Introduction
Lung cancer is the most frequently occurring type of
cancer worldwide and the leading cause of cancer mortality, with a
five-year survival rate ≤15% (1).
Smoking is the primary risk factor for lung cancer. However, lung
cancer develops in <20% of smokers (2). This suggests that environmental and
genetic factors play important roles in lung carcinogenesis. Over
the past few decades, the relationship between environmental and
genetic factors and cancer susceptibility has been widely
investigated. Folate metabolism is considered to be important role
in carcinogenesis through its involvement in the process of DNA
methylation and repair. Methylenetetrahydrofolate reductase (MTHFR)
is located in the short arm of chromosome 1 (1p36.3) and is
involved in DNA methylation and synthesis as an important enzyme in
the folic acid metabolic process. C677T (exon 4) and A1298C (exon
7) are two common polymorphisms in the MTHFR gene, which decrease
enzyme activity compared to the wild-type gene, leading to DNA
repair and methylation disorders and playing an important role in
carcinogenesis (3). Numerous
epidemiological studies investigated the association between MTHFR
C677T and A1298C gene polymorphism and lung cancer predisposition.
However, the results were inconsistent, possibly due to the small
sample size, selection bias or other factors. Meta-analysis is a
valid method to assess disparate results. In 2008, Mao et
al(4) performed a meta-analysis
that included eight articles; however, they did not identify a
significant correlation between the MTHFR polymorphisms and lung
cancer globally. The ethnic lines involved in the meta-analysis
were diverse, including Asians, non-Hispanic whites and Europeans.
MTHFR mutation frequency is diverse between various ethnicities.
The incidence of the C677T homozygous mutant is ∼20% in Chinese,
18% in Italian and 11% in African populations (5). Lung cancer morbidity, histological
type and molecular typing also differ among different ethnic lines.
To the best of our knowledge, there is currently no meta-analysis
available on MTHFR and lung cancer susceptibility in East Asian
populations. Over the past few years, newly published studies were
conducted, most of which involved Japanese, South Korean and
Chinese populations. In order to reduce research bias, we performed
a meta-analysis with the studies involving East Asian populations
of identical race, to investigate the relationship between MTHFR
gene polymorphism and lung cancer susceptibility.
Materials and methods
Publication search
To identify all the studies investigating the
association between MTHFR gene polymorphism and lung cancer
susceptibility, computer searches of literature databases, such as
PubMed, EMBASE, Web of Science, Chinese Biomedicine and CNKI (prior
to November 30, 2012) were performed, using the following MeSH
terms and key words: ‘methylenetetrahydrofolate reductase’ or
‘MTHFR’ and ‘lung’ and ‘cancer’ or ‘carcinoma’. The studies were
read in their entirety and their references were also reviewed to
identify other relevant studies.
Inclusion and exclusion criteria
Eligible studies were required to meet the following
criteria: i) exploration of MTHFR polymorphisms and lung cancer
risk in East Asian population; ii) unrelated case-control or cohort
studies; iii) sufficient published data for estimating an odds
ratio (OR) with 95% confidence intervals (CIs); iv) allelic
distributions in the control group consistent with the
Hardy-Weinberg equilibrium (HWE). Exclusion criteria were as
follows: i) studies that did not provide the population sources of
cases and controls; ii) when more than one study involved the same
sample population, only the best quality one was included.
Data extraction
Information was carefully extracted from the
eligible publications according to the inclusion criteria listed
above. The following data were collected from each study: first
author’s surname, year of publication, country, sample size, source
of controls, genotyping method, number of cases and controls for
each C677T or A1298C genotype, smoking status, histological type
and alcohol consumption.
Statistical analysis
HWE was tested by the Chi-square test. P<0.05 was
considered to indicate a statistically significant difference
(6). Crude ORs with their
corresponding 95% CIs were used to assess the association between
MTHFR C677T and A1298C polymorphisms and lung cancer risk (7). The pooled ORs were estimated for MTHFR
C677T genotypes and MTHFR A1298C genotypes. The heterogeneity
between studies was assessed using the Q statistic (8). When heterogeneity was considered to be
significant (P<0.10), the random-effects model was used. In the
case of non-significant heterogeneity (P≥0.10), the fixed-effects
model was used. We quantified heterogeneity using the I2
metric score (I2<25%, no heterogeneity;
I2=25–50%, moderate heterogeneity; and
I2>50%, significant heterogeneity) (9). Subgroup analyses were also performed
on the basis of the source of controls, smoking status and
histological types. Sensitivity analysis was conducted to determine
whether the exclusion of any one study affected the initial
results. Funnel plots were used to assess publication bias. An
asymmetric plot suggested possible publication bias. Egger’s linear
regression test (10) was performed
to determine the significance of the asymmetry. P<0.05 was
considered to indicate publication bias. Analyses were performed
using Review Manager software version 5.0.
