Introduction
Toll-like receptors (TLRs) bind to microbe
components by recognizing pathogen-associated molecular patterns,
they activate cellular signal transduction pathways, stimulate
innate immune responses and further adjust the adaptive immune
system. TLRs are perceived as a bridge between innate and adaptive
immunity. Research on TLRs has been mainly focused on inflammation,
autoimmune disease and organ transplantation rejection; recently,
the association between TLRs and tumorigenesis has attracted
scientific attention (1).
Among TLRs, TLR9 is the sole family member for
detecting DNA (2). TLR9 was
originally identified as a sensor for bacterial DNA with abundant
unmethylated CpG dinucleotides. However, mammalian DNA of
self-origin, which has a low frequency of unmethylated CpG
dinucleotides, may also stimulate TLR9. This is strongly
underpinned by a recent study reporting that TLR9 recognizes the
sugar backbone 2′-deoxyribose of DNA, but not its bases, suggesting
the nucleotide sequence is not the primary target of TLR9 (2). Therefore, DNA released from damaged
cells may trigger sterile inflammation via TLR9, acting as a
damage-associated molecular pattern molecule.
Accumulating evidence demonstrates that TLR9, which
is mainly expressed on immune cells, is also functionally expressed
on lung cancer cells (1–4). TLR9 signaling may alter the biological
character of lung cancer cells, including promoting proliferation
and enhancing the metastatic potential of tumor cells, indicating
that the activation of TLR signaling in lung cancer cells may
contribute to the progression of lung cancer (3–5). The
A549 human lung adenocarcinoma cell line highly expresses TLR9 and
also exhibits positive expression of the Runt-related transcription
factor 3 (Runx3) (6,7). Runx3, a novel tumor suppressor gene,
was found to be downregulated in gastric, colon and lung cancer
(8–12). CpG-ODN, being a TLR9 agonist, may
rapidly stimulate T and B cells, induce Th1 cytokines [interleukin
(IL)-1, IL-6, IL-12, IL-18, TNF-α and IFN-γ] and promote the
maturation of antigen-presenting cells (APCs), indirectly activate
immune cells and inhibit tumor proliferation. However, the
association between the effect of CpG-ODN on lung cancer cells and
Runx3 expression has not been determined. In this study, we aimed
to elucidate the association between the TLR9 signaling pathway and
Runx3 expression, laying the foundation for further invstigations
on the antitumor mechanism of the TLR9 signaling pathway.
Materials and methods
Cell culture
The A549 human lung adenocarcinoma cell line was
cultured in Dulbecco’s modified Eagle’s medium, supplemented with
100 U/ml penicillin, 100 mg/l streptomycin and 10% fetal bovine
serum (Gibco-BRL, Carlsbad, CA, USA). In order to analyze the
effect of TLR9 on cell proliferation, the cells were stimulated by
CpG-ODN at different concentrations for 2, 4, 6 and 8 h and
collected by centrifugation at 800 × g for 10 min at 4°C. Total RNA
was isolated from cultured cells using an RNA extraction kit
(Takara Bio, Inc., Shiga, Japan) and prepared for polymerase chain
reaction (PCR) amplification.
Reverse transcription-PCR (RT-PCR) and
quantitative PCR (qPCR)
The cells were discharged into TRIzol reagent
(Invitrogen Life Technologies, Carlsbad, CA, USA), total RNA was
isolated by an RNA extraction kit (Takara) and reversed-transcribed
with the ReverTra Ace®qPCR-RT kit (Toyobo, Osaka, Japan)
according to the manufacturer’s instructions. The RT-PCR and qPCR
were performed as previously described (13). The sequences for the primers used
were as follows: β-actin (262 bp), 5′-CACGAAACTACCTTCAACTCC-3′
(forward) and 5′-CACGAAACTACCTTCAACTCC-3′ (reverse); Runx3 (353
bp), 5′-GATGGCAGGCAATGACGA-3′ (forward) and
5′-CATACTCCTGCTTGCTGATC-3′ (reverse). The relative quantification
of mRNA expression was performed with the comparative threshold
cycle method (8).
