Introduction
Esophageal cancer is one of the most common types of
malignant disease, with ~16,980 new cases and 14,710 mortalities in
the United States in 2011 (1).
Esophageal cancer is a multifactorial disease, which is considered
to be a result of complex interactions between environmental and
genetic factors. Diet has been hypothesized to play a role in the
etiology of esophageal cancer. A number of studies have found that
consuming large quantities of red or processed meat is associated
with an increase in the risk of esophageal cancer (2–4).
Aberrant cell proliferation is an important factor
for the development of numerous types of common cancer. Cyclin
families are involved in cell-cycle progression, and in particular,
cyclin D1 (CCND1) is the major regulatory protein that plays a key
role in the transition from G1 to S phase by binding to
cyclin-dependent kinases 4 and 6 to promote the progression of the
cell cycle during cell division (5,6). The
overexpression of CCND1 has always been observed in numerous types
of malignant cancer and indicates a poor clinical outcome (7–9).
Single-nucleotide polymorphisms (SNPs) may change
the functions of the gene and alter the protein expression,
potentially affecting cell proliferation and increase the
susceptibility of developing cancer. The synonymous SNP (rs603965)
of a G to A polymorphism at codon 242 (G870A) in exon 4, is the
most important mutation of the CCND1 gene. The A allele creates a
greater frequency of alternate splicing during transcription, which
was postulated to have a longer half life than the G allele to
bypass the G1/S checkpoint and resulted in an increased CCND1
level, leading to abnormal cell proliferation and circumvention of
apoptosis (10,11).
Although a number of epidemiological studies have
been conducted to assess the association between the CCND1 G870A
polymorphism and esophageal cancer susceptibility, the conclusions
have been inconsistent. Thus far, two related meta-analyses were
conducted by Cai et al (12)
and Zhou et al (13), which
demonstrated various associations between the CCND1 G870A
polymorphism and esophageal cancer risk. Notably, the meta-analyses
by Cai et al (12) only
included eight published studies, and the study by Zhou et
al (13) was conducted with a
focus on Asian populations only. In the present study, 11
case-control studies on the CCND1 G870A polymorphism and esophageal
cancer risk that were previously published were analyzed by
performing a meta-analysis to examine a more specific association
between the CCND1 G870A polymorphism and esophageal cancer risk and
various published observational studies.
Materials and methods
Search strategy
The Pubmed database was searched using the terms
‘CCND1’, ‘cyclin D1’, ‘esophageal cancer’, ‘polymorphism’, and the
combined phrases for all genetic studies on the association between
the CCND1 G870A polymorphism and esophageal cancer risk between
2003 and March 7, 2014. Furthermore, the search was complemented
with a examination of the references of the retrieved studies and
reviews. The following criteria were used to select the studies for
the meta-analysis: i) Observational (case-control or prospective)
studies of the CCND1 G870A polymorphism and esophageal cancer risk;
ii) sufficient published data for estimating an odds ratio (OR)
with a 95% confidence interval (CI); and iii) if studies had partly
or overlapping data, only the largest or most recent sample was
selected, according to Little et al (14). A total of 11 case-control studies,
including 2,111 patients with esophageal cancer and 3,232 controls,
were included in this meta-analysis.
Data extraction
Data were extracted independently by two
investigators (Wen and Hu) from all the selected studies. The data
included the first author’s name, publication data, country of
origin, sources of controls, ethnicity of the study population
(categorized as Asian, Caucasian and Mixed) and number of different
genotype, tumor pathology and Hardy-Weinberg equilibrium (HWE) in
controls.
Statistical analysis
The allele contrast (A vs. G) and codominant (AA vs.
GG, GA vs. GG), dominant [(AA+GA) vs. GG) and recessive models (AA
vs. (GG+GA)] were evaluated using ORs with 95% CI to assess the
strength of the association between the CCND1 G870A polymorphisms
and esophageal cancer risk. Subgroup statistical analyses were
conducted for ethnicity, study design and pathology. Otherwise,
heterogeneity and cumulative analysis were assessed by
χ2-based Q-test (15).
OR estimation was calculated with the fixed-effect model
(Mantel-Haenszel method) when statistical heterogeneity did not
exist (P>0.10) (16). Otherwise,
the random-effects model (DerSimonian and Laird method) was
selected (17). Publication bias
was evaluated by the Begg’s funnel plot and linear regression
asymmetry test by Egger et al (18,19).