Results
Study characteristics
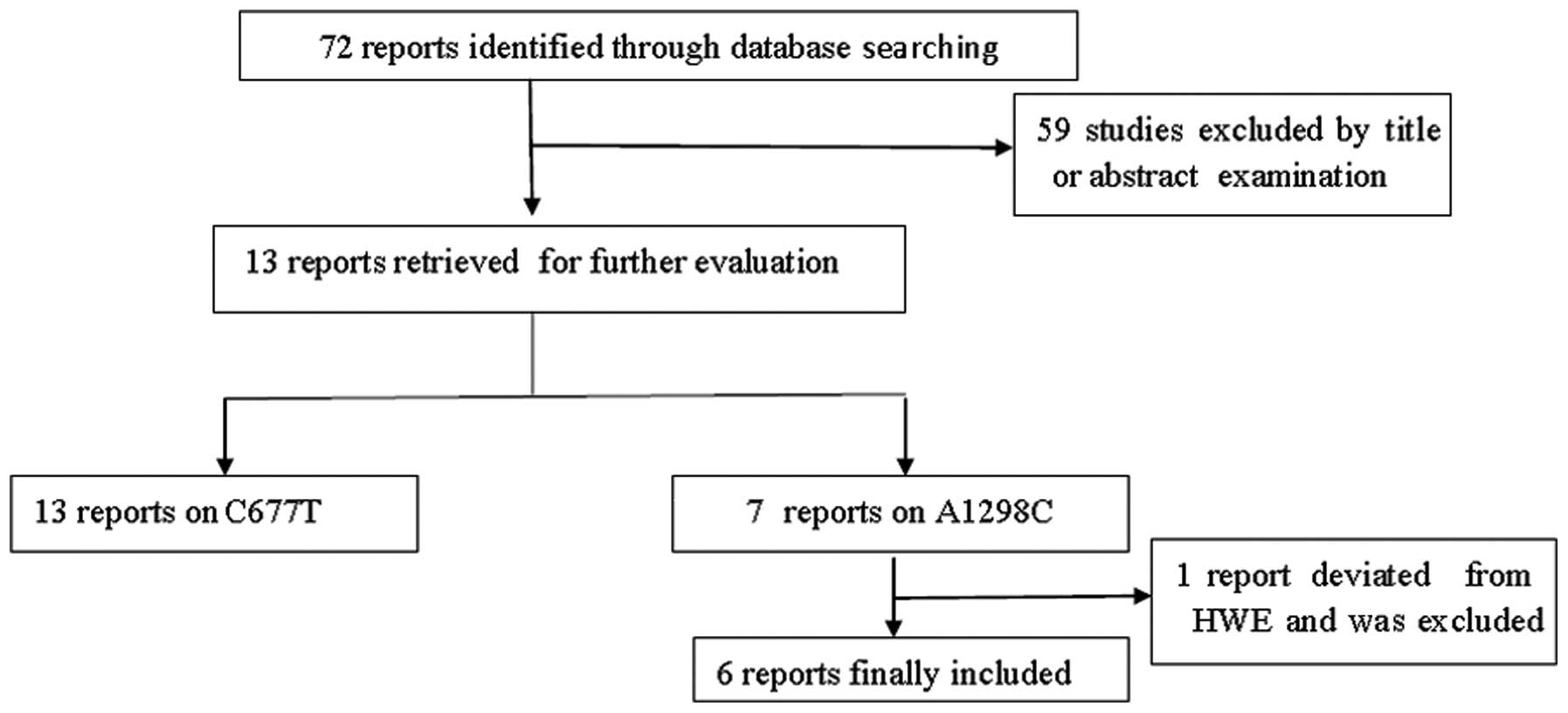
A total of 72 studies were preliminarily retrieved
based on the search terms and 13 studies (11–23)
met the inclusion criteria for detailed analysis following abstract
scanning and full-text assessment (Fig.
1). The possible eligible papers were retrieved by two
independent investigators (X.D. Zhang and W. Li). All eligible
articles provided data on C677T and seven of them also provided
data on A1298C. The main characteristics of the studies are listed
in Table I. However, A1298C allelic
distributions in the control population of one study (21) significantly deviated from HWE.