Western blot analysis
The cells (1×106) were washed with cold
PBS and lysed in 100 μl lysis buffer [150 mmol/l NaCl, 20 mmol/l
Tris-HCl (pH 7.5), 1 mmol/l EDTA, 1 mmol/l EGTA, 1 mmol/l
Na3VO4, 1 mmol/l sodium fluoride, 0.5% DOC,
1% Triton X-100 and 1% Nonidet-P40]. The cell lysates were boiled
with 2X loading buffer for 20 min and analyzed by western blotting.
Mouse anti-human Runx3, mouse anti-human β-actin and anti-NF-κB
were used as primary antibodies (Santa Cruz Biotechnology, Inc.,
Santa Cruz, CA, USA). Following polyacrylamide gel electrophoresis,
the protein was transferred onto a PVDF membrane (Perkin-Elmer,
Waltham, MA, USA). The membrane was incubated with the primary
antibody, followed by incubation with horseradish
peroxidase-conjugated rabbit anti-mouse IgG (Takara Bio, Inc.,
Shiga, Japan). After being thoroughly washed in Tris-Buffered
Saline with Tween-20, the blots were processed for detection of Ag
using an electrochemiluminescence plus western blotting detection
system (GE Healthcare Life Sciences, Pittsburgh, PA, USA). The
Typhoon Molecular Imaging system (GE Healthcare Life Sciences) was
used for scanning and recording the results.
Design of Runx3 siRNA
According to the design principle of RNA
interference target sites, GCCACTTGATTCTGGAGGA was selected as the
Runx3-specific sequence and synthesis of the dsRNA sequence was
performed by Zimmer Medical Int’l Trading Co., Ltd. (Shanghai,
China). The primers used were as follows: Runx3: sense,
5′-GCCACUUGAUUCUGGAGGATT-3′ and antisense,
5′-UCCUCCAGAAUCAAGUGGCTT-3′; control dsRNA, sense,
5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′. All the sequences were labeled with
fluorescence.
Transfection of Runx3 siRNA
The A549 cells in logarithmic growth phase were
cultured in 24-well plates and the transfection was performed when
the cell confluence reached 80%, according to the manufacturer’s
instructions (Lipofectamine®2000, Invitrogen Life
Technologies, Carlsbad, CA, USA). The transfection efficiency was
observed under a fluorescence microscope. The experiment included
four groups: untransfected, transfection reagent control, negative
sequence control and Runx3 siRNA-transfected groups.
Proliferation assay
The cells were divided into A549, A549+CpG,
A549-NC+CpG, A549-siRNA and A549-siRNA+CpG groups. A total of
1×106 cells were seeded into 96-well plates; following
cell adherence to the plate, 10 μl CpG were added to the wells at a
concentration of 5 μl/ml and incubated at 37°C for 1~8 h.
Subsequently, 20 μl (5 mg/ml) MTT were added to each well and the
plate was further incubated for 4 h to deoxidize MTT under
light-blocking conditions. After removal of the MTT dye solution,
the cells were treated with 50 μl DMSO and the absorbance at 490 nm
was measured using the EL×800 UV microplate reader (BioTek,
Winooski, VT, USA). In each experiment and under each condition,
proliferation was assessed in triplicate and the experiments were
repeated at least twice.
Statistical analysis
All the statistical analyses were performed using
GraphPad Prism, software, version 5.0 GraphPad Software, San Diego,
CA, USA). Data are expressed as means ± SD. Comparisons between
groups were performed using the unpaired Student’s t-test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Expression of Runx3 in CpG-ODN-induced
A549 cells
The analysis of the PCR amplification products by
gel electrophoresis revealed a low expression level of Runx3 mRNA
in A549 cells without CpG-ODN, which was increased after the cells
were stimulated by CpG-ODN at concentrations of 2.5, 5 and 10 μg/ml
for 2, 4, 6 and 8 h, respectively (Fig.
1). Similar results were obtained by qPCR. The expression of
Runx3 mRNA was the highest in A549 cells stimulated by CpG-ODN at 5
μg/ml for 8 h, indicating a time-dependent effect (Fig. 1B). Furthermore, the western blot
analysis results indicated that the expression level of Runx3
protein was consistent with the results of RT-PCR (Fig. 1C).