Statistical analysis was performed using STATA versions 10.0 and
11.0 (StataCorp, College Station, TX, USA), and two-sided P-values
(P<0.05) were considered to indicate a statistically significant
difference.
Results
Study characteristics
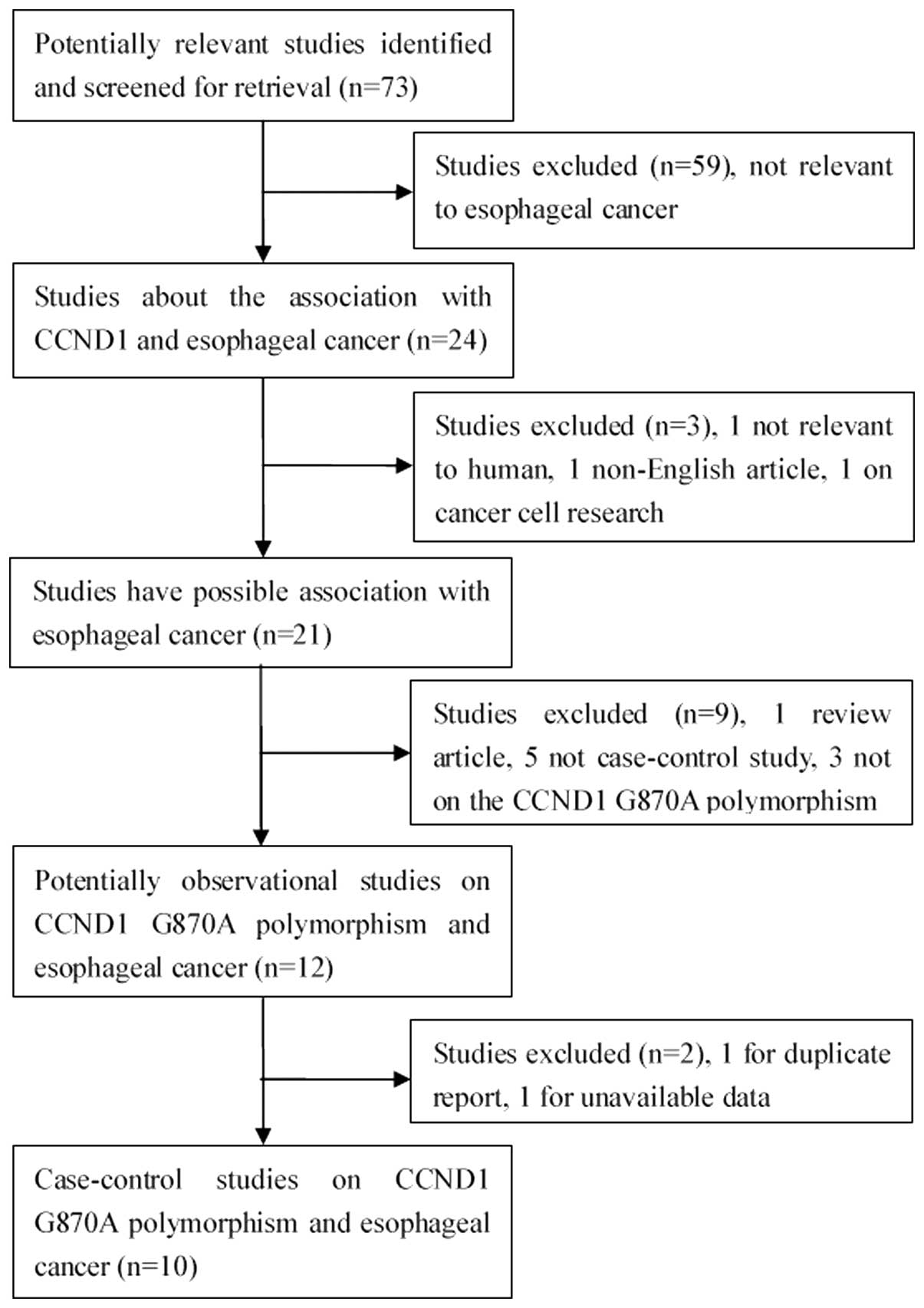
A total of 73 relevant studies were identified
(Fig. 1). Following a careful
review, 10 published studies with 11 case-control studies were
identified, with 2,111 patients esophageal cancer patients and
3,232 controls (20–29). The distribution of the various
genotypes of each study in different populations is shown in
Table I. The diverse genotyping
methods were polymerase chain reaction-restriction fragment length
polymorphism, highly parallel SNP genotyping assay and Taqman
techniques. No study deviated from Hardy-Weinberg equilibrium (HWE)
in control populations.
