Introduction
Breast cancer is the leading cause of mortality
among women worldwide, although the underlying molecular mechanisms
have not been fully elucidated. Estrogens are crucial in breast
cancer, with 60–70% of the cases expressing estrogen receptors
(ERs), predominantly the α-ER, which is encoded by the ESR1
gene (1,2). Therefore, the elucidation of the
mechanisms underlying the effect of estrogens on breast cancer is
of paramount importance.
The CTCFL gene, encoding the CTCFL protein,
also referred to as BORIS (Brother of the Regulator of Imprinting
Sites), has recently emerged as a potential biomarker of female
breast cancer, as it is normally expressed only by male germ cells.
In a previous study, it was demonstrated that the CTCFL gene
is expressed in malignant and non-malignant breast cell lines, as
well as in ~70% of the clinical specimens of breast cancer, but not
in normal breast tissue (3).
CTCFL is a paralogue of CTCF, the gene encoding CTCF,
a ubiquitous 11-zinc finger protein with highly versatile
functions, such as the global three-dimensional genome
organization, including intra- and interchromosomal loop formation
(4–8). CTCF is involved in transcriptional
silencing or activation and may function as an insulator and
chromatin organizer. CTCF shares the 11-zinc finger protein region
with CTCFL and the co-expression of the two genes was previously
demonstrated in breast cancer (9).
However, the expression of CTCFL in breast cancer is
currently highly controversial. Since the first report of its
expression in the majority of clinical breast specimens (3), subsequent studies were highly
divergent, with results ranging from complete absence of
CTCFL expression in breast cancer (10), to its ubiquitous expression in
normal and malignant tissues (11).
A positive correlation between the levels of CTCFL
and ER in breast tumors was previously described (3), suggesting that CTCFL may be under
estrogen regulation. In addition, there exists a coordinated
interaction between CTCF and ER in breast cancer cells, as CTCF
binding to DNA co-localizes with ER sites (12,13);
it is hypothesized that in these sites of co-localization, CTCF may
mark the euchromatic regions, allowing ER to bind and activate or
repress the expression of target genes. Therefore, a
pro-transcriptional role was suggested for CTCF in ER-mediated gene
expression in breast cancer cells (13). CTCF and CTCFL appear to be
associated with estrogens and ER in breast cancer; however, the
knowledge on this subject is currently scant.
The role of estrogens on the regulation of
CTCF and CTCFL gene expression has not yet been
investigated and its determination may help elucidate the biology
of breast cancer. Therefore, the aim of this study was to
investigate the effect of 17β-estradiol (E2) on the CTCF and
CTCFL mRNA expression in MCF7 breast cancer cells, which
represent a suited model for the in vitro study of
estrogenic pathways, as they express ERs.
Materials and methods
Cells
The MCF7 (HTB-22) cell line was obtained from the
American Type Culture Collection (Manassas, VA, USA). This cell
line was derived from the breast adenocarcinoma of a Caucasian
female and was shown to express ERs. The MCF7 cells also express
high CTCF levels (14).
Cell culture conditions
Immediately following their acquisition, the MCF7
cells were propagated by culture in 60-mm polystyrene dishes with
Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10%
fetal bovine serum (FBS), amphotericin and gentamycin, at 37°C in a
5% CO2/95% air atmosphere. In order to determine the
estrogenic effect on CTCF and CTCFL transcription,
the MCF7 cell cultures at high density were incubated for 24 h in
DMEM, with 0.2% human albumin instead of FBS. Subsequently, the
cells were incubated for 20 h with E2
(1,3,5-estratriene-3,17β-diol; Sigma-Aldrich, St. Louis, MO, USA)
at concentrations of 0.01, 0.1 and 1 μM. At the end of the
incubation period, total RNA was obtained with TRIzol reagent
(Invitrogen Life Technologies, Carlsbad, CA, USA) according to the
manufacturer’s instructions. RNA quantitation and purity were
determined by spectrophotometry in a Beckman Coulter DU730
apparatus (Beckman Coulter Inc., Fullerton, CA, USA) at an
absorbance of 260 nm and an absorbance ratio of 260/280 nm,
respectively. The final product was stored at −40°C until use in
quantitative reverse transcription polymerase chain reaction
(qRT-PCR) within the following 3 days.