Inhibitory effect of siRNA on Runx3
expression in A549 cells
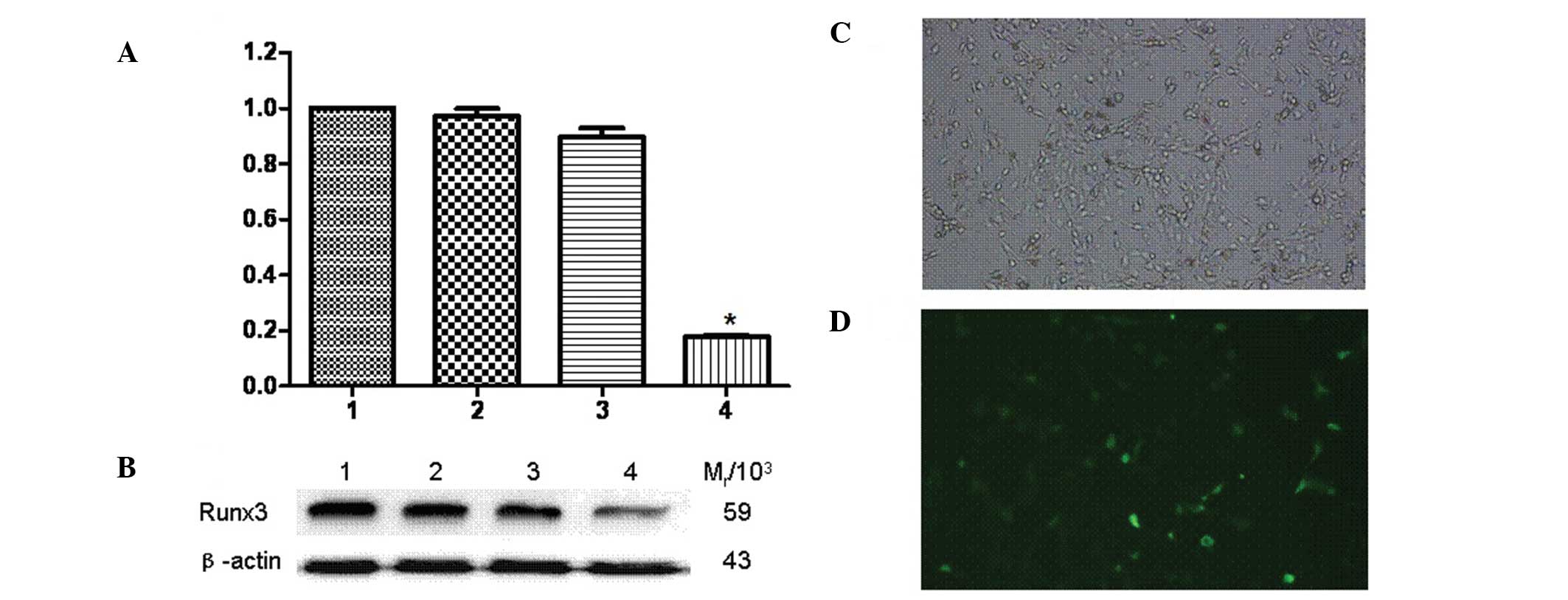
Runx3 siRNA-transfected A549 cells were observed
under a fluorescence microscope and the proportion of fluorescent
staining cells, representing the siRNA transfection rate, was 40%
(Fig. 2C and D). The qPCR results
demonstrated that, compared to the untransfected group, the
expression of the Runx3 gene was inhibited in Runx3
siRNA-transfected A549 cells, but no significant difference was
observed in untransfected cells (Fig.
2A). In addition, the western blot analysis revealed that the
Runx3 protein expression was significantly decreased in transfected
A549 cells compared to that in untransfected cells (Fig. 2B). The relative molecular mass of
protein Runx3 (Mr/103) was 59 and that of
β-actin was 43.