 | Table ICharacteristics of the case-control
studies included in the meta-analysis. |
Table I
Characteristics of the case-control
studies included in the meta-analysis.
| First author | Year | Country/region | Racial descent | Source of
controls | Case, n | Control, n | Genotype
distribution | P-value HWEa | Pathology |
|---|
|
|---|
| Case, n | Control, n |
|---|
|
|
|---|
| GG | GA | AA | GG | GA | AA |
|---|
| Yu | 2003 | China | Asian | Population-based | 321 | 345 | 68 | 157 | 96 | 58 | 177 | 110 | 0.354 | ESCC |
| Zhang | 2003 | China | Asian |
Population-based | 120 | 183 | 11 | 74 | 35 | 38 | 102 | 43 | 0.118 | ESCC |
| Casson | 2005 | Canada | Caucasian | Hospital-based | 56 | 95 | 12 | 27 | 17 | 35 | 52 | 8 | 0.063 | EA |
| Geddert | 2005 | Germany | Caucasian | Hospital-based | 56 | 253 | 16 | 26 | 14 | 63 | 136 | 54 | 0.224 | EA |
| Jain | 2007 | India | Asian | Hospital-based | 151 | 201 | 22 | 76 | 53 | 37 | 111 | 53 | 0.114 | Mixed |
| Akbarib | 2009 | Iran | Caucasian | NA | 279 | 807 | 72 | 126 | 81 | 161 | 376 | 270 | 0.149 | ESCC |
| Akbaric | 2009 | Iran | Caucasian | NA | 465 | 561 | 97 | 238 | 130 | 107 | 290 | 164 | 0.290 | ESCC |
| Liu | 2010 | USA | Caucasian | Hospital-based | 299 | 450 | 79 | 154 | 66 | 128 | 215 | 107 | 0.369 | EA |
| Kurmanov | 2010 | Kazakhstan | Caucasian |
Population-based | 98 | 86 | 19 | 42 | 37 | 24 | 46 | 16 | 0.463 | ESCC |
| Hussain | 2011 | India | Asian |
Population-based | 151 | 151 | 20 | 99 | 32 | 56 | 72 | 23 | 0.986 | ESCC |
| Djansugurova | 2013 | Kazakhstan | Caucasian | Healthy-based | 115 | 100 | 22 | 49 | 44 | 28 | 54 | 18 | 0.363 | ESCC |
Meta-analysis
The main results of the meta-analysis and the
heterogeneity test are shown in Table
II. Overall, there was a significant association between the
CCND1 G870A polymorphism and esophageal cancer risk was observed
for the allele contrast (A vs. G: OR, 1.23; 95% CI, 1.02–1.48;
P=0.029 Pheterogeneity<0.01), codominant (AA vs. GG:
OR, 1.58; 95% CI, 1.06–2.35; P=0.024,
Pheterogeneity<0.01) and recessive models [AA vs. (GG
+ GA): OR, 1.33, 95% CI, 1.03–1.73; P=0.030,
Pheterogeneity<0.01; Fig.
2]. Simultaneously, a borderline significant increased risk was
found in the dominant model [(AA + GA) vs. GG: OR, 1.30, 95% CI,
0.96–1.76; P=0.092, Pheterogeneity<0.01]. No
significant risk effect was found in the subgroup analysis by
ethnicity, study design and pathology.
 | Table IISummary ORs and 95% CI of CCND1 G870A
polymorphism and esophageal cancer risk. |
Table II
Summary ORs and 95% CI of CCND1 G870A
polymorphism and esophageal cancer risk.
