC
functional polymorphism associated with Kawasaki disease
susceptibility and formation of coronary artery aneurysms. Nat
Genet. 40:35–42. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kozyrev SV, Abelson AK, Wojcik J, et al:
Functional variants in the B-cell gene BANK1 are associated with
systemic lupus erythematosus. Nat Genet. 40:211–216. 2008.
View Article : Google Scholar
|
|
8
|
Brenner T, Hamra-Amitay J, Evron T, Boneva
N, Seidman S and Soreq H: The role of readthrough
acetylcholinesterase in the pathophysiology of myasthenia gravis.
FASEB J. 17:214–222. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Karpova AY, Ronco LV and Howley PM:
Functional characterization of interferon regulatory factor 3a
(IRF-3a), an alternative splice isoform of IRF-3. Mol Cell Biol.
21:4169–4176. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li Y, Hu X, Song Y, Lu Z, Ning T, Cai H
and Ke Y: Identification of novel alternative splicing variants of
interferon regulatory factor 3. Biochim Biophys Acta. 1809:166–175.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ren W, Xu HG, Lu C, Jin R, Zou L, Wang Y
and Zhou GP: The characterization of two novel IRF-3 transcripts
starting from intron 2 of the wild type of IRF-3. Mol Biol Rep.
38:4415–4421. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ren W, Zhu LH, Xu HG, Jin R and Zhou GP:
Characterization of a spliced variant of human IRF-3 promoter and
its regulation by the transcription factor Sp1. Mol Biol Rep.
39:6987–6993. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Krüger I, Vollmer M, Simmons DG, Elsässer
HP, Philipsen S and Suske G: Sp1/Sp3 compound heterozygous mice are
not viable: impaired erythropoiesis and severe placental defects.
Dev Dyn. 236:2235–2244. 2007.PubMed/NCBI
|
|
14
|
Bourdon JC, Fernandes K, Murray-Zmijewski
F, et al: p53 isoforms can regulate p53 transcriptional activity.
Genes Dev. 19:2122–2137. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xu HG, Ren W, Lu C and Zhou GP:
Characterization of the human IRF-3 promoter and its regulation by
the transcription factor E2F1. Mol Biol Rep. 37:3073–3080. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu HG, Ren W, Zou L, Wang Y, Jin R and
Zhou GP: Direct repression of the human IRF-3 promoter by E2F1.
Immunogenetics. 63:189–196. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Noda C, Murata T, Kanda T, et al:
Identification and characterization of CCAAT enhancer-binding
protein (C/EBP) as a transcriptional activator for Epstein-Barr
virus oncogene latent membrane protein 1. J Biol Chem.
286:42524–42533. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chang LK, Chung JY, Hong YR, Ichimura T,
Nakao M and Liu ST: Activation of Sp1-mediated transcription by Rta
of Epstein-Barr virus via an interaction with MCAF1. Nucleic Acids
Res. 33:6528–6539. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sankpal UT, Goodison S, Abdelrahim M and
Basha R: Targeting Sp1 transcription factors in prostate cancer
therapy. Med Chem. 7:518–525. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Suzuki T, Kimura A, Nagai R and Horikoshi
M: Regulation of interaction of the acetyltransferase region of
p300 and the DNA-binding domain of Sp1 on and through DNA binding.
Genes Cells. 5:29–41. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhao S, Venkatasubbarao K, Li S and
Freeman JW: Requirement of a specific Sp1 site for histone
deacetylase-mediated repression of transforming growth factor beta
type II receptor expression in human pancreatic cancer cells.
Cancer Res. 63:2624–2630. 2003.
|
|
22
|
Xu HG, Ren W, Zou L, Wang Y, Jin R and
Zhou GP: Transcriptional control of human CD2AP expression: the
role of Sp1 and Sp3. Mol Biol Rep. 39:1479–1486. 2012. View Article : Google Scholar : PubMed/NCBI
|