Effect of Runx3 siRNA transfection on the
proliferation of A549 cells stimulated by CpG-ODN
As shown by the cell growth curve (Fig. 3), the inhibition of cell
proliferation following stimulation by CpG-ODN was markedly
decreased in Runx3 siRNA-transfected A549 cells, almost to the
original cell proliferation state. However, significant inhibition
of A549 cell proliferation was observed in untrasfected A549 cells
following CpG-ODN stimulation.
Discussion
Runx3 is associated with the development and
progression of gastric cancer and silencing of Runx3 in gastric
cancer cells affects the expression of important genes involved in
the metastatic process, including cell adhesion, proliferation and
apoptosis; such silencing may promote peritoneal metastasis. The
expression of Runx3 was found to be significantly reduced in human
gastric mucosa exhibiting intestinal metaplasia and Runx3−/− mouse
gastric epithelial cells bear the potential to differentiate into
Cdx-2 positive intestinal-type cells (14).
CpG-ODN bind to TLR9 expressed on APCs, natural
killer cells and other lymphocytes, mediating anti-infection or
antitumor immune response, which has attracted significant
attention among immunological investigators. Runx3 is a newly
discovered tumor suppressor gene and its expression product may
inhibit the proliferation of tumor cells through the transforming
growth factor-beta (TGF-β) signaling pathway and induce apoptosis
or maintain normal cell growth and development (15–17).
The loss of Runx3 expression may lead to disorders of the TGF-β
signaling pathway and is closely associated with tumorigenesis.
Runx3, as a T-bet collaborative secondary transcription factor, is
also involved in T- and B-lymphocyte differentiation and cytokine
production (18). It was recently
reported that CpG-ODN may directly upregulate the expression of
T-bet in B cells by stimulating the TLR9 pathway as an alternative
signal (19,20). In view of the dual role of Runx3,
which inhibits the proliferation of tumor cells and regulates T and
B cells, it is considered to be an important target for antitumor
immunity.
In the present study, we investigated the expression
level of TLR9 in the A549 cell line and observed the behavior of
A549 cells stimulated by CpG-ODN. The results demonstrated that the
CpG-ODN was able to significantly inhibit the proliferation of A549
cells and upregulate the expression of Runx3 in A549 cells at the
transcriptional as well as the translational level. However, the
expression level of Runx3 was decreased in Runx3 siRNA-transfected
cells and the inhibitory effect of CpG stimulation on cell
proliferation was distinctly reversed by Runx3 siRNA, indicating
that CpG-ODN may inhibit the proliferation of TLR9-positive tumor
cells by regulating the expression of the tumor suppressor gene
Runx3 through the TLR9 signaling pathway. Furthermore, the
upregulation of the Runx3 gene may also induce the expression of
the transcription factor T-bet, promote Th1-type response and
enhance antitumor immunity. It is hypothesized that the induction
of Runx3 via the TLR9 signaling pathway may be of value in
providing a wide range of therapeutic modalities and targets, which
requires further investigation.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation of China (nos. 31270947, 31170849,
81370084 and 81001319), the Natural Science Foundation of Jiangsu
Province (no. BK2011472) and the Postdoctoral Foundation of China
(no. 2013T60508).
References
|
1
|
Chen R, Alvero AB, Silasi DA, Steffensen
KD and Mor G: Cancers take their Toll - the function and regulation
of Toll-like receptors in cancer. Oncogene. 27:225–233. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hazeki K, Uehara M, Nigorikawa K and
Hazeki O: PIKfyve regulates the endosomal localization of CpG
oligodeoxynucleotides to elicit TLR9-dependent cellular responses.
PLoS One. 8:e738942013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schmausser B, Andrulis M, Endrich S,
Muller-Hermelink HK and Eck M: Toll-like receptors TLR4, TLR5 and
TLR9 on gastric carcinoma cells: an implication for interaction
with Helicobacter pylori. Int J Med Microbiol. 295:179–185.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ren T, Wen ZK, Liu ZM, Liang YJ, Guo ZL
and Xu L: Functional expression of TLR9 is associated to the
metastatic potential of human lung cancer cell: functional active
role of TLR9 on tumor metastasis. Cancer Biol Ther. 6:1704–1709.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chang YJ, Wu MS, Lin JT and Chen CC:
Helicobacter pylori-induced invasion and angiogenesis of
gastric cells is mediated by cyclooxygenase-2 induction through
TLR2/TLR9 and promoter regulation. J Immunol. 175:8242–8252. 2005.