| A vs. G | AA vs. GG | GA vs. GG | AA + GA vs. GG | AA vs. GG + GA |
|---|
|
|
|
|
|
|
|---|
|
P-valuea | OR | 95% CI | P-value |
P-valuea | OR | 95% CI | P-value |
P-valuea | OR | 95% CI | P-value |
P-valuea | OR | 95% CI | P-value |
P-valuea | OR | 95% CI | P-value |
P-valuea |
|---|
| Total | 1.23 | 1.02–1.48 | 0.029 |
<0.001b | 1.58 | 1.06–2.35 | 0.024 |
<0.001b | 1.18 | 0.90–1.53 | 0.254 |
<0.001b | 1.30 | 0.96–1.76 | 0.092 |
<0.001b | 1.33 | 1.03–1.73 | 0.030 |
<0.001b |
| Ethnicity |
| Asian | 1.29 | 0.93–1.80 | 0.128 |
0.001b | 1.85 | 0.84–4.09 | 0.129 |
<0.001b | 1.66 | 0.76–3.64 | 0.202 |
<0.001b | 1.74 | 0.79–3.81 | 0.169 |
<0.001b | 1.18 | 0.94–1.47 | 0.149 | 0.228 |
| Caucasian | 1.20 | 0.95–1.53 | 0.135 |
<0.001b | 1.45 | 0.89–2.35 | 0.134 |
<0.001b | 0.96 | 0.81–1.14 | 0.624 | 0.482 | 1.08 | 0.83–1.40 | 0.583 |
0.037b | 1.44 | 0.97–2.14 | 0.070 |
<0.001b |
| Design |
|
Hospital-based | 1.24 | 0.93–1.66 | 0.144 |
0.026b | 1.62 | 0.84–3.10 | 0.150 |
0.013b | 1.12 | 0.86–1.46 | 0.395 | 0.602 | 1.19 | 0.89–1.61 | 0.243 | 0.286 | 1.51 | 0.87–2.61 | 0.143 |
0.008b |
|
Population-based | 1.39 | 0.94–2.04 | 0.096 |
<0.001b | 2.13 | 0.86–5.24 | 0.100 |
<0.001b | 1.68 | 0.74–3.81 | 0.216 |
<0.001b | 1.83 | 0.80–4.17 | 0.150 |
<0.001b | 1.39 | 0.90–2.15 | 0.135 |
0.035b |
| Pathology |
| ESCC | 1.23 | 0.96–1.59 | 0.108 |
<0.001b | 1.58 | 0.92–2.70 | 0.095 |
<0.001b | 1.25 | 0.82–1.91 | 0.307 |
<0.001b | 1.36 | 0.87–2.13 | 0.175 |
<0.001b | 1.28 | 0.93–1.75 | 0.127 | 0.001 |
| EA | 1.25 | 0.82–1.91 | 0.310 |
0.014b | 1.67 | 0.64–4.34 | 0.291 |
0.007b | 1.11 | 0.83–1.49 | 0.469 | 0.396 | 1.18 | 0.77–1.81 | 0.443 | 0.167 | 1.59 | 0.69–3.70 | 0.280 |
0.004b |
Publication bias
The Begg’s funnel plot and Egger’s test were used to
estimate the publication bias in the five models. The shape of the
funnel plots appeared to be symmetrical in the GA vs. GG and (AA +
GA) vs. GG models, but not in the A vs. G, AA vs. GG and AA vs. (GG
+ GA) models (Fig. 3), indicating
that there was a certain amount of publication bias. Egger’s test
was applied to provide further statistical evidence [(P=0.001 for A
vs. G; P<0.001 for AA vs. GG; and P<0.001 for AA vs. (GG +
GA)].
Cumulative and sensitivity analysis
Each study that was involved in the meta-analysis
was deleted separately to assess the influence of the individual
dataset to the pooled ORs. The analysis results demonstrated a
slightly decreased effect each time. In the cumulative
meta-analysis, the results did not become significant until the
last study by Djansugurova et al (29) was accumulated (Fig. 4). Furthermore, the analysis results
demonstrated that there was no significant association when the
study of Djansugurova et al (29) was removed.