The E2 was prepared as a 1×10−3 M stock
solution in absolute ethanol. The controls included ethanol, which
in previous experiments did not exert any effect on the expression
of the genes under investigation. The experiments were performed in
triplicate and repeated three times per biological replica.
qPCR for CTCF and CTCFL mRNA
expression
First, cDNA was synthesized in 20-μl volume
reactions from 1 μg total RNA with the Verso cDNA synthesis kit
(Thermo Fisher Scientific, Waltham, MA, USA). The obtained cDNA (1
μl) was then amplified in 15-μl volume reactions with gene-specific
primers and probes of the Solaris qPCR Gene Expression Assay system
(Thermo Fisher Scientific) according to the manufacturer’s
recommendations. GAPDH was used as a reference gene due to
its good performance, as previously reported (15). The primers had consensus sequences
that recognized all the splice variants of the genes under
investigation and they all exhibited identical temperature
conditions for amplification. qPCR was performed in a Eco
thermocycler (Illumina, San Diego, CA, USA) under the following
conditions: a 15-min step at 95°C to activate polymerase, followed
by 40 cycles at 95°C for DNA denaturation and 60°C for
annealing-extension. The efficiency of the reactions was 99, 95 and
101% for GAPDH, CTCF and CTCFL, respectively.
A formalin-fixed paraffin-embedded specimen of breast cancer
expressing CTCFL was used to determine the efficiency and
specificity of the amplification of this gene, as its expression in
MCF7 cells was not detected under any conditions, as described
below.
Statistical analysis
The comparative data were analyzed with REST 2009
software (Qiagen GmbH, Hilden, Germany) employing 6,000
randomizations. P<0.05 was considered to indicate a
statistically significant difference.
Methylation analysis of the CTCFL
promoter
Genomic DNA (1 μg) was bisulfite-modified with the
Imprint DNA Modification kit (MOD50; Sigma-Aldrich) and eluted in a
final volume of 20 μl, following the manufacturer’s protocol. The
modified DNA was stored at −40°C and used within 1 week.
Methylation-specific PCR (MSP) for the CTCFL promoter was
performed with 1 μl of the modified DNA in a final reaction volume
of 25 μl, containing 12.5 μl of GoTaq Master Mix (Promega Inc.,
Madison, WI, USA), 0.5 μl of each forward and reverse primers
described elsewhere (16), and 10.5
μl of water.
The PCR amplification consisted of 35 cycles of
denaturation at 95°C for 45 sec, annealing at 55°C for 45 sec and
extension at 72°C for 45 sec. The amplification products were
analyzed by agarose gel electrophoresis with ethidium bromide
staining.
Estrogen response elements in CTCF and
CTCFL promoters
Data mining for localization of the ER binding sites
in the human CTCF and CTCFL promoter was performed
using the online software LASAGNA-Search, developed by the
Department of Computer Science and Engineering, University of
Connecticut, Storrs, CT (17)
available at http://biogrid.engr.uconn.edu/lasagna_search/.
Results
Effect of E2 on CTCF and CTCFL mRNA
expression
In order to investigate the effects of E2 on the
mRNA transcription of CTCF and CTCFL, qRT-PCR was
performed. The MCF7 cells exhibited a basal mRNA transcription as
expected. E2 exerted a statistically significant downregulating
effect to 0.68 the value of the control. E2 at concentrations of
0.01 and 0.01 μM also exerted a downregulating effect, although it
was of no statistical significance (Table I).
 | Table IEffect of E2 on CTCF
transcription in MCF7 cells. |
Table I
Effect of E2 on CTCF
transcription in MCF7 cells.
| E2 (μM) | Relative
expression | 95% CI | P-value |
|---|
| 0.01 | 0.834 | 0.588–1.177 | 0.201 |
| 0.1 | 0.830 | 0.368–1.662 | 0.177 |
| 1 | 0.684 | 0.343–0.857 | 0.000 |
By contrast, basal transcription of CTCFL in
MCF7 cells was not detected and incubation with E2 did not result
in CTCFL mRNA upregulation under any hormone concentration.