Thirteen studies (7,409 cases and 6,331 controls) on C677T
genotypes, among which there were six articles (2,049 cases and
2,851 controls) on A1298C genotypes, were included in our
meta-analysis. The included studies had all been conducted on East
Asian populations. Controls of four studies (11,12,18,19)
were derived from hospital-based participants and controls of nine
studies (13–17,20–23)
were derived from population-based participants.
 | Table IData of methylenetetrahydrofolate
reductase (MTHFR) polymorphisms included in this study. |
Table I
Data of methylenetetrahydrofolate
reductase (MTHFR) polymorphisms included in this study.
| Authora | Year of
publication | Country | Test methods | MTHFR genotype | HWE of control | Cases (n) | Controls (n) | Source of
controls | Refs. |
|---|
| Ma et al | 2012 | China | PCR-RFLP | C677T | 0.83 | 120 | 60 | Hospital-based | (11) |
| Kiyohara et
al | 2011 | Japan | PCR-RFLP | C677T | 0.62 | 379 | 462 | Hospital-based | (12) |
| | | | A1298C | 0.63 | | | | |
| Cui et al | 2011 | China | Real-time PCR | C677T | 0.48 | 641 | 438 | Population-based | (13) |
| Cheng et
al | 2011 | China | PCR-RFLP | C677T | 0.77 | 178 | 180 | Population-based | (14) |
| Cui et al | 2011 | Korean | PCR-RFLP real-time
PCR | C677T | 0.14 | 1,700 | 3,938 | Population-based | (15) |
| Yang et
al | 2010 | China | DNA MassArray | C677T | 0.52 | 120 | 165 | Population-based | (16) |
| Liu et al | 2009 | China | PCR-RFLP | C677T | 0.67 | 716 | 358 |
Population-based | (17) |
| | | | A1298C | 0.49 | | | | |
| Liu et
al | 2008 | China | SNP gene chips | C677T | 0.48 | 517 | 500 | Hospital-based | (18) |
| | | | A1298C | 0.62 | | | | |
| Suzuki et
al | 2007 | Japan | Real-time PCR | C677T | 0.17 | 1,030 | 515 | Hospital-based | (19) |
| | | | A1298C | 0.51 | | | | |
| Jin et
al | 2007 | China | PCR-RFLP | C677T | 0.83 | 100 | 100 |
Population-based | (20) |
| | | | A1298C | 0.73 | | | | |
| Zhang et
al | 2005 | China | PCR-RFLP | C677T | 0.15 | 500 | 505 |
Population-based | (21) |
| | | | A1298C |
0.01a | | | | |
| Shen et
al | 2005 | China | Real-time PCR | C677T | 0.12 | C677T | C677T |
Population-based | (22) |
| | | | A1298C | 0.51 | 111 | 116 | | |
| | | | | | A1298C | A1298C | | |
| | | | | | 109 | 114 | | |
| Jeng et
al | 2003 | China | PCR-RFLP | C677T | 0.44 | 232 | 59 |
Population-based | (23) |
Quantitative synthesis
There was significant heterogeneity (P<0.0001,
I2=71%) among the 13 studies on C677T. To eliminate
heterogeneity, we subgrouped the 13 studies based on the sources of
controls; however, the heterogeneity was not eliminated. The pooled
ORs were estimated using the random-effects model. The results
demonstrated a significantly increased risk of lung cancer in the
MTHFR 677TT compared to the C677CC/CT genotype (OR=1.24; 95% CI,
1.01–1.52) (Fig. 2). No statistical
significance was observed between other C677T gene models and lung
cancer (Table II). In a subgroup of
hospital-based controls, the C677TT genotype exhibited a
significantly increased risk of lung cancer compared to the C677CC
genotype (OR=3.01; 95% CI, 1.07–8.46). There was no significant
association for population-based controls (OR=1.02; 95% CI,
0.55–1.89). Two published studies on MTHFR polymorphism and lung
cancer risk in smokers provided detailed data. In the stratified
analysis, no association was found between MTHFR C677TT
polymorphism, compared to C677CC/CT, and the risk of lung cancer,
regardless of the smoking status. However, the study indicated that
the MTHFR 677TT genotype is associated with a significant increase
in the risk of lung squamous carcinoma, compared to C677CC/CT
(OR=1.53; 95% CI, 1.09–2.14), whereas no association was found
between the MTHFR C677TT genotype and the risk of lung
adenocarcinoma (Table III).
Stratified analysis based on alcohol consumption and folate intake
was not performed, since no sufficient data were reported. We
performed a sensitivity analysis by sequentially omitting
individual studies. As regards the C677T genotype, there was no
significant deviation from the initial ORs and no significant
association between C677T polymorphism and the risk of lung cancer
was observed. As regards the A1298C polymorphism, heterogeneity was
not observed among the six studies. There was no significant
relationship between A1298C polymorphism and lung cancer risk by
the fixed-effects model (Table II).