Discussion
CCND1 has been mapped to chromosome 11q13, encoding
a key cell cycle regulatory protein with 295 amino acids. CCND1
regulates the transition from the G1 to S phase during cell
division. A high activity of CCND1 results in the premature cell
passage through the G1-S transition, which leads to the generation
of unrepaired DNA damage and the accumulation of genetic errors
(30). The protein overexpression
of CCND1 has been found in numerous types of cancer, and has also
been regarded as the malignant characterization of cancer. There
are various polymorphisms in CCND1, but the G to A mutation is well
known and does not result in any amino acid alteration within the
protein sequence. However, the CCND1 A allele results in an
alternatively spliced transcript of CCND1 with a longer half life
than the CCND1 G allele. This mutation helps the variant cell pass
through the G1/S checkpoint easily and results in abnormal
perforation, leading to cancer development (31). Findings of previous studies have
shown that the CCND1 A allele may increase the risk of breast,
prostate, colorectal and other types of cancer in different
ethnicities (32–36).
In 2003, Yu et al (20) conducted the first study between the
CCND1 G870A polymorphism and esophageal cancer, however, no
significant association was found in a Chinese population. Thus
far, conflicting conclusions on the association of the CCND1 G870A
polymorphism and esophageal cancer susceptibility exist. The study
by Zhang et al (21) found
an increased risk for developing esophageal cancer with the CCND1
870A allele, and ~2.0-fold increased risk was found among the AA
genotype compared to the GG and GA genotypes in a Northern Chinese
population. The study by Casson et al (22) found that the apparently elevated
risk of esophageal cancer was associated with the AA genotype
compared to the GG genotype (OR, 5.99; 95% CI, 1.89–18.96) in a
Canadian population. In the study by Jain et al (24), the AA genotype was marginally
associated with esophageal cancer (OR, 1.5; 95% CI, 0.98–2.4), and
there was a higher risk in the upper location (OR, 3.8; 95% CI,
1.6–9.3) in an Indian population. Kurmano et al (26) reported a significantly increased
association in Kazakhstan with the variant homozygous AA genotype
(OR, 2.66; 95% CI, 1.35–5.24). Hussain et al (27) also indicated that the Indian
individuals carrying the GA and AA genotype had a 2.8-fold
increased risk for the development of esophageal cancer, and the
higher risk was observed in individuals with smoking and drinking
habits. Conversely, the study by Akbari et al (28) found that the G allele was associated
with a 1.5-fold increased risk of esophageal cancer under the
recessive model (OR, 1.50; 95% CI, 1.14–2.16; P=0.02). Furthermore,
no significant association between the CCND1 G870A polymorphism and
esophageal cancer in a German population was found in the study by
Geddert et al (23). A
similar conclusion was also found in the study by Liu et al
in an American population (25),
and Djansugurova et al (29)
in Kazakhstan. Recently, two meta-analyses were conducted by Cai
et al (37) and Zhou et
al (38), which demonstrated
different conclusions in regards to the CCND1 G870A polymorphism
with limited published data. Therefore, it is assumed that the
reason for the contrary results may be the sample sizes. Thus far,
a number of other studies (28,38)
regarding this focus have been published. Therefore, the
meta-analysis was performed to clarify the results on the
association. In the meta-analysis, including 11 case-control
studies with 2,111 esophageal cancer patients and 3,232 controls,
certain possible risks were explored between the potential function
of the CCND1 G870A polymorphism and esophageal cancer.
There were several limitations for the analysis in
the present study. Firstly, the results were based on the
unadjusted estimates with unavailable original data of these
collected studies, which limited the evaluation with certain
covariates, including age, smoking, drinking and other
environmental factors. Secondly, the sample size was relatively
small in the analysis, which may have induced the bias of the
results and the disability of drawing more detailed conclusions.
Thirdly, the controls of several studies were hospital-based
individuals with other diseases, which may result in specific
selection biases. Fourthly, a certain amount of publication bias
was always present until the subgroup analyses were conducted,
similar to the sensitivity analysis. These deviations influenced
the preciseness and reliability of the results. Additionally, the
majority of the included studies were conducted on Caucasians and
Asians, but there were no studies of African populations. Therefore
the variation in ethnicity may have generated biases.
Taken together, despite these limitations, the
meta-analysis suggests that the CCND1 G870A polymorphism is a
potential elevated risk in the development of esophageal cancer.
These findings may be helpful in increasing the understanding of
the CCND1 G870A polymorphism in the etiology of esophageal cancer.
In the future, large and well-designed case-control studies are
required to verify these findings.