Therefore, the possible downregulating effects of E2 on
CTCFL expression could not be assessed.
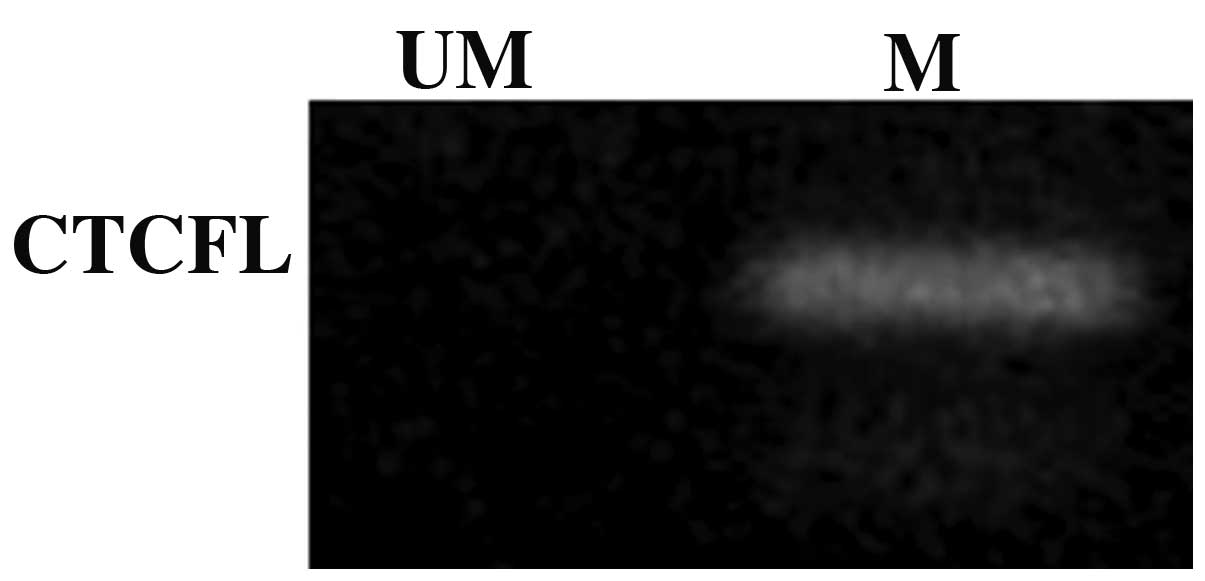
Analysis of methylation of the CTCFL
promoter
To further investigate the underlying mechanisms
responsible for the lack of expression of the CTCFL gene,
the methylation state of its promoter was determined by MSP under
basal conditions and only the methylated promoter was detected
(Fig. 1), which was suggestive of
gene silencing.
Data mining for estrogen response
elements in the CTCF and CTCFL promoters
As estrogen response elements have not been
described in the CTCF and CTCFL promoters, a search
for consensus sequences was performed with the LASAGNA-Search
software. Only a consensus sequence for CTCF was identified
in the minus strand, 247 bp upstream from the transcription start
site; this sequence was GTCCGCTTGACCT. By contrast, consensus
sequences for CTCFL were not identified.
Discussion
ER are the driving transcription factors in the
majority of breast cancers; when coupled to estrogens they activate
or inhibit genes involved in cell cycle progression and cell
survival during malignant transformation. CTCF is linked to the ER
biology through interactions that have not yet been fully
elucidated. CTCFL is a CTCF paralogue, which is expressed in breast
cancer. To date, a role for estrogens in the regulation of CTCF and
CTCFL expression has not been reported. In this study, the effect
of E2 on the transcription of the CTCF and CTCFL
genes in the MCF7 breast cancer cell line was investigated.