There was also no significant association in the subgroup analysis
according to the control source. We did not perform a stratified
analysis based on smoking status, histological type, alcohol
consumption and folate intake, since no sufficient data were
provided by A1298C studies. The relationships between the A1298C
polymorphisms and the risk of lung cancer were not altered during
the sensitivity analysis.
 | Table IIMeta-analysis of
methylenetetrahydrofolate reductase (MTHFR) polymorphisms and risk
of lung cancer. |
Table II
Meta-analysis of
methylenetetrahydrofolate reductase (MTHFR) polymorphisms and risk
of lung cancer.
| Genetic models | OR (95% CI) | I2
(%) | P-value |
|---|
| C677T | | | |
| TT vs. CC/CT | | | |
| All | 1.24
(1.01–1.52) | 71 | 0.0001 |
|
Hospital-based | 1.39
(0.85–2.27) | 85 | 0.0002 |
|
Population-based | 1.20
(0.90–1.51) | 63 | 0.005 |
| TT vs. CC | | | |
| All | 1.44
(0.86–2.39) | 94 | 0.00001 |
|
Hospital-based | 3.01
(1.07–8.46) | 94 | 0.00001 |
|
Population-based | 1.02
(0.55–1.89) | 94 | 0.00001 |
| TT/CT vs. CC | | | |
| All | 1.15
(0.84–1.56) | 92 | 0.00001 |
|
Hospital-based | 1.26
(0.90–1.74) | 77 | 0.004 |
|
Population-based | 1.08
(0.71–1.63) | 93 | 0.00001 |
| A1298C | | | |
| CC vs. AA/AC | | | |
| All | 1.18
(0.88–1.59) | 0 | 0.48 |
|
Hospital-based | 1.24
(0.88–1.76) | 0 | 0.44 |
|
Population-based | 1.02
(0.57–1.82) | 25 | 0.26 |
| CC vs. AA | | | |
| All | 1.18
(0.88–1.59) | 0 | 0.45 |
|
Hospital-based | 1.24
(0.87–1.75) | 4 | 0.35 |
|
Population-based | 1.04
(0.58–1.86) | 20 | 0.29 |
| CC/AC vs. AA | | | |
| All | 1.02
(0.90–1.15) | 0 | 0.76 |
|
Hospital-based | 1.01
(0.87–1.17) | 22 | 0.28 |
|
Population-based | 1.04
(0.90–1.15) | 0 | 0.76 |
 | Table IIIStratified analyses based on smoking
and histology type of the association between
methylenetetrahydrofolate reductase (MTHFR) C677T polymorphisms (TT
vs. CC/CT) and lung cancer. |
Table III
Stratified analyses based on smoking
and histology type of the association between
methylenetetrahydrofolate reductase (MTHFR) C677T polymorphisms (TT
vs. CC/CT) and lung cancer.
| Cases
| Controls
| | | |
|---|
| Subgroup
analyses | TT | CC/CT | TT | CC/CT | OR (95% CIs) | P-value | Refs. |
|---|
| Ever smoker | 104 | 498 | 72 | 661 | 1.38
(0.59–3.21) | 0.46 | (12,17) |
| Never smoker | 55 | 246 | 42 | 220 | 1.28
(0.78–2.10) | 0.33 | (12,17) |
| Squamous
carcinoma | 103 | 131 | 137 | 392 | 1.53
(1.09–2.14) | 0.01 | (11,13,14) |
| Adenocarcinoma | 88 | 142 | 137 | 392 | 2.13
(0.90–5.00) | 0.08 | (11,13,14) |