Acknowledgements
The authors gratefully acknowledge the support of
the subjects who participated in the present study. The study was
partly supported by the Foundation of Ministry of Education of
Hubei Province (grant no. D20142407), Foundation of Hubei
University of Medicine (grant no. 2013GPY07) and Taihe Hospital
(grant nos. EBM2013006 and EBM2013031).
References
|
1
|
Siegel R, Ward E, Brawley O and Jemal A:
Cancer statistics, 2011: the impact of eliminating socioeconomic
and racial disparities on premature cancer deaths. CA Cancer J
Clin. 61:212–236. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Keszei AP, Schouten LJ, Goldbohm RA and
van den Brandt PA: Red and processed meat consumption and the risk
of esophageal and gastric cancer subtypes in The Netherlands Cohort
Study. Ann Oncol. 23:2319–2326. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Steevens J, Schouten LJ, Goldbohm RA and
van den Brandt PA: Vegetables and fruits consumption and risk of
esophageal and gastric cancer subtypes in the Netherlands cohort
study. Int J Cancer. 129:2681–2693. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bravi F, Edefonti V, Randi G, et al:
Dietary patterns and the risk of esophageal cancer. Ann Oncol.
23:765–770. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sherr CJ: Cancer cell cycles. Science.
274:1672–1677. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhao J, Li L, Wei S, et al:
Clinicopathological and prognostic role of cyclin D1 in esophageal
squamous cell carcinoma: a meta-analysis. Dis Esophagus.
25:520–526. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kaminagakura E, Werneck da Cunha I, Soares
FA, Nishimoto IN and Kowalski LP: CCND1 amplification and protein
overexpression in oral squamous cell carcinoma of young patients.
Head Neck. 33:1413–1419. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Troncone G, Volante M, Iaccarino A, et al:
Cyclin D1 and D3 overexpression predicts malignant behavior in
thyroid fine-needle aspirates suspicious for Hurthle cell
neoplasms. Cancer. 117:522–529. 2009.PubMed/NCBI
|
|
9
|
Abramson VG, Troxel AB, Feldman M, et al:
Cyclin D1b in human breast carcinoma and coexpression with cyclin
D1a is associated with poor outcome. Anticancer Res. 30:1279–1285.
2010.PubMed/NCBI
|
|
10
|
Betticher DC, Thatcher N, Altermatt HJ,
Hoban P, Ryder WD and Heighway J: Alternate splicing produces a
novel cyclin D1 transcript. Oncogene. 11:1005–1011. 1995.PubMed/NCBI
|
|
11
|
Li Z, Jiao X, Wang C, et al: Alternative
cyclin D1 splice forms differentially regulate the DNA damage
response. Cancer Res. 70:8802–8811. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cai W, Wang ZT, Zhong J and Zhang Y: Lack
of association between Cyclin D1 gene G870A polymorphism and
esophageal cancer: evidence from a meta-analysis. Genet Mol Res.
12:6636–6645. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou X, Gu Y and Zhang SL: Association
between p53 codon 72 polymorphism and cervical cancer risk among
Asians: a HuGE review and meta-analysis. Asian Pac J Cancer Prev.
13:4909–4914. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Little J, Bradley L, Bray MS, et al:
Reporting, appraising, and integrating data on genotype prevalence
and gene-disease associations. Am J Epidemiol. 156:300–310. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lau J, Ioannidis JP and Schmid CH:
Quantitative synthesis in systematic reviews. Ann Intern Med.
127:820–826. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mantel N and Haenszel W: Statistical
aspects of the analysis of data from retrospective studies of
disease. J Natl Cancer Inst. 22:719–748. 1959.PubMed/NCBI
|
|
17
|
DerSimonian R and Laird N: Meta-analysis
in clinical trials. Control Clin Trials. 7:177–188. 1986.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Harbord RM, Egger M and Sterne JA: A
modified test for small-study effects in meta-analyses of
controlled trials with binary endpoints. Stat Med. 25:3443–3457.
2006. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Egger M, Davey Smith G, Schneider M and
Minder C: Bias in meta-analysis detected by a simple, graphical
test. BMJ. 315:629–634. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu C, Lu W, Tan W, et al: Lack of
association between CCND1 G870A polymorphism and risk of esophageal
squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev.