View Article : Google Scholar
|
|
6
|
Li QL, Kim HR, Kim WJ, et al:
Transcriptional silencing of the RUNX3 gene by CpG hypermethylation
is associated with lung cancer. Biochem Biophys Res Commun.
314:223–228. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kato N, Tamura G, Fukase M, Shibuya H and
Motoyama T: Hypermethylation of the RUNX3 gene promoter in
testicular yolk sac tumor of infants. Am J Pathol. 163:387–391.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xu Y, Gao J, Su Z, et al: Downregulation
of Hlx closely related to the decreased expressions of T-bet and
Runx3 in patients with gastric cancer may be associated with a
pathological event leading to the imbalance of Th1/Th2. Clin Dev
Immunol. 2012:9498212012.
|
|
9
|
Kang KA, Kim KC, Bae SC and Hyun JW:
Oxidative stress induces proliferation of colorectal cancer cells
by inhibiting RUNX3 and activating the Akt signaling pathway. Int J
Oncol. 43:1511–1516. 2013.PubMed/NCBI
|
|
10
|
Li M, Tan SY, Zhang J and You HX: Effects
of paeonol on intracellular calcium concentration and expression of
RUNX3 in LoVo human colon cancer cells. Mol Med Rep. 7:1425–1430.
2013.PubMed/NCBI
|
|
11
|
Lim J, Duong T, Do N, Do P, Kim J, Kim H,
El-Rifai W, Ruley HE and Jo D: Antitumor activity of cell-permeable
RUNX3 protein in gastric cancer cells. Clin Cancer Res. 19:680–690.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ito K: Tumor suppressive functions of
RUNX3 in gastric carcinogenesis. J Jpn Biochem Soc. 84:278–282.
2012.(In Japanese).
|
|
13
|
Yang P, Qiu G, Wang S, Su Z, Chen J, Wang
S, Kong F, Lu L, Ezaki T and Xu H: The mutations of Th1
cell-specific T-box transcription factor may be associated with a
predominant Th2 phenotype in gastric cancers. Int J Immunogenet.
37:111–115. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yano T, Ito K, Fukamachi H, Chi XZ, Wee
HJ, Inoue K, Ida H, Bouillet P, Strasser A, Bae SC and Ito Y: The
RUNX3 tumor suppressor upregulates Bim in gastric epithelial cells
undergoing transforming growth factor beta-induced apoptosis. Mol
Cell Biol. 26:4474–4488. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fainaru O, Woolf E, Lotem J, Yarmus M,
Brenner O, Goldenberg D, Negreanu V, Bernstein Y, Levanon D, Jung S
and Groner Y: Runx3 regulates mouse TGF-beta-mediated dendritic
cell function and its absence results in airway inflammation. EMBO
J. 23:969–979. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li J, Kleeff J, Guweidhi A, Esposito I,
Berberat PO, Giese T, Buchler MW and Friess H: RUNX3 expression in
primary and metastatic pancreatic cancer. J Clin Pathol.
57:294–299. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Miyazono K, Suzuki H and Imamura T:
Regulation of TGF-beta signaling and its roles in progression of
tumors. Cancer Sci. 94:230–234. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Djuretic IM, Levanon D, Negreanu V, Groner
Y, Rao A and Ansel KM: Transcription factors T-bet and Runx3
cooperate to activate Ifng and silence Il4 in T helper type 1
cells. Nat Immunol. 8:145–153. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu N, Ohnishi N, Ni L, Akira S and Bacon
KB: CpG directly induces T-bet expression and inhibits IgG1 and IgE
switching in B cells. Nat Immunol. 4:687–693. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rosa R, Damiano V, Formisano L, Nappi L,
Marciano R, Veneziani BM, De Placido S and Bianco R: Combination of
a Toll-like receptor 9 agonist with everolimus interferes with the
growth and angiogenic activity of renal cell carcinoma.
Oncoimmunology. 2:e251232013. View Article : Google Scholar : PubMed/NCBI
|