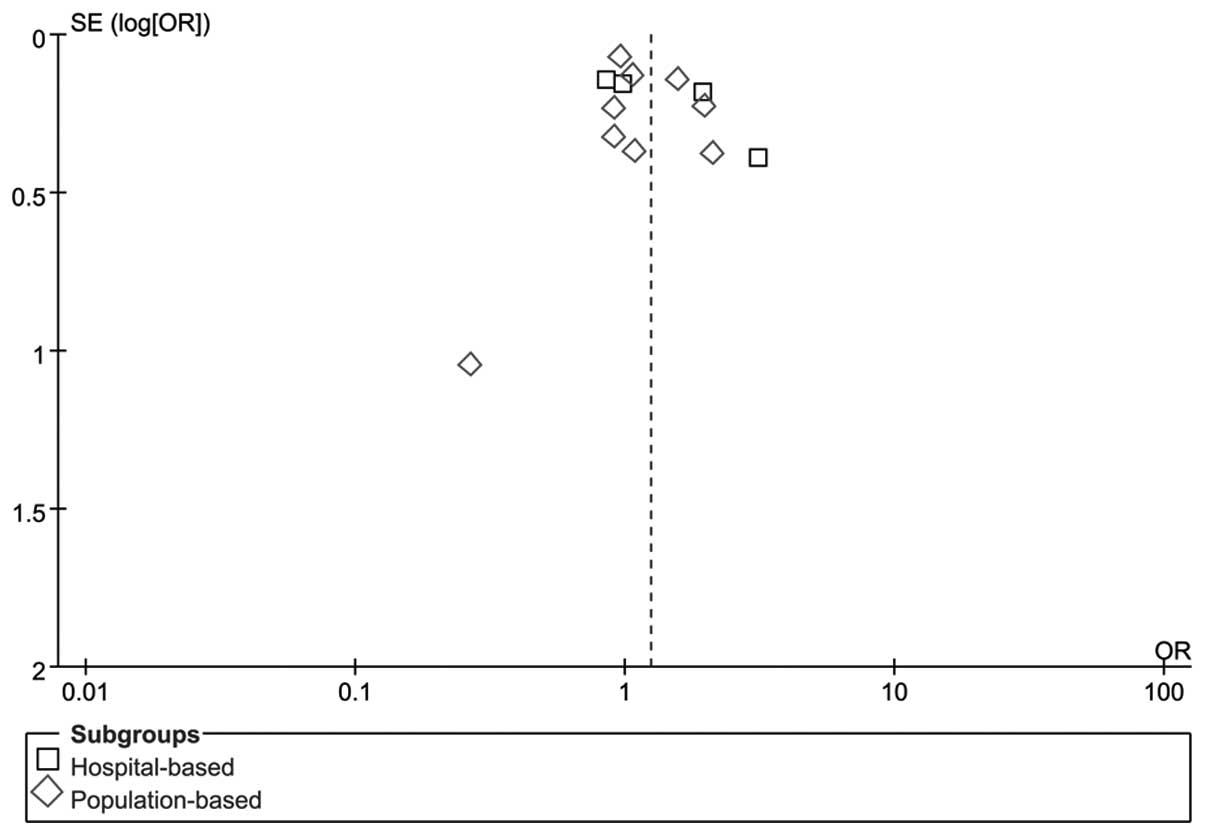
Publication bias
RevMan funnel plots and Egger’s tests were performed
to assess publication bias. The shapes of the funnel plots did not
reveal any obvious sign of asymmetry for C677TT vs. CC/CT (Fig. 3). Furthermore, Egger’s test was used
to provide statistical evidence of funnel plot symmetry. The
results revealed no evidence of publication bias (P=0.33). No
evidence of publication bias was observed in other gene models. The
results suggested that the data provided by these meta-analyses are
relatively reliable.
Discussion
Epidemiological studies have demonstrated that
environmental and genetic factors may contribute to the development
of the vast majority of tumors. Previously published studies
demonstrated that MTHFR polymorphisms were associated with breast
(24), colon (25), gastric (26) and other types of cancer. MTHFR may
play an important role in folate metabolism as a key enzyme, by
irreversibly catalyzing the generation of
5,10-methylenetetrahydrofolate (5,10-CH2-THF) to
5-methylenetetrahydrofolate (5-CH3-THF), which is a
methyl group donor for DNA repair and synthesis. The lack of a
methyl group leads to genome-wide hypomethylation, which affects
the stability of the genome and may easily lead to malignant
transformation of normal cells (27). Genome-wide hypomethylation is an
important characteristic of cancer cells. 5,10-CH2-THF
is a substrate of the thymidylate synthetase enzyme, is involved in
purine synthesis in the methylation of deoxyuridine monophosphate
(dUMP) nucleotide to deoxythymidine monophosphate (dTMP) and
contributes to DNA synthesis. In the case of dTMP shortage, it is
replaced by dUMP in DNA synthesis, leading to DNA double-strand
breaks and resulting in tumorigenesis (28). MTHFR C677T and A1298C are two common
genetic polymorphisms. The MTHFR variant genotype C677TT decreases
enzymatic activity by 70% (3) and
C677CT by 40% (29). MTHFR A1298C
polymorphism also reduces the specific activity of the enzyme,
although to a lesser extent. Decreased enzymatic activity of MTHFR
affects folate metabolism. Folate facilitates the provision of
methyl groups that are required for intracellular methylation
reactions and DNA repair. Thus, MTHFR polymorphism may increase the
risk of lung cancer, as well as of other malignancies (30). Published data from recent studies
demonstrated a significant correlation between MTHFR gene
polymorphism and the risk of lung cancer (11–14,17,20–23,31–33).
However, other studies failed to demonstrate such a correlation
(15,16,18,19,34–36).