We did not detect any basal CTCFL
transcription in MCF7 cells. There are discordant reports regarding
CTCFL expression in this cell line; Hines et
al(10) and Vatolin et
al(18) did not detect any
CTCFL expression by conventional RT-PCR and/or qPCR methods.
By contrast, Renaud et al(19) reported CTCFL expression in
MCF7 cells measured by qPCR and northern blot analysis. In
addition, D’Arcy et al(3)
detected CTCFL expression by RT-PCR, western blot analysis
and immunostaining. The inconsistency between those studies may be
attributed to the heteroclonal nature of MCF7 cells; this cell line
was established in 1973 and MCF7 cells have since been widely
distributed worldwide, resulting in different stocks exhibiting
clonal heterogeneity (20). In the
studies that were in agreement with the present study, the source
of MCF7 cells was not specified by Vatolin et al(18), who mentioned that some of the cell
lines they used were obtained from ATTC, whereas Hines et
al(10) mentioned that the cell
lines were obtained from ATCC. As regards studies in disagreement
with the present study, D’Arcy et al(3) acknowledge M. O’Hare and B. Gusterson
for providing breast cell lines, whereas Renaud et
al(19) did not mention the
source. Therefore, the clonal heterogeneity of the MCF7 cells
appears to be a plausible explanation for the divergence in
CTCFL expression results.
Basal expression of CTCF was found as
expected, according to previously reported findings (14). CTCF is a ubiquitously expressed
regulator of fundamental cellular events, including transcription,
intra- and interchromosomal interactions and chromatin structure
(7). The ENCODE project (21) unveiled the significance of CTCF in
long-range chromatin interactions. A total of 77,811 distinct
binding sites for CTCF were identified across 19 cell types, which
underlines the importance of this factor in maintaining genome
integrity (22), although up- and
downregulation of its expression by diverse stimuli have been
reported; for example, CTCF overexpression has been
associated with resistance to apoptosis in breast cancer cell lines
(14) and its downregulation has
been reported in epithelial ovarian cancer (23). In this study, a downregulating
effect on CTCF transcription by E2 was documented; thus,
further studies are required to investigate whether this hormone
modulates CTCF transcription in vivo, directly or
indirectly.
The results of the promoter methylation analysis of
CTCFL are in agreement with the results obtained from the
gene transcription analysis depicted above, as the methylation of
the CTCFL promoter is indicative of gene silencing.
Furthermore, the data mining for consensus sequences
of estrogen response elements in the promoters was in agreement
with the results, as only a sequence for CTCF was
identified, suggesting that it is a target for ERs, unlike
CTCFL. However, distant estrogen response elements in the
genes and long-range interactions, as well as indirect effects of
E2 on the two genes, cannot be excluded.
In conclusion, this study demonstrated that E2
downregulated CTCF mRNA expression in MCF7 cells, which did
not exhibit basal transcription of CTCFL, whereas E2 did not
exert any upregulating effects on CTCFL mRNA. These results
suggest that there is an independent association between ER
positivity and CTCFL expression in breast cancer. However,
further investigations on this subject are required.
Acknowledgements
E.P.D.C. received a fellowship as a research
assistant from the Council of Science and Technology of the State
of Durango (COCYTED), Durango, Mexico.
References
|
1
|
Russo J and Russo IH: The role of estrogen
in the initiation of breast cancer. J Steroid Biochem Mol Biol.
102:89–96. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Castoria G, Migliaccio A, Giovannelli P
and Auricchio F: Cell proliferation regulated by estradiol
receptor: Therapeutic implications. Steroids. 75:524–527. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
D’Arcy V, Pore N, Docquier F, et al:
BORIS, a paralogue of the transcription factor, CTCF, is aberrantly
expressed in breast tumours. Br J Cancer. 98:571–579.
2008.PubMed/NCBI
|
|
4
|
Phillips JE and Corces VG: CTCF: master
weaver of the genome. Cell. 137:1194–1211. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ohlsson R, Lobanenkov V and Klenova E:
Does CTCF mediate between nuclear organization and gene expression?