12:1762003.PubMed/NCBI
|
|
21
|
Zhang J, Li Y, Wang R, et al: Association
of cyclin D1 (G870A) polymorphism with susceptibility to esophageal
and gastric cardiac carcinoma in a northern Chinese population. Int
J Cancer. 105:281–284. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Casson AG, Zheng Z, Evans SC, et al:
Cyclin D1 polymorphism (G870A) and risk for esophageal
adenocarcinoma. Cancer. 104:730–739. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Geddert H, Kiel S, Zotz RB, et al:
Polymorphism of p16 INK4A and cyclin D1 in adenocarcinomas of the
upper gastrointestinal tract. J Cancer Res Clin Oncol. 131:803–808.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jain M, Kumar S, Lal P, Tiwari A, Ghoshal
UC and Mittal B: Role of BCL2 (ala43thr), CCND1 (G870A) and FAS
(A-670G) polymorphisms in modulating the risk of developing
esophageal cancer. Cancer Detect Prev. 31:225–232. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu G, Cescon DW, Zhai R, et al: p53
Arg72Pro, MDM2 T309G and CCND1 G870A polymorphisms are not
associated with susceptibility to esophageal adenocarcinoma. Dis
Esophagus. 23:36–39. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kurmano BK, Djansugurova L, Bersimba R, et
al: TP53 Arg72Pro and CCND1 A870G polymorphisms and esophageal
cancer risk. J Life Sci. 4:16–20. 2010.
|
|
27
|
Hussain S, Yuvaraj M, Thakur N, et al:
Association of cyclin D1 gene polymorphisms with risk of esophageal
squamous cell carcinoma in Kashmir Valley: a high risk area. Mol
Carcinog. 50:487–498. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Akbari MR, Malekzadeh R, Shakeri R, et al:
Candidate gene association study of esophageal squamous cell
carcinoma in a high-risk region in Iran. Cancer Res. 69:7994–8000.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Djansugurova LB, Perfilyeva AV, Zhunusova
GS, Djantaeva KB, Iksan OA and Khussainova EM: The determination of
genetic markers of age-related cancer pathologies in populations
from Kazakhstan. Front Genet. 4:702013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hall M and Peters G: Genetic alterations
of cyclins, cyclin-dependent kinases, and Cdk inhibitors in human
cancer. Adv Cancer Res. 68:67–108. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Solomon DA, Wang Y, Fox SR, et al: Cyclin
D1 splice variants. Differential effects on localization, RB
phosphorylation, and cellular transformation. J Biol Chem.
278:30339–30347. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sergentanis TN and Economopoulos KP:
Cyclin D1 G870A polymorphism and breast cancer risk: a
meta-analysis comprising 9,911 cases and 11,171 controls. Mol Biol
Rep. 38:4955–4963. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Comstock CE, Augello MA, Benito RP, et al:
Cyclin D1 splice variants: polymorphism, risk, and isoform-specific
regulation in prostate cancer. Clin Cancer Res. 15:5338–5349. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yaylim-Eraltan I, Arikan S, Yildiz Y, et
al: The influence of cyclin D1 A870G polymorphism on colorectal
cancer risk and prognosis in a Turkish population. Anticancer Res.
30:2875–2880. 2010.PubMed/NCBI
|
|
35
|
Tsai MH, Tsai CW, Tsou YA, Hua CH, Hsu CF
and Bau DT: Significant association of cyclin D1 single nucleotide
polymorphisms with oral cancer in taiwan. Anticancer Res.
31:227–231. 2011.PubMed/NCBI
|
|
36
|
Akkiz H, Bayram S, Bekar A, Akgöllü E and
Ozdil B: Cyclin D1 G870A polymorphism is associated with an
increased risk of hepatocellular carcinoma in the Turkish
population: case-control study. Cancer Epidemiol. 34:298–302. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cai W, Wang ZT, Zhong J and Zhang Y: Lack
of association between Cyclin D1 gene G870A polymorphism and
esophageal cancer: evidence from a meta-analysis. Genet Mol Res.
12:6636–6645. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhuo W, Zhang L, Wang Y, Zhu B and Chen Z:
Cyclin D1 G870A polymorphism is a risk factor for esophageal cancer
among Asians. Cancer Invest. 30:630–636. 2012. View Article : Google Scholar : PubMed/NCBI
|