The meta-analysis may provide comprehensive evaluation and
quantitive analysis of inconsistent results from individual studies
of identical aim. We conducted the meta-analysis to elucidate the
association of MTHFR C677T and A1298C polymorphisms with
susceptibility to lung cancer in East Asian populations. The
results indicate a significantly increased risk of lung cancer
associated with the MTHFR 677TT genotype compared to the MTHFR
677CC/CT genotypes, but indicated no positive relationship between
other MTHFR C677T genetic models or the A1298 genetic model and the
risk of lung cancer, when the studies were pooled together. The
harmful substances in tobacco may cause DNA damage, leading to gene
mutations, thus increasing the risk of lung and esophageal cancer,
as well as of other malignancies; by contrast, the adequate intake
of folate significantly reduces the risk of lung cancer (22). Therefore, folate metabolism and
smoking are crucial in the occurrence of lung cancer. In the
stratified meta-analysis based on smoking status, we did not
observe a positive relationship between MTHFR C677T gene
polymorphism and lung cancer risk, regardless of the smoking
status. The results suggested that smoking was not a predominant
factor affecting the association between MTHFR C677T polymorphisms
and lung cancer risk. Of note, only two published studies provided
sufficient data to assess the relationship between MTHFR
polymorphism and lung cancer risk in the presence of smoking.
Therefore, the results are not considered reliable. In the
stratified analysis based on histological types, we observed that
the MTHFR 677TT genotype is associated with a significant increase
in the risk of lung squamous carcinoma compared to C677CC/CT, but
is negative in lung adenocarcinoma. The results suggest that MTHFR
C677T polymorphisms may be related to histological type. We were
not able to perform a stratified analysis based on alcohol
consumption and folate intake, due to the insufficient data.
Heterogeneity may affect the interpretation of the
meta-analysis results. There was significant heterogeneity for
C677T among the 13 studies. Heterogeneity is generally divided into
clinical, methodological and statistical heterogeneity (37), which may lead to a meta-analysis
heterogeneity. Clinical heterogeneity includes differences in the
studied populations, sample selection (e.g., source of control
populations) and other risk factors. Methodological heterogeneity
may be mostly due to differences in trial design and quality.
Statistical heterogeneity is due to the variation of the treatment
effects estimated among different trials. Among these, clinical
heterogeneity is the most common reason for meta-analysis
heterogeneity. The identification of the source of heterogeneity is
one of the most important aims of a meta-analysis. Therefore, we
selected East Asian populations of identical race, leading similar
lifestyles and exposed to similar environmental factors, in order
to perform this meta-analysis. We also stratified the studies
according to the source of controls. Subsequently, a subgroup
analysis stratified according to the source of control population
demonstrated a significant heterogeneity as well, suggesting that
the source of the control population contributed only slightly to
the overall heterogeneity. In the subgroup analysis based on the
source of the control population, we observed a significantly
increased risk of lung cancer associated with the C677TT genotype,
compared to the C677CC genotype, in a subgroup of hospital-based
controls. However, we were unable to reach definitive conclusions
due to the small sample size of this study and the complexity of
the individuals included in the control groups, who may suffer from
other diseases associated with MTHFR mutations. Therefore, more
detailed information should be obtained to evaluate the
relationship among them. Heterogeneity may be due to the source of
control population including, not only hospital patients, but the
general population as well; subjects in the general population may
suffer from other diseases associated with MTHFR gene polymorphism,
such as hypertension (38),
cardiovascular and cerebrovascular diseases (39).
Certain limitations of this meta-analysis should be
acknowledged. Firstly, some individuals in the control group are
likely to suffer from diseases associated with MTHFR gene
polymorphism. Secondly, some individuals in the control group may
develop lung cancer years later, although they were negative for
signs of lung cancer at the time of the investigation. Thirdly, we
were not able to conduct a subgroup analysis according to age,
folate intake and alcohol consumption, due to the sparsity of
available data on lung cancer subtype. Finally, subgroup analyses
according to smoking status and histological type were performed,
but the sample size was insufficient to support the results.
Despite these limitations, the meta-analysis included a substantial
number of cases and controls from the studies on the association
between MTHFR polymorphism and lung cancer risk, which greatly
enhanced the statistical power of the analysis and provided enough
evidence for the authors to draw a reliable conclusion.
Furthermore, no publication bias was identified in the present
meta-analysis, which enhances the reliability of the meta-analysis
results.
In conclusion, this meta-analysis indicated that
MTHFR 677T polymorphism has a significant association with lung
cancer risk. However, since the number of the studies included in
our meta-analysis was limited, large-sample studies using
standardized unbiased methods, definitively diagnosed lung cancer
patients, more detailed individual data and well-matched controls
should be conducted. Moreover, lung cancer development results from
the interaction of genetic and environmental factors. Future
studies should consider gene-gene and gene-environment interactions
in lung cancer. Further investigations on MTHFR polymorphisms and
lung cancer susceptibility may elucidate the relationship between
the two and provide a theoretical basis for lung cancer
prevention.