Bioessays. 32:37–50. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Botta M, Haider S, Leung IX, Lio P and
Mozziconacci J: Intra- and inter-chromosomal interactions correlate
with CTCF binding genome wide. Mol Syst Biol. 6:4262010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Handoko L, Xu H, Li G, et al:
CTCF-mediated functional chromatin interactome in pluripotent
cells. Nat Genet. 43:630–638. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nakahashi H, Kwon KR, Resch W, et al: A
genome-wide map of CTCF multivalency redefines the CTCF code. Cell
Rep. 3:1678–1689. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Loukinov DI, Pugacheva E, Vatolin S, et
al: BORIS, a novel male germ-line-specific protein associated with
epigenetic reprogramming events, shares the same 11-zinc-finger
domain with CTCF, the insulator protein involved in reading
imprinting marks in the soma. Proc Natl Acad Sci USA. 99:6806–6811.
2002. View Article : Google Scholar
|
|
10
|
Hines WC, Bazarov AV, Mukhopadhyay R and
Yaswen P: BORIS (CTCFL) is not expressed in most human breast cell
lines and high grade breast carcinomas. PLoS One. 5:e97382010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jones TA, Ogunkolade BW, Szary J, et al:
Widespread expression of BORIS/CTCFL in normal and cancer cells.
PLoS One. 6:e223992011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dutertre M, Gratadou L, Dardenne E, et al:
Estrogen regulation and physiopathologic significance of
alternative promoters in breast cancer. Cancer Res. 70:3760–3770.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ross-Innes CS, Brown GD and Carroll JS: A
co-ordinated interaction between CTCF and ER in breast cancer
cells. BMC Genomics. 12:5932011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Docquier F, Farrar D, D’Arcy V, et al:
Heightened expression of CTCF in breast cancer cells is associated
with resistance to apoptosis. Cancer Res. 65:5112–5122. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lanoix D, Lacasse AA, St-Pierre J, Taylor
SC, Ethier-Chiasson M, Lafond J and Vaillancourt C: Quantitative
PCR pitfalls: the case of the human placenta. Mol Biotechnol.
52:234–243. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hong JA, Kang Y, Abdullaev Z, et al:
Reciprocal binding of CTCF and BORIS to the NY-ESO-1 promoter
coincides with derepression of this cancer-testis gene in lung
cancer cells. Cancer Res. 65:7763–7774. 2005.PubMed/NCBI
|
|
17
|
Lee C and Huang CH: LASAGNA-Search: an
integrated web tool for transcription factor binding site search
and visualization. Biotechniques. 54:141–153. 2013.PubMed/NCBI
|
|
18
|
Vatolin S, Abdullaev Z, Pack SD, et al:
Conditional expression of the CTCF-paralogous transcriptional
factor BORIS in normal cells results in demethylation and
derepression of MAGE-A1 and reactivation of other cancer-testis
genes. Cancer Res. 65:7751–7762. 2005.
|
|
19
|
Renaud S, Pugacheva EM, Delgado MD, et al:
Expression of the CTCF-paralogous cancer-testis gene, brother of
the regulator of imprinted sites (BORIS), is regulated by three
alternative promoters modulated by CpG methylation and by CTCF and
p53 transcription factors. Nucleic Acids Res. 35:7372–7388. 2007.
View Article : Google Scholar
|
|
20
|
Nugoli M, Chuchana P, Vendrell J, et al:
Genetic variability in MCF-7 sublines: evidence of rapid genomic
and RNA expression profile modifications. BMC Cancer. 3:132003.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
ENCODE Project Consortium. Bernstein BE,
Birney E, Dunham I, et al: An integrated encyclopedia of DNA
elements in the human genome. Nature. 489:57–74. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang H, Maurano MT, Qu H, et al:
Widespread plasticity in CTCF occupancy linked to DNA methylation.
Genome Res. 22:1680–1688. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Link PA, Zhang W, Odunsi K and Karpf AR:
BORIS/CTCFL mRNA isoform expression and epigenetic
regulation in epithelial ovarian cancer. Cancer Immun.
13:62013.
|