Acknowledgements
This study was supported by the
National Natural Science Foundation of China (no. 81172234).
References
|
1
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J and
Thun M: Cancer statistics. CA Cancer J Clin. 59:225–249. 2009.
|
|
2
|
Schwartz AG: Lung cancer: family history
matters. Chest. 130:936–937. 2006.PubMed/NCBI
|
|
3
|
van der Put NM, Gabreels F, Stevens EM, et
al: A second common mutation in the methylenetetrahydrofolate
reductase gene: an additional risk factor for neural-tube defects?
Am J Hum Genet. 62:1044–1051. 1998.PubMed/NCBI
|
|
4
|
Mao RF, Fan Y, Jin Y, Bai J and Fu S:
Methylenetetrahydrofolate reductase gene polymorphisms and lung
cancer: a meta-analysis. J Hum Genet. 53:340–348. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Curtin K, Bigler J, Slattery ML, Caan B,
Potter JD and Ulrich CM: MTHFR C677T and A1298C polymorphisms:
diet, estrogen, and risk of colon cancer. Cancer Epidemiol
Biomarkers Prev. 13:281–292. 2004.PubMed/NCBI
|
|
6
|
Guo SW and Thompson EA: Performing the
exact test of Hardy-Weinberg proportion for multiple alleles.
Biometrics. 48:361–372. 1992. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Woolf B: On estimating the relation
between blood group and disease. Ann Hum Genet. 19:251–253. 1995.
View Article : Google Scholar
|
|
8
|
Zintzaras E and Ioannidis JP:
Heterogeneity testing in meta-analysis of genome searches. Genet
Epidemiol. 28:123–137. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Higgins JP and Thompson SG: Quantifying
heterogeneity in a meta-analysis. Stat Med. 21:1539–1558. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Egger M, Davey Smith G, Schneider M and
Minder C: Bias in meta-analysis detected by a simple, graphical
test. BMJ. 315:629–634. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ma QL, Li YF, Ji M, Yang KY, Wang JY, Li S
and Huang YC: The relationship of Methylenetetrahydrofolate
reductase SNP677C_T and lung cancer susceptibility. Chin J
Clinicians (Electronic Edition). 6:213–215. 2012.(In Chinese).
|
|
12
|
Kiyohara C, Horiuchi T, Takayama K and
Nakanishi Y: Methylenetetrahydrofolate reductase polymorphisms and
interaction with smoking and alcohol consumption in lung cancer
risk: a case-control study in a Japanese population. BMC Cancer.
11:4592011. View Article : Google Scholar
|
|
13
|
Cui LH, Yu Z, Zhang TT, Shin MH, Kim HN
and Choi JS: Influence of polymorphisms in MTHFR 677 C→T, TYMS
3R→2R and MTR 2756 A→G on NSCLC risk and response to platinum-based
chemotherapy in advanced NSCLC. Pharmacogenomics. 12:797–808.
2011.
|
|
14
|
Cheng Z, Wang W, Song YN, et al:
Investigation of the relationship between C677T polymorphisms of
methylenetetrahydrofolate reductase and lung cancer risk. Chin J
Tuberc Respir Dis. 34:57–58. 2011.(In Chinese).
|
|
15
|
Cui LH, Shin MH, Kim HN, et al:
Methylenetetrahydrofolate reductase C677T polymorphism in patients
with lung cancer in a Korean population. BMC Med Genet. 12:282011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang XX, Li FX, Yi J, Li X, Sun JZ and Hu
NY: An association of methylenetetrahydrofolate reductase reductase
C677T and gastric cancer, colorectal cancer and lung cancer
susceptibility. Guangdong Med J. 31:2375–2377. 2010.(In
Chinese).
|
|
17
|
Liu CS, Tsai CW, Hsia TC, et al:
Interaction of methylentetrahydrofolate reductase genotype and
smoking habit in Taiwanese lung cancer patients. Cancer Genomics
Proteomics. 6:325–329. 2009.PubMed/NCBI
|
|
18
|
Liu H, Jin G, Wang H, et al: Association
of polymorphisms in one-carbon metabolizing genes and lung cancer
risk: a case-control study in Chinese population. Lung Cancer.
61:21–29. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Suzuki T, Matsuo K, Hiraki A, et al:
Impact of one-carbon metabolism-related gene polymorphisms on risk
of lung cancer in Japan: a casecontrol study. Carcinogenesis.
28:1718–1725. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jin C, Zhang YH, Peng MF, Li WD, Ma L and
Chen WS: Study of the relationship between C677T, A1298C gene
polymorphisms of methylenetetrahydrofolate reductase and lung
cancer. Chin Clin Oncol. 12:671–675. 2007.
|
|
21
|
Zhang XM, Miao XP, Tan W, Qu SN, Sun T,
Zhou YF and Lin DX: Association between genetic polymorphisms in
methylentetrahydrofolate reductase and risk of lung cancer. Acta
Academiae Medicinae Sinicae. 27:700–703. 2005.(In Chinese).
|
|
22
|
Shen M, Rothman N, Berndt SI, et al:
Polymorphisms in folate metabolic genes and lung cancer risk in
Xuan Wei, China. Lung Cancer. 49:299–309. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jeng YL, Wu MH, Huang HB, et al: The
methylenetetrahydrofolate reductase 677C→T polymorphism and lung
cancer risk in a Chinese population. Anticancer Res. 23:5149–5152.
2003.
|
|
24
|
Hosseini M, Houshmand M and Ebrahimi A:
MTHFR polymorphisms and breast cancer risk. Arch Med Sci.
7:134–137. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hubner RA and Houlston RS: MTHFR C677T and
colorectal cancer risk: a meta-analysis of 25 populations. Int J
Cancer. 120:1027–1035. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zintzaras E: Association of
methylenetetrahydrofolate reductase (MTHFR) polymorphisms with
genetic susceptibility to gastric cancer: a meta-analysis. J Hum
Genet. 51:618–624. 2006. View Article : Google Scholar
|
|
27
|
Issa JP: DNA methylation as a therapeutic
target in cancer. Clin Cancer Res. 13:1634–1637. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim YI: Folate and carcinogenesis:
evidence, mechanisms, and implications. J Nutr Biochem. 10:66–88.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Weisberg IS, Jacques PF, Selhub J, et al:
The 1298A→C polymorphism in methylenetetrahydrofolate reductase
(MTHFR): in vitro expression and association with homocysteine.
Atherosclerosis. 156:409–415. 2001.
|
|
30
|
Stern LL, Mason JB, Selhub J and Choi SW:
Genomic DNA hypomethylation, a characteristic of most cancers, is
present in peripheral leukocytes of individuals who are homozygous
for the C677T polymorphism in the methylenetetrahydrofolate
reductase gene. Cancer Epidemiol Biomarkers Prev. 9:849–853.
2000.
|
|
31
|
Arslan S, Karadayi S, Yildirim M, Ozdemir
O and Akkurt I: The association between methylene-tetrahydrofolate
reductase gene polymorphism and lung cancer risk. Mol Biol Rep.
38:991–996. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hung R, Hashibe M, McKay J, et al:
Folate-related genes and the risk of tobacco-related cancers in
Central Europe. Carcinogenesis. 28:1334–1340. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shi Q, Zhang Z, Li G, Pillow PC, Hernandez
LM, Spitz MR and Wei Q: Sex differences in risk of lung cancer
associated with methylene-tetrahydrofolate reductase polymorphisms.
Cancer Epidemiol Biomarkers Prev. 14:1477–1484. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Siemianowicz K, Gminski J, Garczorz W,
Slabiak N, Goss M, Machalski M and Magiera-Molendowska H:
Methylenetetrahydrofolate reductase gene C677T and A1298C
polymorphisms in patients with small cell and non-small cell lung
cancer. Oncol Rep. 10:1341–1344. 2003.PubMed/NCBI
|
|
35
|
Heijmans BT, Boer JM, Suchiman HE, et al:
A common variant of the methylenetetrahydrofolate reductase gene
(1p36) is associated with an increased risk of cancer. Cancer Res.
63:1249–1253. 2003.PubMed/NCBI
|
|
36
|
Shen H, Spitz MR, Wang LE, Hong WK and Wei
Q: Polymorphisms of methylene-tetrahydrofolate reductase and risk
of lung cancer: a case-control study. Cancer Epidemiol Biomarkers
Prev. 10:397–401. 2001.PubMed/NCBI
|
|
37
|
Higgins JPT and Green S: Cochrane Handbook
for Systematic Reviews of Interventions Version 510 (updated March
2011). The Cochrane Collaboration. 2011, Available at https://handbook-5-1.cochrane.org.
|
|
38
|
Golledge J and Norman PE: Relationship
between two sequence variations in the gene for peroxisome
proliferator-activated receptor-gamma and plasma homocysteine
concentration. Health in men study. Hum Genet. 123:35–40. 2008.
View Article : Google Scholar
|
|
39
|
Sazci A, Ergul E, Tuncer N, Akpinar G and
Kara I: Methylenetetrahydrofolate reductase gene polymorphisms are
associated with ischemic and hemorrhagic stroke: Dual effect of
MTHFR polymorphisms C677T and A1298C. Brain Res Bull. 71:45–50.
2006. View Article : Google Scholar : PubMed/NCBI
|